Abstract
Connexins are the transmembrane proteins that form gap junctions between adjacent cells. The function of the diverse connexin molecules is related to their tissue-specific expression and highly dynamic turnover. Although multiple connexins have been previously reported to compensate for each other's functions, little is known about how connexins influence their own expression or intracellular regulation. Of the three vertebrate lens connexins, two connexins, connexin43 (Cx43) and connexin46 (Cx46), show reciprocal expression and subsequent function in the lens and in lens cell culture. In this study, we investigate the reciprocal relationship between the expression of Cx43 and Cx46. Forced depletion of Cx43, by tumor-promoting phorbol ester 12-O-tetradecanoylphorbol-13-acetate, is associated with an up-regulation of Cx46 at both the protein and message level in human lens epithelial cells. An siRNA-mediated down-regulation of Cx43 results in an increase in the level of Cx46 protein, suggesting endogenous Cx43 is involved in the regulation of endogenous Cx46 turnover. Overexpression of Cx46, in turn, induces the depletion of Cx43 in rabbit lens epithelial cells. Cx46-induced Cx43 degradation is likely mediated by the ubiquitin-proteasome pathway, as (i) treatment with proteasome inhibitors restores the Cx43 protein level and (ii) there is an increase in Cx43 ubiquitin conjugation in Cx46-overexpressing cells. We also present data that shows that the C-terminal intracellular tail domain of Cx46 is essential to induce degradation of Cx43. Therefore, our study shows that Cx43 and Cx46 have novel functions in regulating each other's expression and turnover in a reciprocal manner in addition to their conventional roles as gap junction proteins in lens cells.
Keywords: Connexin, Gap Junctions, Proteasome, Protein Degradation, Protein Turnover, connexin43, connexin46, connexin50
Introduction
Gap junctions are intercellular aqueous channels composed of transmembrane proteins called connexins (1). The gap junction-dependent or independent functions of connexins are important in the regulation of several cellular processes, including growth, proliferation, differentiation, protection, and cell death (2, 3). Distinct expression patterns and highly dynamic turnover rates are the key components that regulate tissue-specific activity of different connexin molecules. The expression and turnover of connexins are fine-tuned balances of several processes such as gene expression, mRNA stability, protein synthesis and transport, and degradation (4, 5). Connexin turnover and function is also modulated by several intrinsic and extrinsic factors, including intra- and extracellular pH, various phosphorylation events, cellular status, and chemical reagents such as the tumor-promoting phorbol ester, TPA3 (6–10).
One tissue that relies on the gap junction-mediated communication for normal function and growth is the vertebrate lens. The lens is naturally avascular and, therefore, gap junction-mediated functions play a major role in maintaining proper homeostasis and transparency of the lens. The vertebrate lens endogenously expresses three connexin proteins required for proper lens development and function, connexin43 (Cx43), connexin46 (Cx46), and connexin50 (Cx50) (11–17). These connexins show differential spatial distributions that are related to their specific functions at different regions of the lens. Cx43 and Cx50 are mainly expressed at the lens epithelium and are required to maintain gap junction activity at the surface of the lens (17, 18). However, Cx46 is preferentially expressed in the differentiating fiber cells and therefore is the major functioning connexin in the lens nuclear region (15, 19). During lens epithelial-to-fiber cell differentiation Cx43 is substantially down-regulated and Cx46 is significantly up-regulated. This reciprocal expression between Cx43 and Cx46 is critical for maintaining connexin-mediated communication in lens tissue; however, the underlying molecular mechanism of this reciprocal relationship remains to be clarified.
Previously, it was observed that Cx43 and Cx46 are reciprocally expressed in normal breast tissue and tumor xenografts (24), as well as Y79 retinoblastoma xenografts (25). Cx43 is well known for tumor suppressor functions and has been previously reported to be down-regulated in breast tumor when compared with normal breast tissue (20–23). Two prior studies demonstrated that Cx46 is significantly up-regulated in tumor xenografts and functions as a proto-oncogene favoring tumor growth while at an early hypoxic stage (24, 25). Therefore, the reciprocal expression between these two connexins plays a crucial role in the regulation of cell proliferation in tumorigenesis as well as in lens development. Multiple connexins expressed in the same tissue have been previously reported to compensate for each other's function (4, 26, 27), however, little is known about how one connexin can control the expression and turnover of another connexin.
