Abstract
Tumor overexpression of urokinase-type plasminogen activator (uPA) and its specific inhibitor SerpinE1 (plasminogen activator inhibitor type-1) correlates with poor prognosis and increased metastatic potential. Conversely, tumor expression of uPA and another specific inhibitor, SerpinB2 (plasminogen activator inhibitor type-2), are associated with favorable outcome and relapse-free survival. It is not known how overexpression of these uPA inhibitors results in such disparate outcomes. A possible explanation may be related to the presence of a proposed low density lipoprotein receptor (LDLR)-binding motif in SerpinE1 responsible for mitogenic signaling via ERK that is absent in SerpinB2. We now show that complementation of such a LDLR-binding motif in SerpinB2 by mutagenesis of two key residues enabled high affinity binding to very LDLR (VLDLR). Furthermore, the VLDLR-binding SerpinB2 form behaved in a manner indistinguishable from SerpinE1 in terms of enhanced uPA-SerpinB2 complex endocytosis and subsequent ERK phosphorylation and cell proliferation; that is, the introduction of the LDLR-binding motif to SerpinB2 was necessary and sufficient to allow it to acquire characteristics of SerpinE1 associated with malignancy. In conclusion, this study defines the structural elements underlying the distinct interactions of SerpinE1 versus SerpinB2 with endocytic receptors and how differential VLDLR binding impacts on downstream cellular behavior. This has clear relevance to understanding the paradoxical disease outcomes associated with overexpression of these serpins in cancer.
Keywords: Cell Surface Receptor, ERK, Protein-Protein Interactions, Serpin, Tumor, Cancer, Plasminogen Activator Inhibitor Type-1, Plasminogen Activator Inhibitor Type-2
Introduction
SerpinE1 (plasminogen activator inhibitor type-1, PAI-1)4 and SerpinB2 (plasminogen activator inhibitor type-2, PAI-2) are both efficient inhibitors of the urokinase-type plasminogen activator (uPA), a key enzyme in the tissue remodeling process. Combined tumor overexpression of uPA, its receptor (uPAR), and SerpinE1 (serine protease inhibitor E1) is strongly correlated with poor patient prognosis (1–3) and metastatic potential (4–8). Consequently, uPA and SerpinE1 expression is recommended as an independent prognostic indicator in node-negative breast cancer (9). On the contrary, tumor expression of SerpinB2 is correlated with increased relapse-free survival in breast cancer (and possibly other cancer types) (10). Further, the relationship between outcome and high SerpinB2 levels in node-negative breast cancer is only relevant if uPA levels are also elevated (1, 11), suggesting that the inhibitory relationship between uPA and SerpinB2 mediates this functionality.
Binding of uPA to uPAR has been reported to induce the activation of numerous cell type-specific signaling pathways, such as MAPK/ERK, JAK/STAT, Src, focal adhesion kinase, and Rac, thereby affecting cell adhesion, migration, differentiation, and apoptosis (12). Because uPAR is not a transmembrane protein, being linked to the cell surface by a glycosylphosphatidylinositol anchor, uPA/uPAR signaling is mediated by integrins and several other adaptor molecules, including members of the low density lipoprotein receptor (LDLR) superfamily (12–15). Both SerpinE1 and SerpinB2 efficiently inhibit uPA in solution and at the cell surface (16–18) through the formation of covalent, irreversible enzyme-substrate complexes. Following the formation of an inhibitory complex at the cell surface, uPA-Serpin complexes are rapidly cleared via LDLR-mediated endocytosis (18–21). Inhibition of uPA by SerpinE1 has previously been shown to modulate uPA/uPAR signaling, resulting in sustained activation of the MAPK/ERK signaling pathway in both MCF-7 and SGC7901 cells (22, 23), leading to increased cell growth (23) and expression of MMP-2 and MMP-9 (22) in these cell lines, respectively. Webb et al. (23) demonstrated that this promitogenic activity was dependent on the direct, high affinity interaction between the SerpinE1 moiety of the uPA-SerpinE1 complex and the very low density lipoprotein receptor (VLDLR). We have previously demonstrated that SerpinB2 does not initiate these signaling events following uPA inhibition and VLDLR-mediated endocytosis (21), and SerpinB2 (unlike SerpinE1) does not bind directly to LDLRs (20, 21). Additionally, the affinity and mechanism of interaction between uPA-SerpinB2 complex and LDLRs is markedly different from that of uPA-SerpinE1 (20, 21, 24). Numerous studies have identified several positively charged residues within (Lys69, Arg76, and Lys80) and nearby (Lys88, Arg118, and Lys122) α-helix D of SerpinE1 that contribute to the high affinity binding of uPA-SerpinE1 with LDLRs (24–27). In addition, α-helix D of SerpinE1 contains a proposed LDLR minimal binding motif consisting of two basic residues (Arg76 and Lys80) separated by two to five residues and N-terminally flanked by hydrophobic residues (Leu75 and Tyr79) (28) (see Fig. 1A). Our previous analysis suggested that the decreased affinity of uPA-SerpinB2 may be due to an incomplete LDLR-binding motif within α-helix D of SerpinB2, in which the first hydrophobic/basic amino acid pair is conserved, whereas the second pair is replaced by serine residues (21) (see Fig. 1A). Indeed, previous studies using a mutant form of SerpinE1 with an incomplete LDLR-binding motif (R76E) observed decreased LRP1 (low density lipoprotein receptor related protein-1) and VLDLR binding affinity and hence functional outcomes (21, 23, 25). We hypothesized that SerpinB2 may inhibit and clear cell surface uPA (and hence proteolytic activity) without influencing the promitogenic signaling pathways activated via SerpinE1, thus accounting for the differential prognosis associated with the expression of these serpins in tumors (10, 21). Herein, we show that VLDLR and LRP1 binding can be introduced to SerpinB2 by complementation of the LDLR-binding motif within α-helix D. This has profound impact on the functionality of SerpinB2.
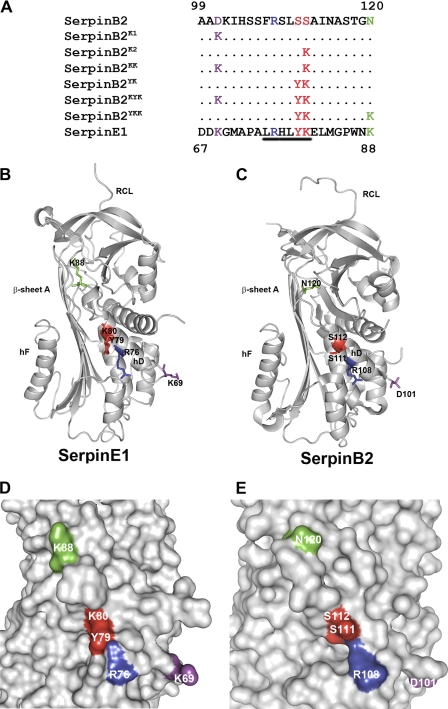
FIGURE 1.
Structural modeling of SerpinE1 and SerpinB2. A, amino acid sequence alignment of the area in and adjacent to α-helix D of SerpinE1 and SerpinB2. The proposed LDLR minimal binding motif in SerpinE1 is underlined. Residues reported to be important for the interaction between SerpinE1 and LDLRs (24–27), and the corresponding residues in SerpinB2 are colored. The colored residues in SerpinB2 were mutated as indicated, resulting in the single and compound mutant forms that were utilized in this study. All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone. B–E, ribbon representation (B and C) and surface representations (D and E) of the relative locations of SerpinE1 residues implicated in LDLR binding and the corresponding residues in SerpinB2. Analysis was performed using PyMol, SerpinE1 (Protein Data Bank code 9PAI) (30), and SerpinB2 ΔCD-loop (Protein Data Bank code 2ARQ) (31).
EXPERIMENTAL PROCEDURES
Materials
QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA). The Alexa 488 labeling kit and Alexa 488 quenching polyclonal antibody were from Invitrogen (Carlsbad, CA). Z-Gly-Gly-Arg-AMC was from Calbiochem (San Diego, CA). HMW-uPA was from American Diagnostica (Stamford, CT). Protease inhibitor mixture, glutamine, transferrin, and selenium were from Sigma-Aldrich. SerpinE1 14-1B stable mutant and purified human receptor-associated protein (RAP) were from Molecular Innovations (Novi, MI). Antibodies to ERK and phosphorylated ERK were from Cell Signaling Technology (Danvers, MA). The recombinant human VLDLR ligand-binding region (V1–V7) was a gift from Prof. Dieter Blass (University of Vienna, Vienna, Austria). Human purified placental LRP1 was a kind gift from Prof. Phillip Hogg (University of New South Wales, Sydney, Australia). Both VLDLR and LRP1 were ≥95% pure as determined by SDS-PAGE (data not shown).
Cell Lines
The human breast adenocarcinoma (MCF-7) and human prostate carcinoma (PC-3) cell lines were obtained from American Type Culture Collection (Manassas, VA) distributed by Cryosite (Sydney, Australia). All of the cell lines were routinely cultured as previously described (21, 29). The cells were routinely tested for the absence of mycoplasma contamination and reconfirmed for the presence of VLDLR and absence of LRP1 (MCF-7) or vice versa (PC-3) as well as for the presence of uPAR by immunofluorescence staining as previously described (20, 21). Both cell lines also express negligible levels of either Serpin (20, 21).
Protein Structure Modeling
Molecular modeling and figure preparation was performed using structural coordinates obtained from x-ray crystal structures of the relaxed conformations of SerpinE1 (30) (Protein Data Bank code 9PAI) and SerpinB2 ΔCD-loop (31) (Protein Data Bank code 2ARQ) and PyMol (DeLano Scientific, Palo Alto, CA).
Site-directed Mutagenesis of SerpinB2 ΔCD-loop
Residues of SerpinB2 were replaced with the corresponding residues of SerpinE1 on the pQE9/SerpinB2 ΔCD-loop backbone (29), with the QuikChange site-directed mutagenesis kit using the following mutagenic primers (paired with their complements): D101K, 5′-GAAGTGGGAGCCGCTGCAAAGAAAATCCATTCATCCTTCC-3′; S111Y, 5′-CATTCATCCTTCCGCTCTCTCTATAAGGCAATCAATGCATCCAC-3′; S112K, 5′-CATCCTTCCGCTCTCTCAGCAAGGCAATCAATGCATCCACAGG-3′; and N120K, 5′-CAATCAATGCATCCACAGGGAAGTATTTACTGGAAAGTGTCAATAAG-3′. Each construct was confirmed by DNA sequencing before expression in Escherichia coli and purification as previously described (29). The SerpinB2 ΔCD-loop backbone was used for all SerpinB2 forms because removal of the CD-loop of SerpinB2 allows for easier purification of protein without compromising the inhibitory activity or LDLR binding affinity of SerpinB2 (29).
Preparation of uPA-Serpin Complexes uPA
Serpin complexes were prepared as previously described (23). Briefly, uPA and SerpinE1 or SerpinB2 forms were incubated at a 1:1 molar ratio for 30 min at 37 °C. Fractionation of samples of complexes by SDS-PAGE confirmed the presence of uPA-Serpin complexes with minimal/negligible residual-free protease or Serpin (data not shown).
Surface Plasmon Resonance
Surface plasmon resonance was performed essentially as previously described (20, 21, 29). VLDLR or LRP1 were loaded onto CM5 BIAcore sensor chips to a level of ∼2000 response units. Analytes were diluted into BIAcore running buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm CaCl2, 0.005% Tween 20) and run over the chip at 20 μl/min. Between samples, the sensor surface was regenerated with 100 mm H3PO4. The data were analyzed using BIAevaluation software (Version 4), using a blank cell as the reference cell. Where multiple CM5 chips were used, the results were validated against control chips and samples to ensure reproducibility.
Far-UV Circular Dichroism Spectrometry
The proteins were buffer-exchanged by dialysis into 10 mm sodium phosphate immediately prior to analysis and diluted to a concentration of 0.4 mg/ml. Far-UV circular dichroism spectrometry was performed using a J-810 spectropolarimeter (Jasco, Easton, MD) equipped with a thermoelectric temperature control at 25 °C. The spectra were collected from 190 to 240 nm at 0.1-nm intervals, with each spectrum representing the average of 10 scans and a sample of 10 mm sodium phosphate serving as a reference.