In the present study, we have investigated the reciprocal relationship between the expression of the two connexin proteins, Cx43 and Cx46. We show that TPA forced depletion of Cx43 protein and this is associated with an up-regulation of Cx46. The up-regulation of Cx46 is both at the mRNA and protein level and occurs via a protein kinase C (PKC)-dependent pathway in human lens epithelial cells (HLECs). Additionally, siRNA-mediated down-regulation of Cx43 results in an increase of Cx46 indicating that endogenous Cx43 is involved in the control of Cx46 protein levels. Furthermore, we show that overexpression of Cx46 causes a reduction of Cx43 protein. Overexpression of Cx46 increases Cx43 ubiquitination and therefore mediates its degradation by a proteasome-dependent pathway. We also provide evidence that Cx46 membrane localization is not necessary for induction of degradation and that only the cytoplasmic C-terminal tail domain of Cx46 is required to induce degradation of Cx43. Our results provide mechanistic insight into the regulation of Cx43 and Cx46 in lens cells and offers explanation of the reciprocal relationship observed unnaturally in tumor xenografts and naturally in the vertebrate lens.
EXPERIMENTAL PROCEDURES
Pharmacological Inhibitors
TPA and 4α-phorbol-12,13-didecanoate (PDD) were purchased from Calbiochem (La Jolla, CA). N-Acetyl-Leu-Leu-Nle-CHO (ALLN), MG132, leupeptin, chloroquine, the PKC inhibitor GF109203X, and the MAPK inhibitor U0126 were purchased from Sigma-Aldrich.
Cell Culture
To assure the results of this publication both human and rabbit lens epithelial cells were used. Immortalized HLECs and rabbit lens epithelial NN1003A cells (NN) were cultured as described previously (24). Briefly, cells were cultured in low glucose DMEM (Invitrogen) supplemented with 10% premium fetal bovine serum (Atlanta Biologicals, Atlanta, GA) to at least 70% confluence prior to use for experimentation. Cells were grown in a humidified atmosphere of 5% CO2 and 37 °C. All cell culture plastics were purchased from Midwest Scientific (St. Louis, MO).
Cloning and Transfection
The coding region of rat Cx43 (Gja1, NM_012567.2, 382 residues), rat Cx46 (Gja3, NM_024376.1, 416 residues), or rat Cx50 (Gja8, NM_153465.2, 440 residues) was subcloned into the pEGFP-N3 vector (Clontech, Mountain View, CA) to create C-terminal-tagged EGFP fusion proteins. Plasmid transfection was mediated by LipofectamineTM 2000 (Invitrogen) at 70–80% confluency according to the manufacturer's suggestions. The transfected cells were grown in the presence of 1 mg/ml G418 for 3–4 weeks for the selection of positive colonies. Then, the positive colonies were isolated and grown in media containing 500 μg/ml G418 thereafter.
To generate the Cx46 C-terminal (CT) tail domain deletion mutant (Cx46-dCT), wild-type Cx46 was truncated at amino acid 225 and subcloned into pEGFP-N3. Amino acids 225–416 of wild-type Cx46 was subcloned into pEGFP-N3 to generate an expression vector that expresses only the CT tail domain of Cx46 fused to EGFP. Wild-type rat Cx46 cDNA was also subcloned into the pQE-TriSystem vector (Qiagen, Valencia, CA) to generate a C-terminal 5×-His-tagged protein. The expression of Cx43-EGFP, Cx46-EGFP, Cx50-EGFP, Cx46-dCT-EGFP, Cx46Tail-EGFP, and Cx46-His were checked by Western blot analyses using anti-GFP and anti-his tag antibodies, respectively.
Whole Cell Homogenate Preparations
Cells were washed three times with cold phosphate-buffered saline (PBS), collected by scraping from plates, and centrifuged at 4000 rpm for 5 min at 4 °C. The cell pellets were lysed in ice-cold 1× RIPA buffer (10 mm Tris-HCl (pH 7.5),150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Nonidet P-40, 1% sodium deoxycholate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin) containing 1 mm PMSF. Cell lysates were sonicated for 10 s (three times), and the protein concentration of each sample was measured using the Bio-Rad Protein Assay.
Western Blot and Antisera
Western blot analyses were performed as previously described (24, 25, 28). Mouse anti-N-terminal-Cx43 (anti-Cx43 NT) was purchased from Fred Hutchinson Cancer Center (Seattle, WA); rabbit anti-phospho-Cx43 (Ser-368) was from Cell Signaling Technologies (Danvers, MA); rabbit anti-C-terminal-Cx43 (anti-Cx43 CT, 1:5000) and mouse anti-β-actin (1:10000) were from Sigma-Aldrich; rabbit anti-Cx46 (1:2000) was from U.S. Biologicals (Swampscott, MA); mouse anti-Cx50 (1:4000) was from Zymed Laboratories Inc.-Invitrogen (San Franscisco, CA); and mouse anti-EGFP antibody (1:5000) was purchased from Clontech. Mouse anti-pentahis tag (1:1000) was purchased from Qiagen (Valencia, CA). Mouse anti-ubiquitin antibody was purchased from Calbiochem (EMD Biosciences, OH). Rabbit anti-C-terminal-Cx43 (anti-Cx43 CT) was used to detect Cx43 if not mentioned otherwise.