Activity Assay
Several substrate and uPA concentrations were used to find the optimum range and to set the gain on a Fluorostar Optima fluorescence plate reader (BMG Labtech). SerpinB2 samples were diluted in reaction buffer (20 mm Hepes, pH 7.6, 100 mm NaCl, 0.5 mm EDTA, 0.01% (v/v) Tween 20) and mixed with fluorogenic substrate, Z-Gly-Gly-Arg-AMC, in 180 μl of reaction buffer. After a brief preincubation at 37 °C, HMW-uPA (final concentration, 0.675 nm) was added to start the reaction, and fluorescence emission was measured immediately at 37 °C. All of the assays were performed in triplicate, and the values were corrected by subtracting the background (reaction buffer plus substrate only).
Endocytosis Assay
Cell internalization of Alexa488-labeled uPA alone or in complex with either SerpinE1 or SerpinB2 forms was conducted in the absence or presence of the LDLR antagonist RAP (to confirm the involvement of LDLRs in this process) essentially as described previously (21, 29).
Analysis of ERK Activation
Activation of ERK was analyzed as described previously (23). MCF-7 cells were grown in 12-well plates to ∼60% confluency and serum-starved for 12 h. The cells were incubated with 10 nm uPA or uPA-Serpin complexes for the time periods indicated. The cells were lysed with radioimmune precipitation assay buffer (50 mm Hepes, pH 7.4, 100 mm NaCl, 1% (v/v) Nonidet P-40, 2 mm EDTA, 0.4 mg/ml sodium orthovanadate, 5 mg/ml dithiothreitol, 1 ml/L protease inhibitor mixture), electrophoresed on 12% SDS-PAGE gels, transferred to PVDF membranes, and probed with antibodies that detect phosphorylated and total ERK.
Cell Proliferation Assay
Cell proliferation was examined in real time using the xCELLigence RTCA DP System (Roche Applied Science). The xCELLigence system allows continuous quantitative monitoring of cellular behavior including proliferation by measuring electrical impedance. MCF-7 cells were seeded at 5000 cells/well into E-Plate 16-well plates and cultured for 24 h. The medium was replaced with 100 μl of serum-free RPMI containing 300 μg/ml glutamine, 5 μg/ml transferrin, and 38 nm selenium in the presence of 10 nm uPA, uPA-Serpin complexes, or medium alone. Proliferation was continuously monitored every 15 min over a time period of 72 h. Data analysis was carried out using RTCA Software 1.2.1 supplied with the instrument.
Statistical Analyses
Statistical significance of treatment groups as compared with control groups was determined using an unpaired Student's t test (GraphPad Prism V 5.1; GraphPad Software, San Diego, CA). p values of <0.05 were considered statistically significant.
RESULTS
Structural Comparison Indicates Absence of a Consensus LDLR-binding Motif in SerpinB2
The surface charge and topology of the region defining the consensus LDLR-binding motif on α-helix D of SerpinE1 are clearly different from the corresponding region of SerpinB2 (Fig. 1, B–E). Hence, the differential binding to LDLRs and subsequent functional effects of SerpinE1 and SerpinB2 may be due to an incomplete LDLR-binding motif within SerpinB2 (10). To directly investigate this, we substituted residues of SerpinB2 with those corresponding to either the LDLR-binding motif within α-helix D of SerpinE1 and/or others previously identified as important for LDLR binding within or adjacent to α-helix D of SerpinE1 (24–26, 32) (Fig. 1, A–C). These residues were introduced into SerpinB2 ΔCD-loop in a stepwise manner to generate relevant single or compound mutants defined as indicated in Fig. 1A. Substitution of Ser111 and Ser112 in SerpinB2 with tyrosine and lysine, respectively (corresponding to Tyr79 and Lys80 in SerpinE1), to generate SerpinB2YK completed a potential LDLR minimal binding motif in SerpinB2 (Fig. 1A). SerpinB2YK and other mutants retained relative secondary structure and full inhibitory activity (supplemental Figs. S1 and S2).
Biochemical Characterization of LDLR-binding SerpinB2 Mutants
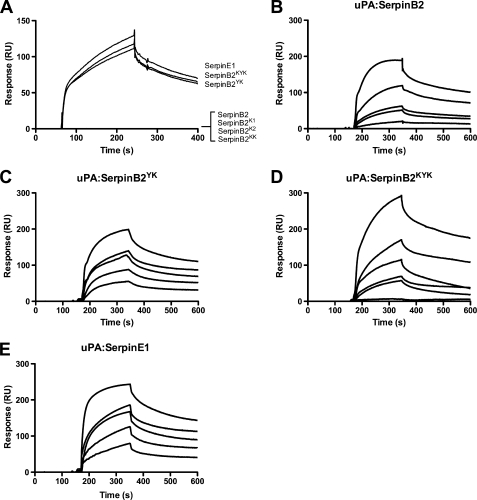
The panel of SerpinB2 mutants described in Fig. 1A were first assessed for their ability to bind directly to VLDLR using surface plasmon resonance methodology (Fig. 2). As previously demonstrated (21), SerpinB2 alone did not interact with VLDLR (Fig. 2A). VLDLR binding was also not observed for SerpinB2 mutants with incomplete LDLR minimal binding motifs (i.e. SerpinB2K1, SerpinB2K2, or SerpinB2KK) (Fig. 2A). Urokinase-independent binding to VLDLR was only achieved by SerpinB2 mutants containing a complete LDLR minimal binding motif (i.e. SerpinB2YK and SerpinB2KYK) (Fig. 2A). Dose-dependent kinetic analysis gave affinity constants (KD) of 37.3 and 50.1 nm for SerpinB2YK and SerpinB2KYK, respectively (Table 1).
FIGURE 2.
Surface plasmon resonance analysis of the interaction between SerpinB2 constructs, SerpinE1, and VLDLR. A, screening of SerpinB2 mutants for VLDLR binding. Surface plasmon resonance sensograms showing the interaction between 100 nm SerpinB2 mutants or SerpinE1 and immobilized VLDLR. The data shown are representative of at least three independent experiments. B–E, surface plasmon resonance analysis of the dose-dependent interaction between uPA complexed SerpinB2 mutants or uPA-SerpinE1 and VLDLR. The data presented here are representative of at least three experiments with analyte concentrations from 100 to 6.25 nm. All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone.
TABLE 1.
Kinetic parameters of the interaction between SerpinB2 forms used in this study and the endocytosis receptor VLDLR
The values were determined using surface plasmon resonance. The binding data were fitted using BIAevaluation 4.0 software. The binding model chosen represents that with the lowest χ2 value. (The values are the averages ± S.E., n ≥ 3). All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone.
| Analyte | Binding model | ka | kd | KD | χ2 |
|---|---|---|---|---|---|
| m−1s−1 | s−1 | nm | |||
| SerpinB2 | No binding | ||||
| SerpinB2K1 | No binding | ||||
| SerpinB2K2 | No binding | ||||
| SerpinB2KK | No binding | ||||
| SerpinB2YK | 1:1 | 6.77 × 104 (±2.68 × 104) | 1.99 × 10−3 (±0.76 × 10−3) | 37.3 (±7.0) | 0.97 |
| SerpinB2KYK | 1:1 | 6.68 × 104 (±2.91 × 104) | 4.31 × 10−3 (±2.22 × 10−3) | 50.1 (±10.2) | 1.02 |
| SerpinE1 | 1:1 | 8.98 × 104 (±2.1 × 104) | 3.51 × 10−3 (±1.97 × 10−3) | 45.3 (±11.7) | 5.9 |
| uPA-SerpinB2 | 1:1 | 4.16 × 105 (±1.80 × 105) | 1.36 × 10−3 (±0.23 × 10−3) | 4.30 (±0.56) | 3.28 |
| uPA-SerpinB2YK | Heterogeneous analyte | 4.78 × 104 (±0.38 × 104) | 3.15 × 10−5 (±1.07 × 10−5) | 1.35 (±0.41) | 7.6 |
| 1.09 × 105 (±0.51 × 105) | 5.69 × 10−3 (±1.35 × 10−3) | 70.1 (±24.9) | |||
| uPA-SerpinB2KYK | Heterogeneous analyte | 2.01 × 104 (±0.22 × 104) | 3.7 × 10−5 (±2.11 × 10−5) | 1.53 (±0.43) | 4.3 |
| 1.66 × 105 (±0.33 × 105) | 3.35 × 10−3 (±1.05 × 10−3) | 51.2 (±12.9) | |||
| uPA-SerpinE1 | Heterogeneous analyte | 9.01 × 104 (±1.5 × 104) | 1.65 × 10−5 (±0.19 × 10−3) | 1.48 (±0.30) | 9.4 |
| 3.55 × 105 (±0.33 × 105) | 0.98 × 10−3 (±0.44 × 10−3) | 49.5 (±12.6) |
Kinetic analyses of VLDLR interactions with uPA-SerpinB2 (Fig. 2B and Table 1) or uPA-SerpinB2K2 and uPA-SerpinKK (data not shown) confirmed the single-site binding model anticipated for uPA complexes incorporating non-VLDLR binding SerpinB2 forms (21). In contrast, the interactions between VLDLR and uPA complexes incorporating the VLDLR binding SerpinB2YK or SerpinB2KYK forms were best described by a heterogeneous analyte model with two independent, competing binding sites on the complex with KD values of 1.35 and 70.1 nm for uPA-SerpinB2YK and 1.53 and 51.2 nm for uPA-SerpinB2KYK (Fig. 2, C and D, and Table 1). As previously observed (21), uPA-SerpinE1 also best fits this binding model for interaction with VLDLR (Fig. 2E) and gave KD values similar to those observed for uPA-SerpinB2YK and uPA-SerpinB2KYK (Table 1).
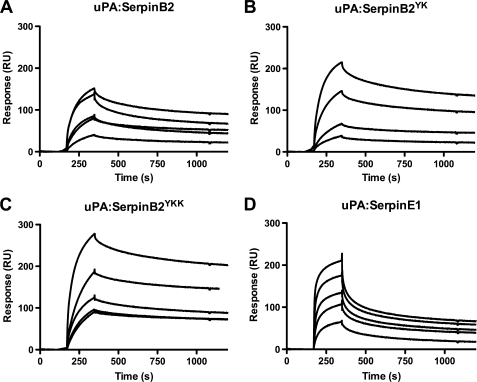
We have previously compared the differential binding interactions of uPA-SerpinE1 and uPA-SerpinB2 with a related LDLR family member, LRP1 (20). Again, although SerpinB2 alone did not bind (data not shown), kinetic analyses of uPA-SerpinB2 binding to LRP1 confirmed the single site binding model interaction (KD = 1.02 nm) (Fig. 3A and Table 2). Interestingly, unlike that seen for VLDLR binding, the interaction between the uPA-SerpinB2YK complex and LRP1 displayed kinetics different from those observed for uPA-SerpinE1 (uPA-SerpinE1 KD = 0.252 and 2.13 nm; uPA-SerpinB2YK KD = 0.808 and 57.8 nm) (Fig. 3B and Table 2). Because Skeldal et al. (24) found that Lys88 of SerpinE1 (referred to as Lys90 in their study) is critical for the interaction between uPA-SerpinE1 and LRP1 (but not VLDLR), we generated an additional mutant (SerpinB2YKK) replacing the asparagine at position 120 (corresponding to Lys88 of SerpinE1) with lysine (Fig. 1A). The uPA-SerpinB2YKK complex displayed strikingly similar LRP1 binding affinities to uPA-SerpinE1, with the high and lower affinity interactions (KD = 0.128 and 2.72 nm) not significantly different from those of uPA-SerpinE1 (Table 2). However, it should be noted that individual ka and kd values were significantly different (Fig. 3, C and D, and Table 2), indicating subtle differences in interaction mechanics that may reflect variations in local surface potential and/or steric effects. Importantly, these differences had negligible impact on overall binding affinities and downstream functional outcomes (see below).
FIGURE 3.
Surface plasmon resonance analysis of the interaction between uPA-Serpin complexes and LRP1. Surface plasmon resonance analysis of the dose-dependent interaction between LRP1 and uPA complexed SerpinB2 mutants as indicated (A–C) and uPA complexed SerpinE1 (D). The data presented here are representative of at least three experiments with analyte concentrations from 100 to 6.25 nm. All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone.