PCR
Total RNA was isolated from control or TPA-treated HLECs or NN1003A wild-type cells and NN1003A cells stably overexpressing Cx46-EGFP or EGFP (empty vector) using an RNeasy Mini Kit (Qiagen). Reverse transcription (RT-PCR) was carried out using SuperScriptTM III First Strand RT-PCR kit (Invitrogen), according to the manufacturer's protocol, to generate cDNA. The cDNAs obtained from rabbit NN1003A cells were dilution optimized prior to being amplified by PCR using primers specific for rabbit Cx43 or β-actin. The cDNAs generated from HLECs samples were subjected to PCR with human Cx46, Cx43, or β-actin cDNA-specific primers.
The sequences for the primers used are given as follows: human β-actin (forward, 5′-GGCATCCACGAAACTACCTT-3′; reverse, 5′-AGCACTGTGTTGGCGTACAG-3′); rabbit β-actin (forward, 5′-GAAATCGTGCGTGACATTAAG-3′; reverse, 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGGGC-3′); human Cx43 (forward, 5′-GTGCCTGAACTTGCCTTTTC-3′; reverse, 5′-CCCTCCAGCAGTTGAGTAGG-3′); rabbit Cx43 (forward, 5′-GATGAGCAGTCTGCCTTTCGT-3′; reverse, 5′-CGTTGACACCATCAGTTTGG-3′); and human Cx46 (forward, 5′-ACCGCACGTGTGAAAGGAAT-3′; reverse, 5′-GGAGTGCTCCTGTGCATTTT-3′).
Real-time quantitative PCR (qRT-PCR) was performed using a Bio-Rad iQ iCycler Detection System with SYBR Green SuperMix (Bio-Rad). Reactions were performed in a total volume of 25 μl (12.5 μl of 2× SYBR buffer SuperMix, 1 μl of each primer at 10 μm concentration, and 10.5 μl of the previously dilution-optimized reverse-transcribed cDNA template).
qRT-PCR protocols for each primer set were optimized using four serial 5× dilutions of template cDNA obtained from mouse lenses (obtained from a previous study). The protocols used are as follows: denaturation (95 °C for 5 min) followed by amplification repeated 40 times (95 °C for 15 s, 60 °C for 45 s). A melt curve analysis was performed following every run to ensure a single amplified product for every reaction. All reactions were carried out in triplicate for every sample. The same reference standard dilution series was repeated on every experimental plate.
Immunofluorescent Labeling, Fluorescence, and Confocal Microscopy
HLECs and NN1003A cells were grown on glass coverslips in 6-well plates until 80% confluency. Treatments were carried out per experimental protocol. Cells were fixed with 4% formaldehyde in PBS for 20 min at room temperature, quenched with 50 mm glycine, washed with PBS, permeabilized with 0.05% Triton X-100 in PBS for 30 min, washed, and blocked with 5% BSA (blocking solution) in PBS for at least 1 h at room temperature. Cells were then treated overnight at 4 °C with rabbit anti-Cx43CT antibody (1:250) or rabbit anti-Cx46 antibody (1:250) in blocking solution with constant gentle rocking. After washing three times for 5 min each in PBS, slides were labeled for 10 min in the dark at room temperature with 500 nm DAPI in PBS to stain nuclei. Finally, after three more washes for 5 min each in PBS, slides were mounted in Prolong Gold Antifade Reagent (Invitrogen-Molecular Probes, Eugene, OR) for at least 24 h prior to visualization. The slides were viewed with a confocal microscope (LSM 510 Meta, Carl Zeiss, Göttingen, Germany) or by a fluorescence microscope (Axiovert 200M, Carl Zeiss). For fluorescence studies involving transiently transfected cells, NN1003A or HLECs cells were transfected with plasmids encoding Cx46-EGFP, and mutants for no more than 48 h prior to formaldehyde fixation. Monoclonal anti-58K/FTCD antibody (1:50, Sigma-Aldrich) was used in conjunction with donkey anti-mouse-Alexa Fluor 594 antibody (1:800, Invitrogen) to mark the Golgi apparatus in HLECs.