TABLE 2.
Kinetic parameters of the interaction between Serpin forms used in this study and the endocytosis receptor LRP1
The values were determined using surface plasmon resonance. The binding data were fitted using BIAevaluation 4.0 software. The binding model chosen represents that with the lowest χ2 value. (The values are the averages ± S.E., n ≥ 3). All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone.
| Analyte | Binding model | ka | kd | KD | χ2 |
|---|---|---|---|---|---|
| m−1s−1 | s−1 | nm | |||
| uPA-SerpinE1 | Heterogeneous analyte | 9.52 × 104 (±2.89 × 104) | 2.68 × 10−5 (±0.84 × 10−5) | 0.252 (±0.087) | 8.31 |
| 6.09 × 105 (±1.56 × 105) | 1.30 × 10−3 (±0.33 × 10−3) | 2.13 (±0.56) | |||
| uPA-SerpinB2 | 1:1 | 6.74 × 104 (±4.53 × 104) | 6.71 × 10−4 (±1.32 × 10−4) | 1.02 (±0.54) | 3.41 |
| uPA-SerpinB2YK | Heterogeneous analyte | 1.38 × 105 (±0.48 × 105) | 1.9 × 10−5 (±0.87 × 10−5) | 0.808 (±0.246) | 3.35 |
| 1.24 × 105 (±0.44 × 105) | 8.06 × 10−3 (±7.41 × 10−3) | 57.8 (±39.0) | |||
| uPA-SerpinB2YKK | Heterogeneous analyte | 3.43 × 105 (±1.34 × 105) | 3.68 × 10−5 (±1.67 × 10−5) | 0.128 (±0.081) | 7.32 |
| 1.97 × 105 (±1.39 × 105) | 1.10 × 10−2 (±1.55 × 10−2) | 2.72 (±1.51) |
Introduction of LDLR Binding to SerpinB2 Increased uPAR-mediated Endocytosis
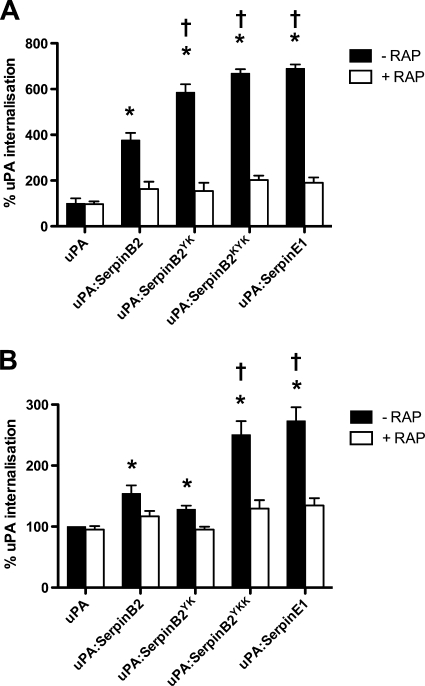
We previously demonstrated that the affinity of uPA-Serpin complex for LDLRs is a key determinant of its rate of endocytosis (21), and we again observed significantly more uPA-SerpinB2 endocytosis (∼4-fold, relative to uPA alone) by MCF-7 cells in an LDLR antagonist (RAP)-sensitive manner over a 30-min time period (Fig. 4A). Given that VLDLR is the only LDLR family member expressed by MCF-7 cells, these data reflect the increased affinity of the complex for VLDLR (Table 1). Accordingly, the RAP-sensitive endocytosis of uPA-SerpinB2YK was increased to a level comparable with that of uPA-SerpinE1 (∼7-fold compared with uPA alone) (Fig. 4A). The uPA-SerpinB2KYK complex had no significant impact on endocytosis over and above that of uPA-SerpinB2YK (Fig. 4A). This result reflected the similar VLDLR binding affinities of these two complexes (Table 1) and further confirms that completion of the LDLR-binding motif is necessary and sufficient for enhanced endocytosis via VLDLR.
FIGURE 4.
Increased LDLR affinity leads to increased internalization of uPA-Serpin complexes by cancer cells. A, MCF-7 cells were incubated in the presence or absence of RAP (200 nm) for 15 min at 37 °C, prior to analysis of internalization of Alexa 488-labeled uPA by fluorescence quenching assay. The values are the means ± S.E. (n = 3). B, PC-3 cells were incubated in the presence or absence of RAP (200 nm) for 15 min at 37 °C prior to analysis of internalization of Alexa 488-labeled uPA by fluorescence quenching assay. The values are the means ± S.E. (n = 3). *, p < 0.005 compared with relative uPA treatment; †, p < 0.005 compared with relative uPA-SerpinB2 treatment. All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone.
Further investigation of endocytosis in an LRP1-dependent cell model (PC-3) showed that the LDLR-binding motif alone was not sufficient for enhanced endocytosis via LRP1. Although a small but significant increase in RAP-sensitive uPA-SerpinB2 endocytosis was observed compared with uPA alone, no significant difference in internalization was observed between uPA-SerpinB2 and uPA-SerpinB2YK (Fig. 4B). However, the levels of uPA-SerpinB2YKK and uPA-SerpinE1 endocytosis were similarly significantly enhanced ∼2.5-fold compared with uPA alone and ∼1.6-fold compared with uPA-SerpinB2 (Fig. 4B). These functional data are entirely consistent with the biochemical data describing the differential interactions of uPA-SerpinB2YK versus uPA-SerpinB2YKK with LRP1 (Table 2).