Plasma Membrane Fractionation
The fractionation of plasma membrane proteins from untransfected or transfected NN1003A cells was carried out using a QproteomeTM Plasma Membrane Protein kit (Qiagen) according to the manufacturer's instruction manual. In brief, 1 × 107 cells were collected by scraping, centrifuged, and resuspended in 2 ml of Lysis Buffer PM. Then cells were centrifuged again at 450 × g for 5 min, and cell pellets were resuspended in 500 μl of Lysis Buffer PM containing protease inhibitor solution (1:100). Lysis Solution PL (2.5 μl) was then added to the resuspended cells, and, following an incubation period for 15 min at 4 °C, cells were disrupted using a 21-gauge needle and syringe. The cell lysates were then centrifuged at 12,000 × g for 20 min to remove the intact cells, cell debris, and nuclei. The supernatants that contained cytosolic proteins, microsomes, Golgi vesicles, and plasma membrane were incubated with 20 μl of Binding Ligand PBL, a ligand that is specific for the molecules on the plasma membrane, for 1 h at 4 °C. The ligand-plasma membrane complexes were then precipitated using Strep-Tactin magnetic beads and washed with Wash Buffer PW. The plasma membrane proteins were next eluted with Elution Buffer PME. The eluted fractions were quantitated by using the Bio-Rad Protein Assay and analyzed by Western blotting.
Immunoprecipitation
Untransfected NN1003A cells (control) or cells stably overexpressing EGFP, Cx46-EGFP, or Cx50-EGFP were grown in 100-mm dishes to ∼90% confluency. NN1003A cells were also transiently transfected with Cx50-GFP plasmid for 36 h. All cells were harvested, and whole cell homogenates were prepared as described above. The total protein concentration in the whole cell lysates were quantified by Bio-Rad Protein Assay. To pre-clear the whole cell lysates, 0.25 μg of rabbit nonspecific IgG and 20 μl of protein A/G-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were added to the 750 μg of each whole cell lysate. The agarose bead-IgG complexes were removed by centrifugation at 3000 rpm. Supernatants were then incubated with 5 μg of rabbit anti-Cx43CT antibody for overnight at 4 °C. After the overnight incubation, the supernatants were further incubated with 20 μl of protein A/G-Agarose beads for 4 h at 4 °C. The beads were collected by centrifugation at 3000 rpm, washed three times with 1× RIPA buffer with protease inhibitors, and the samples were boiled for 5 min in 2× denaturing electrophoresis sample buffer. The samples were then analyzed by Western blot with anti-ubiquitin and anti-Cx43 antibodies.
siRNA Transfection
HLECs were grown in 60-mm dishes. When confluency reached 70%, cells were treated with 50 nm of either anti-Cx43 siRNA (Qiagen) or AllStars Negative Control siRNA (Qiagen) and 20 μl of HiPerFect transfection reagent (Qiagen). Cells were harvested after 24 and 48 h of transfection, and whole cell homogenates were prepared.
Statistical Analyses
Origin software (MicroCal Inc., Northampton, MA) was used to generate statistical analyses and associated data graphs. The level of significance (see the asterisks in the figure legends) was considered at p < 0.05 using paired t test analyses. All data are presented as mean ± S.E. of at least three independent experiments.
RESULTS
TPA Causes Depletion of Cx43 and an Increase in Cx46 Protein Levels in HLECs
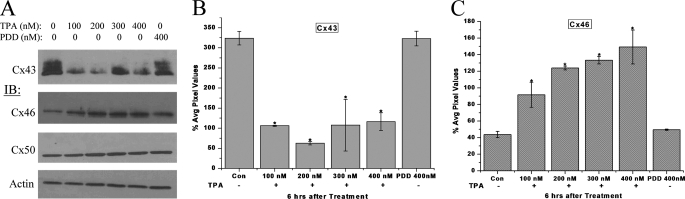
Lens epithelial cells express three connexin proteins, Cx43, Cx46, and Cx50. These three connexins are phosphoproteins and are phosphorylated in response to TPA-stimulated PKC activation (29, 30). Previous studies have also shown that TPA causes a loss of Cx43 gap junction plaques from the cell surface and a reduction in the Cx43 protein level in many cell types (31–33). Therefore, to investigate the reciprocal relationship of Cx43 and Cx46 expression, we first treated HLECs with TPA to initiate a forced degradation of Cx43 and then examined the effect on Cx46 and Cx50 by Western blotting. As revealed by Western blotting, Cx43 protein levels were depleted after 6 h of TPA treatment and not PDD treatment, a non-active TPA analog (Fig. 1, A and B). Surprisingly, the protein levels of Cx46 increased in response to TPA treatment and not PDD treatment (Figs. 1A and 1C), whereas the Cx50 protein levels showed no change (Fig. 1A) with either treatment. TPA treatment caused a significant depletion of Cx43 protein levels at all doses and a major increase in Cx46 protein levels in a dose-dependent manner.
FIGURE 1.