Introduction of VLDLR Binding to SerpinB2 Induces Mitogenic Signaling in MCF-7 Cells
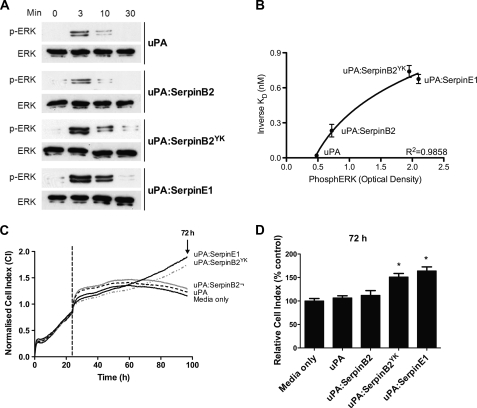
As observed previously (21, 23), the increased ERK phosphorylation in breast cancer cells by uPA-SerpinE1 compared with uPA is not observed in cells treated with uPA-SerpinB2 (Fig. 5A), and this effect is potentially caused by a lack of a LDLR minimal binding motif in SerpinB2. The importance of an intact LDLR minimal binding motif in facilitating this activity was confirmed, because stimulation of MCF-7 cells with uPA-SerpinB2YK initiated a significant increase in ERK phosphorylation (∼3.5-fold within 3 min, relative to uPA alone) (Fig. 5A). The kinetics of ERK phosphorylation was similar to that observed for uPA-SerpinE1. Furthermore, a correlation (R2 = 0.9790) was observed between VLDLR affinity and ERK phosphorylation after 3 min of treatment (Fig. 5B).
FIGURE 5.
uPA-SerpinB2YK induces promitogenic signaling pathways and increased cell proliferation in MCF-7 cells similar to those of uPA-SerpinE1. A, MCF-7 cells were cultured in serum-free medium for 12 h and then exposed to 5 nm uPA, uPA-SerpinB2, uPA-SerpinB2YK, or uPA-SerpinE1 for the indicated times. The cell lysates were analyzed for phosphorylated and total ERK by Western blotting. B, levels of phosphorylated ERK after 3 min of stimulation with 10 nm uPA, uPA-SerpinB2, uPA-SerpinB2YK, or uPA-SerpinE1 were correlated with the affinity of each ligand for VLDLR (Table 1). The data were fitted to a logarithmic equation using GraphPad Prism v5.00 (GraphPad Software, San Diego, CA). C, MCF-7 cells were cultivated in E-plates with RPMI with 5% FCS for 24 h, before changing to serum-free medium containing 10 nm uPA, uPA-SerpinB2, uPA-SerpinB2YK, uPA-SerpinE1, or medium only, and cell proliferation was monitored over 72 h in real time using an xCELLigence RTCA DP system. The electronic readout of cell sensor impedance is displayed continuously as the cell index (CI) normalized to the compound addition time point (indicated by a vertical dotted line). The cell index value at each time point is defined as (Rn − Rb)/Rb, where Rn is the electrode impedance of wells with cells, and Rb is the background of wells with media alone. The curves are the means of four replicates. D, bar graph showing CI values 72 h after the addition of proteins as percentages of medium only treatment (72-h treatment time point is indicated on electronic readout by arrow in C). The values are the means ± S.E. (n = 4). *, p < 0.005 compared with medium only control. All of the SerpinB2 forms were constructed on the SerpinB2 ΔCD-loop backbone.
A downstream functional outcome of sustained ERK phosphorylation in MCF-7 cells stimulated by uPA-SerpinE1 is induction of proliferation (21, 23). Thus, we measured the effect of uPA-SerpinB2YK on MCF-7 cell proliferation in real time using the xCELLigence system (Fig. 5, C and D). Cell indices for uPA-SerpinB2YK and uPA-SerpinE1 treatments were distinct from those observed for cells incubated with uPA or uPA-SerpinB2 (Fig. 5C). Clear proliferative effects were evident over an extended time frame following the addition of either uPA-SerpinB2YK or uPA-SerpinE1, as reflected in an apparent logarithmic phase growth pattern (Fig. 5C). This resulted in a significant (∼1.6-fold) increase in cell index relative to controls (media, uPA, or uPA-SerpinB2) after 72 h (Fig. 5D). Interestingly, cell indices following uPA-SerpinB2YK or uPA-SerpinE1 treatment were initially slightly lower than those obtained following treatment with uPA or uPA-SerpinB2. No difference was observed between cells incubated with uPA or uPA-SerpinB2 (Fig. 5, C and D), with both reaching a plateau between 12 and 36 h post-treatment before declining, suggesting a decrease in cell number.
DISCUSSION
This study provides the first molecular characterization of key structural elements that define the differential interactions of SerpinE1 and SerpinB2 with LDLR receptors and their functional implications for downstream mitogenic signaling and cell proliferation. Completion of the LDLR-binding motif in SerpinB2 to match a corresponding motif in SerpinE1 complemented SerpinB2 activity with the promitogenic function of SerpinE1 (when complexed with uPA). Thus, we suggest that the high LDLR binding affinity and endocytosis rate of uPA-SerpinE1 are part of the repertoire of effects attributable to the promitogenic function of SerpinE1. This partly explains why, despite a shared serpin inhibitory function, high tumor levels of uPA and SerpinE1 promote tumor progression, whereas high levels of SerpinB2 are protective (10).
We and others have previously reported the existence of a strong correlation between the affinity of uPA-Serpin complexes for VLDLR and uPA-Serpin endocytosis (21, 23) and suggested that VLDLR affinity may be the rate-limiting determinant of uPA/uPAR clearance from the cell surface (21). In comparison with uPA-SerpinE1, the uPA-SerpinB2YK complex displayed similar binding affinity to VLDLR and was internalized via this endocytosis receptor in a similar manner. The importance of the LDLR-binding motif in increasing the VLDLR binding affinity and enhanced endocytosis was confirmed by the lack of a significant impact of uPA-SerpinB2KYK over and above that of uPA-SerpinB2YK. Because members of the LDLR family have overlapping yet distinct ligand specificities (33), the presence of the LDLR minimal binding motif alone was not sufficient to introduce binding to LRP1. Skeldal et al. (24) reported that Lys88 was critical for the interaction of SerpinE1 with LRP1. Consistent with this, modification of Asn120 in SerpinB2 (corresponding to Lys88 in SerpinE1), in conjunction with a complete LDLR minimal binding motif, produced a SerpinB2 form (i.e. SerpinB2YKK) with LRP1 binding kinetics and LRP1-mediated endocytosis similar to those of uPA-SerpinE1. These observations confirm that LDLR affinity is the rate-limiting step of uPA-Serpin endocytosis.