TPA causes depletion of Cx43 and an increase in Cx46 protein levels. A, whole cell homogenates of HLECs treated with the indicated doses of TPA for 6 h were analyzed by Western blotting with anti-Cx43, -Cx46, and -Cx50 antibodies. β-Actin was used as loading control. PDD, the inactive structural analog of TPA, was used as a negative control. B and C, quantitative comparison (densitometric analysis) of the detected protein levels of Cx43 (pixel intensity of multiple Cx43 bands detected were calculated together), and Cx46 respectively, in different treatments were normalized to the levels of loading control β-actin. The 0-h treatment is regarded as a control. All bands were digitized by UN-SCAN-It gel software. The average pixel values for each Cx band were calculated, normalized, and plotted in percent of loading control. Although Cx43 protein levels decreased significantly in response to TPA treatment, Cx46 protein levels increased significantly at the same time. The asterisks indicate significantly different amounts of protein levels in comparison to the control (p < 0.05). No significant changes in the Cx50 protein levels were detected (histogram not shown). Data are representative of three independent experiments.
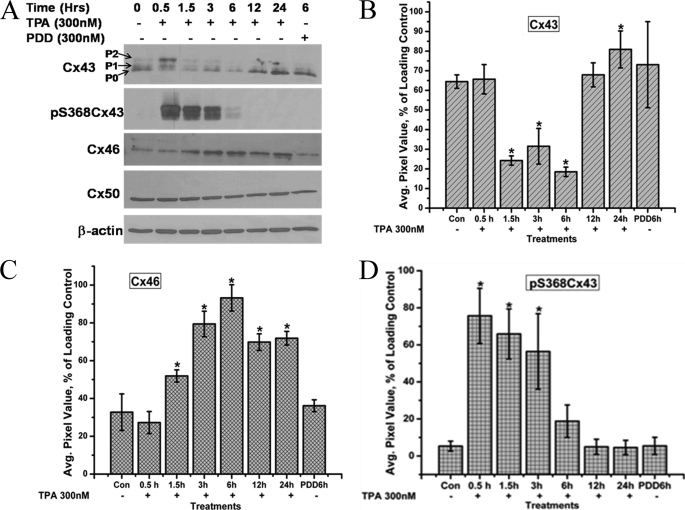
Phosphorylation of Cx43 has been studied extensively. In gap junctional communication-competent cells, Cx43 usually forms three major bands in SDS-PAGE based on its phosphorylation status on different residues (34, 35). The fastest migrating Cx43 band is termed Cx43-P0, and the other two slower migrating forms are termed Cx43-P1 and Cx43-P2, respectively (Fig. 2A). In the TPA time course study, 300 nm TPA treatment induced hyperphosphorylation of Cx43 within 30 min, seen as an increase in the Cx43-P1 and Cx43-P2 bands, and a loss of the Cx43-P0 band (Fig. 2A). The TPA-induced hyperphosphorylation led to the depletion of Cx43 protein levels up to 6 h. At 12 h, the level of Cx43-P2 band was reduced and the levels of Cx43-P0 and Cx43-P1 recovered with the overall Cx43 protein levels becoming comparable with the control levels (Fig. 2, A and B). The transient nature of the phosphorylation and the involvement of PKC in the phosphorylation of Cx43 were confirmed by Western blotting with the pS368Cx43 antibody, which recognizes only the Cx43 forms phosphorylated at the serine 368 residue. Ser-368, present in the C-terminal tail domain of Cx43, has been shown to be phosphorylated by PKC (36), and our laboratory has previously reported the interaction between PKCγ and Cx43 in response to TPA, growth factors (37), and oxidative stress (28) as well as PKCγ involvement in lens connexin regulation (38). The transient depletion of total Cx43 protein levels was concomitant with the phosphorylation at the Ser-368 residue (Fig. 2, A and D), suggesting that the phosphorylation of Cx43 at this residue by PKC leads to the degradation of Cx43 protein as reported previously (29). The phospho-S368-Cx43 isoforms (P1 and P2) were not detected after 12 h when the Cx43-P0 isoform recovered.
FIGURE 2.
Time-course analysis of connexin protein and phosphorylation levels in response to treatment with 300 nm TPA treatment. A, whole cell homogenates from HLECs treated with 300 nm TPA for the indicated time periods were analyzed by Western blotting with anti-Cx43, phospho-ser368Cx43, Cx46, and Cx50 antibodies. β-Actin was used as loading control. PDD treatment was used as a negative control of TPA treatment. B–D, quantitative comparison of the detected protein levels of Cx43 (pixel intensity of multiple Cx43 bands detected were calculated together), Cx46 and phosphor-S368Cx43, respectively, in different treatments normalized to the levels of loading control β-actin. The asterisks indicate significantly different amounts of protein levels in comparison to the control (p < 0.05). Data are representative of three independent experiments.
Significantly higher levels of Cx46 were detected after 1.5 h of TPA treatment (Fig. 2, A and C) with the highest levels of Cx46 detected after 6 h, which coincides with the maximum reduced levels of Cx43. There was no visible change in the levels of Cx50 protein throughout the 24 h of TPA treatment (Fig. 2A). These data are consistent with a PKC-mediated phosphorylation and subsequent depletion of Cx43 protein levels. The increase in Cx46 protein levels in response to TPA treatment is a novel finding and was studied further.
TPA-induced Changes in the Localization of Cx43 but Not Cx46 in HLECs
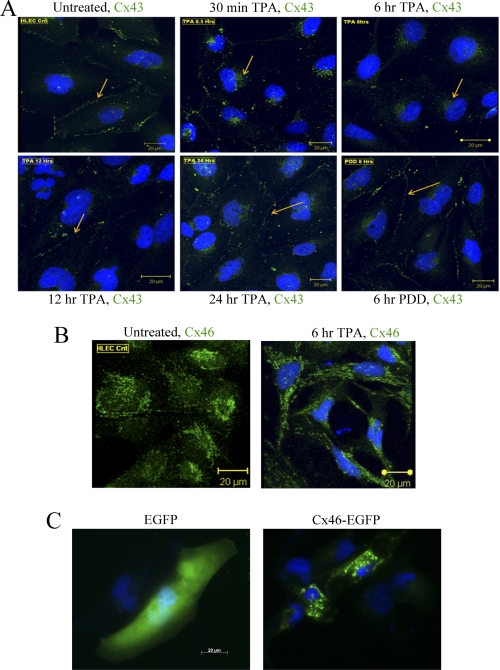
Previously, we have localized Cx43 mostly to the apposed membranes of neighboring cells in HLECs (37). Here we examined the fate of Cx43 after TPA-induced phosphorylation and depletion using confocal microscopy. In agreement with our previous findings, Cx43 was found mostly in the apposed cell membranes forming punctate gap junction plaques and some cytoplasmic occurrence surrounding the nuclei (Fig. 3A). Within 30 min of 300 nm TPA treatment, most of the Cx43 was internalized from the plasma membrane with an increase in the intracellular localization. TPA treatment for 6 h caused a near complete loss of Cx43 immunofluorescence staining except for a faint signal in the perinuclear area (Fig. 3A). After 12 h of TPA treatment, Cx43 gap junction plaques reappeared on the apposed plasma membranes. Further, after 24 h, the Cx43 staining was comparable to the control cells. These data are in accordance with the amounts of Cx43 proteins detected at different time points in the Western blotting experiments.
FIGURE 3.
TPA-induced changes in localization of Cx43 and Cx46 in HLECs. A, HLECs were treated with or without 300 nm TPA for different time periods as indicated in the figure and labeled with anti-Cx43 antibody (green). DAPI (blue) was used to stain nuclei. Arrows point to the gap junction plaques at the membrane or in the cytosol. B, control HLECs or TPA-treated (300 nm, 6 h) HLECs were labeled with anti-Cx46 antibody (green) and DAPI (blue). Cx46 staining was found predominantly in the intercellular compartments of HLEC cells. C, fluorescence microscopy of HLEC cells transiently expressing either EGFP (green) or Cx46-EGFP (green) with DAPI staining of the nuclei (blue). Again, Cx46-EGFP localized to the perinuclear region.
Although Cx46 is a known gap junction protein in the lens, a few studies have reported the localization of Cx46 to the perinuclear region in bone osteoblastic cells (39) and lung adenoma (40). Immunofluorescence using anti-Cx46 antibody detected Cx46 in the intracellular compartments of HLECs (Fig. 3B). Even after 6 h of 300 nm TPA treatment, Cx46 continued to localize to the intracellular and perinuclear compartments with more punctate staining. Additionally, by using the intrinsic fluorescence of the Cx46-EGFP fusion protein, continued perinuclear localization of Cx46-EGFP was seen in HLECs (Fig. 3C). Thus, Cx46 distribution differs from Cx43 localization inside HLECs with or without TPA treatment.
TPA Causes an Increase in Cx46 mRNA Levels
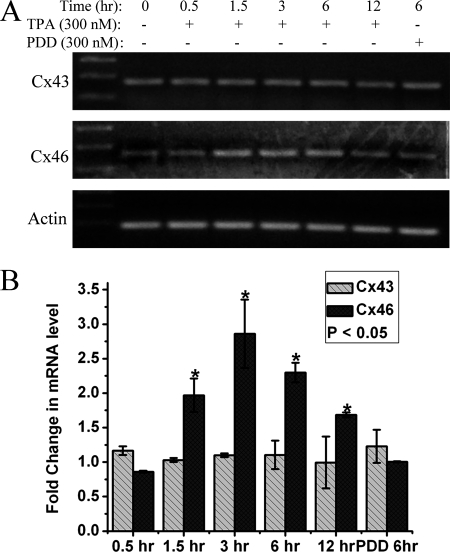
To investigate whether the reciprocal regulation of Cx43 and Cx46 is at the protein level or message level, reverse transcription was performed to check the mRNA levels of Cx43 and Cx46 in response to TPA treatment at different time intervals. Both RT-PCR (Fig. 4A) and qRT-PCR (Fig. 4B) revealed that, although there was no significant change in Cx43 message levels, there was a ∼3-fold increase in Cx46 message levels in response to 3 h of 300 nm TPA treatment. The observed increase in Cx46 message was time-dependent, after reaching a maximum 3-fold increase over the control cells, it continued to decrease until 12 h, although still significantly higher than control cells. This increase in Cx46 message corresponded with a similar amount of increase in Cx46 protein levels in response to TPA treatment. This suggests that TPA influences the up-regulation of Cx46 at the message level.