Webb et al. (23) demonstrated that the affinity of uPA-Serpin complexes for VLDLR is correlated with sustained activation of ERK and increased cell proliferation in MCF-7 cells. We have now shown that SerpinB2 function can be complemented with the VLDLR binding and downstream functional activity associated with SerpinE1 by completion of the LDLR minimal binding motif within α-helix D. As such, it is clear that VLDLR affinity is the key property underlying the differences in ERK activation and subsequent effects on cell proliferation between uPA-SerpinE1 and uPA-SerpinB2. Given this, it was unnecessary to investigate the impact of uPA-SerpinB2KYK (or other SerpinB2 mutants generated with incomplete LDLR-binding motifs) on mitogenic signaling, because the VLDLR binding affinity of this complex was indistinguishable from that of uPA-SerpinB2YK. Direct binding of SerpinE1 to LRP1 has also been demonstrated to activate the JAK/STAT pathway, leading to increased cell motility (34). However, because the prognostic data for SerpinE1 is only significant in tumors also expressing uPA (1–3, 35), the impact of complementing SerpinB2 with LRP1 binding on the activation of this pathway was not investigated in this study.
Although high affinity binding of uPA-Serpin complexes to VLDLR is clearly vital for the induction of downstream ERK activation, it is important to note that high affinity binding of any ligand to VLDLR will not necessarily induce the same response. This is particularly highlighted by the high affinity interaction of RAP with VLDLR (KD = 0.7 nm) (36), which fails to induce downstream ERK activation and cell proliferation (21, 23). Therefore, the rapid formation of a multimolecular complex consisting of VLDLR, a uPA-Serpin complex, uPAR, and any associated integrins or coreceptors is likely necessary for the induction of this signal, not merely the recognition of a high affinity ligand by VLDLR alone.
Other factors that may impact on the prognostic differences between overexpression of SerpinE1 versus SerpinB2 in cancer include the complex interplay of interactions between uPA/uPAR, SerpinE1, and other ligands and coreceptors, through which SerpinE1 modulates adhesion and migration. For example, in the context of cell adhesion, SerpinE1 competes with uPAR and integrins for vitronectin binding (10, 37, 38), but SerpinB2 does not bind vitronectin (39). Importantly, the vitronectin-binding site(s) in SerpinE1 are distinct from the LDLR minimal binding motif (40), and the SerpinB2 constructs used in this study did not bind to vitronectin or impact on cell adhesion (data not shown). This excludes any influence of modulation of cell adhesion on the proliferative effects of SerpinB2YK on a VLDLR-dependent cell line and implies that these effects are a direct response of the increased affinity for VLDLR. Regardless, the absence of an LDLR minimal binding motif in the SerpinB2 moiety of uPA-SerpinB2 complex suggests a mechanism where, unlike SerpinE1, extracellular SerpinB2 could inhibit and clear cellular uPA, thereby preventing plasmin-mediated ECM degradation, without inducing cell proliferation.
In conclusion, this study clearly demonstrates that VLDLR affinity is the critical determinant underlying the disparate roles of SerpinE1 and SerpinB2 in mitogenic signaling and cell proliferation. Understanding the molecular basis of such physiological roles provides a mechanistic basis for the association of these proteins with disease severity and outcome and provides a sound rationale for potential therapeutic development.
Supplementary Material
This work was financially supported by an Australian Postgraduate Award (to B. J. C.), by the University of Wollongong through the Centre for Medical Bioscience, by a University of Wollongong Small Grant, and by Cancer Institute New South Wales Fellowships (to M. R., D. N. S., and D. R. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs S1 and S2.
- PAI
- plasminogen activator inhibitor
- LDLR
- low density lipoprotein receptor
- uPA
- urokinase plasminogen activator
- uPAR
- urokinase plasminogen activator receptor
- VLDLR
- very low density lipoprotein receptor
- RAP
- receptor-associated protein.
REFERENCES
- 1. Foekens J. A., Peters H. A., Look M. P., Portengen H., Schmitt M., Kramer M. D., Brünner N., Jänicke F., Meijer-van Gelder M. E., Henzen-Logmans S. C., van Putten W. L., Klijn J. G. (2000) Cancer Res. 60, 636–643 [PubMed] [Google Scholar]
- 2. Duffy M. J. (2004) Curr. Pharm. Des. 10, 39–49 [DOI] [PubMed] [Google Scholar]
- 3. Choong P. F., Nadesapillai A. P. (2003) Clin. Orthop. Relat. Res. 415, S46–S58 [DOI] [PubMed] [Google Scholar]
- 4. Duffy M. J., Maguire T. M., McDermott E. W., O'Higgins N. (1999) J. Surg. Oncol. 71, 130–135 [DOI] [PubMed] [Google Scholar]
- 5. Scherrer A., Wohlwend A., Kruithof E. K., Vassalli J. D., Sappino A. P. (1999) Br. J. Haematol. 105, 920–927 [DOI] [PubMed] [Google Scholar]
- 6. Schmitt M., Wilhelm O. G., Reuning U., Krüger A., Harbeck N., Lengyel E., Graeff H., Gänsbacher B., Kessler H., Bürgle M., Stürzebecher J., Sperl S., Magdolen V. (2000) Fibrinolysis Proteol. 14, 114–132 [Google Scholar]
- 7. Weigelt B., Peterse J. L., van't Veer L. J. (2005) Nat. Rev. Cancer 5, 591–602 [DOI] [PubMed] [Google Scholar]