FIGURE 4.
TPA causes an increase in Cx46 mRNA levels. A, reverse transcription PCR and B, quantitative real-time PCR was completed as described under “Experimental Procedures.” section. Both A and B show that, although the Cx43 message level remains constant throughout the different time periods indicated, Cx46 message was up-regulated up to 3-fold in response to TPA treatment. β-Actin was used as a housekeeping control gene. Quantitative real-time PCR data in B is plotted as the -fold change in message over the control untreated HLECs message level. The asterisks indicate significantly different amounts of message levels in comparison to the control (p < 0.05). Data are representative of three independent experiments.
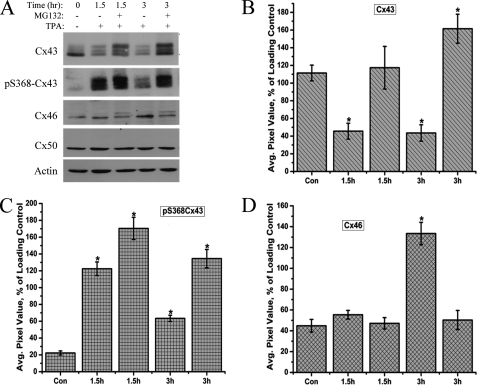
Proteasome Inhibitor MG132 Blocks the TPA-induced Depletion of Cx43 Protein
To verify the involvement of the proteasomal degradation pathway in the TPA-induced depletion of Cx43 protein, HLECs were treated with the proteasomal inhibitor MG132 for 30 min followed by co-incubation with TPA for the indicated time periods as illustrated in Fig. 5A. The TPA-induced loss of the Cx43 protein was completely counteracted by MG132, and at the 3-h time point, a significantly higher amount of Cx43 protein was detected (Fig. 5, A and B). Western blotting with the pS368Cx43 antibody showed that phosphorylation of Cx43 at the Ser-368 residue was still intact in the presence of the proteasomal inhibitor and, over time, the pS368Cx43 form accumulated in the MG132-treated samples (Fig. 5C). Under the treatment of MG132, Cx43 mostly remained in the P1 and P2 status at the time points investigated (Fig. 5A). These data suggest that PKC-mediated phosphorylation of Cx43 at the Ser-368 residue is involved in the proteasome-mediated degradation of Cx43 as previously suggested (31).
FIGURE 5.
Proteasome inhibitor MG132 blocks the TPA-induced depletion of Cx43 protein. A, HLECs were treated with or without MG132 (10 μm) for 30 min and coincubated with TPA (300 nm) for the time points indicated. Equal amounts of whole cell lysates were analyzed by Western blotting with Cx43, phospho-S368Cx43, Cx46, and Cx50 antibodies. β-Actin was used as loading control. B–D, densitometric analysis of the detected protein levels of Cx43 (pixel intensity of multiple Cx43 bands detected were calculated together) (B), Cx46 (C), and phospho-S368-Cx43 (D) in different treatments normalized to the levels of loading control, β-actin. The 0-h treatment is regarded as a control. The asterisks indicate significantly different amounts of protein levels in comparison to the control (p < 0.05). No significant change in the Cx50 protein levels was observed. Data are representative of three independent experiments.
Interestingly, whereas the Cx46 protein level increased in response to TPA treatment, pretreatment with MG132 blocked this up-regulation of Cx46 at the time points investigated (Fig. 5, A and D). No significant change in the Cx50 protein level was detected. This suggests that the up-regulation of Cx46 synthesis is possibly dependent on the decrease in Cx43 protein levels. Pretreatment with MG132 resulted in the detection of a slow moving band (upper band) for Cx46 suggesting that it might possibly be a phospho-Cx46 band, which accumulates in the presence of the proteasomal inhibitor. Taken together, these data suggest that TPA influences the up-regulation of Cx46 protein levels through the action of Cx43 degradation but does not clarify the up-regulation of Cx46 mRNA observed in our studies. Degradation of Cx43 could influence the up-regulation or stability of Cx46 mRNA in an unknown manner.