- 8. Look M. P., van Putten W. L., Duffy M. J., Harbeck N., Christensen I. J., Thomssen C., Kates R., Spyratos F., Fernö M., Eppenberger-Castori S., Sweep C. G., Ulm K., Peyrat J. P., Martin P. M., Magdelenat H., Brünner N., Duggan C., Lisboa B. W., Bendahl P. O., Quillien V., Daver A., Ricolleau G., Meijer-van Gelder M. E., Manders P., Fiets W. E., Blankenstein M. A., Broët P., Romain S., Daxenbichler G., Windbichler G., Cufer T., Borstnar S., Kueng W., Beex L. V., Klijn J. G., O'Higgins N., Eppenberger U., Jänicke F., Schmitt M., Foekens J. A. (2002) J. Natl. Cancer Inst. 94, 116–128 [DOI] [PubMed] [Google Scholar]
- 9. Harris L., Fritsche H., Mennel R., Norton L., Ravdin P., Taube S., Somerfield M. R., Hayes D. F., Bast R. C., Jr. (2007) J. Clin. Oncol. 25, 5287–5312 [DOI] [PubMed] [Google Scholar]
- 10. Croucher D. R., Saunders D. N., Lobov S., Ranson M. (2008) Nat. Rev. Cancer 8, 535–545 [DOI] [PubMed] [Google Scholar]
- 11. Bouchet C., Hacène K., Martin P. M., Becette V., Tubiana-Hulin M., Lasry S., Oglobine J., Spyratos F. (1999) J. Clin. Oncol. 17, 3048–3057 [DOI] [PubMed] [Google Scholar]
- 12. D'Alessio S., Blasi F. (2009) Front. Biosci. 14, 4575–4587 [DOI] [PubMed] [Google Scholar]
- 13. Preissner K. T., Kanse S. M., May A. E. (2000) Curr. Opin. Cell Biol. 12, 621–628 [DOI] [PubMed] [Google Scholar]
- 14. Resnati M., Pallavicini I., Wang J. M., Oppenheim J., Serhan C. N., Romano M., Blasi F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1359–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strickland D. K., Ranganathan S. (2003) J. Thromb. Haemostasis 1, 1663–1670 [DOI] [PubMed] [Google Scholar]
- 16. Astedt B., Lecander I., Ny T. (1987) Fibrinolysis 1, 203–208 [Google Scholar]
- 17. Ellis V., Wun T. C., Behrendt N., Rønne E., Danø K. (1990) J. Biol. Chem. 265, 9904–9908 [PubMed] [Google Scholar]
- 18. Al-Ejeh F., Croucher D., Ranson M. (2004) Exp. Cell Res. 297, 259–271 [DOI] [PubMed] [Google Scholar]
- 19. Argraves K. M., Battey F. D., MacCalman C. D., McCrae K. R., Gåfvels M., Kozarsky K. F., Chappell D. A., Strauss J. F., 3rd, Strickland D. K. (1995) J. Biol. Chem. 270, 26550–26557 [DOI] [PubMed] [Google Scholar]
- 20. Croucher D., Saunders D. N., Ranson M. (2006) J. Biol. Chem. 281, 10206–10213 [DOI] [PubMed] [Google Scholar]
- 21. Croucher D. R., Saunders D. N., Stillfried G. E., Ranson M. (2007) Biochem. J. 408, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Y., Liu Z., Tian J., Zong Y., Yang P., Qu S. (2010) FEBS Lett. 584, 3469–3473 [DOI] [PubMed] [Google Scholar]
- 23. Webb D. J., Thomas K. S., Gonias S. L. (2001) J. Cell Biol. 152, 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skeldal S., Larsen J. V., Pedersen K. E., Petersen H. H., Egelund R., Christensen A., Jensen J. K., Gliemann J., Andreasen P. A. (2006) FEBS J. 273, 5143–5159 [DOI] [PubMed] [Google Scholar]
- 25. Stefansson S., Muhammad S., Cheng X. F., Battey F. D., Strickland D. K., Lawrence D. A. (1998) J. Biol. Chem. 273, 6358–6366 [DOI] [PubMed] [Google Scholar]
- 26. Horn I. R., van den Berg B. M., Moestrup S. K., Pannekoek H., van Zonneveld A. J. (1998) Thromb. Haemostasis 80, 822–828 [PubMed] [Google Scholar]
- 27. Rodenburg K. W., Kjoller L., Petersen H. H., Andreasen P. A. (1998) Biochem. J. 329, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jensen G. A., Andersen O. M., Bonvin A. M., Bjerrum-Bohr I., Etzerodt M., Thøgersen H. C., O'Shea C., Poulsen F. M., Kragelund B. B. (2006) J. Mol. Biol. 362, 700–716 [DOI] [PubMed] [Google Scholar]
- 29. Cochran B. J., Gunawardhana L. P., Vine K. L., Lee J. A., Lobov S., Ranson M. (2009) BMC Biotechnol. 9, 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aertgeerts K., De Bondt H. L., De Ranter C. J., Declerck P. J. (1995) Nat. Struct. Biol. 2, 891–897 [DOI] [PubMed] [Google Scholar]
- 31. Di Giusto D. A., Sutherland A. P., Jankova L., Harrop S. J., Curmi P. M., King G. C. (2005) J. Mol. Biol. 353, 1069–1080 [DOI] [PubMed] [Google Scholar]
- 32. Ehrlich H. J., Gebbink R. K., Keijer J., Pannekoek H. (1992) J. Biol. Chem. 267, 11606–11611 [PubMed] [Google Scholar]
- 33. Strickland D. K., Gonias S. L., Argraves W. S. (2002) Trends Endocrinol. Metab. 13, 66–74 [DOI] [PubMed] [Google Scholar]
- 34. Degryse B., Neels J. G., Czekay R. P., Aertgeerts K., Kamikubo Y., Loskutoff D. J. (2004) J. Biol. Chem. 279, 22595–22604 [DOI] [PubMed] [Google Scholar]
- 35. Ohba K., Miyata Y., Kanda S., Koga S., Hayashi T., Kanetake H. (2005) J. Urol. 174, 461–465 [DOI] [PubMed] [Google Scholar]
- 36. Battey F. D., Gåfvels M. E., FitzGerald D. J., Argraves W. S., Chappell D. A., Strauss J. F., 3rd, Strickland D. K. (1994) J. Biol. Chem. 269, 23268–23273 [PubMed] [Google Scholar]
- 37. Stefansson S., McMahon G. A., Petitclerc E., Lawrence D. A. (2003) Curr. Pharm. Des. 9, 1545–1564 [DOI] [PubMed] [Google Scholar]
- 38. Deng G., Royle G., Wang S., Crain K., Loskutoff D. J. (1996) J. Biol. Chem. 271, 12716–12723 [DOI] [PubMed] [Google Scholar]
- 39. Mikus P., Urano T., Liljeström P., Ny T. (1993) Eur. J. Biochem. 218, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 40. Jensen J. K., Wind T., Andreasen P. A. (2002) FEBS Lett. 521, 91–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.