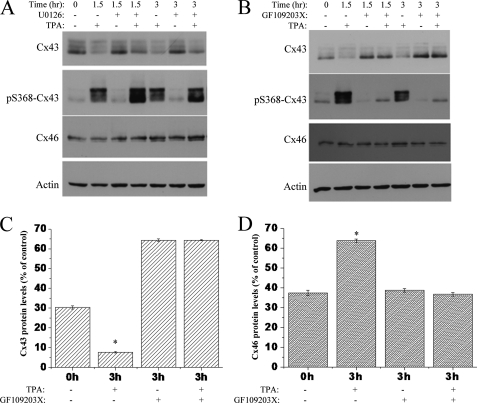
TPA-induced Degradation of Cx43 and an Increase in Cx46 Are Counteracted by a PKC Inhibitor
We next determined the signaling pathways that are involved in the TPA-induced Cx43 loss and Cx46 up-regulation. Treatment with the MEK-1 (MAPK pathway) inhibitor U0126 prior to the incubation with TPA for 1.5 or 3 h did not block the decrease in total Cx43 or pS368Cx43 phosphorylation (Fig. 6A). Pretreatment with U0126 was also not able to prevent the TPA-induced increase in Cx46 protein that occurred concomitantly with total Cx43 degradation and Ser-368 phosphorylation after 3 h of TPA treatment (Fig. 6A). However, Fig. 6B shows that the effects of TPA on Cx43 and Cx46 were completely counteracted by the PKC inhibitor GF109203X at 1.5 and 3 h of treatment. Pretreatment with GF109203X blocked the TPA-induced degradation of Cx43 and also decreased phosphorylation at Ser-368 of Cx43. Although TPA alone caused an increase in Cx46 protein after 3 h, pretreatment with GF109203X showed no such increase (Fig. 6, B and D). Together, these results indicated that the PKC pathway is involved in the TPA-induced degradation of Cx43 and the subsequent up-regulation of Cx46 in HLECs.
FIGURE 6.
TPA-induced degradation of Cx43 and increase in Cx46 protein were counteracted by the PKC inhibitor GF109203X. HLECs were grown to 70% confluency and treated with or without MEK-1 inhibitor U0126 (5 μm) for 30 min followed by TPA (300 nm) (A) or with or without GF109203X (10 μm) (B) for 30 min followed by TPA (300 nm), for the indicated time periods. Cell lysates were prepared and immunoblotted with Cx43, Ser-368-Cx43, Cx46, and β-actin antibodies. Notice the decrease in total Cx43 after treatment with TPA alone. Pre-treatment with the PKC inhibitor blocked the Cx43 degradation and Ser-368 phosphorylation after 1.5 and 3 h of TPA incubation. The increase of Cx46 protein level after 3 h of TPA treatment was also prevented by pre-treatment with GF109203X. These effects were not observed for the treatment with MEK-1 inhibitor. Quantitative comparison (densitometric analysis) of detected Cx43 (C) and Cx46 (D) protein levels (relative to β-actin loading control) in the presence of TPA with or without GF109203X are shown. Data are represented as mean ± S.E. The asterisk indicates significant statistical difference (p < 0.05).
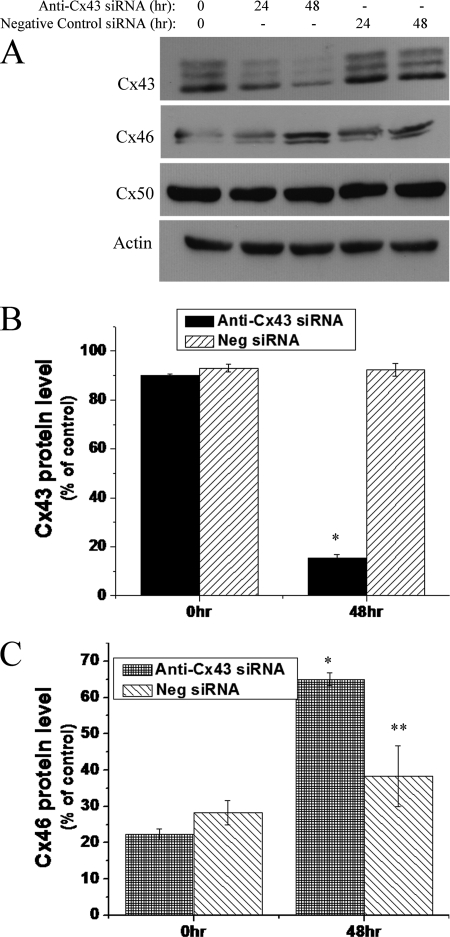
siRNA-mediated Knockdown of Endogenous Cx43 Results in Increased Cx46 Protein Levels in HLECs
Pretreatment with the PKC inhibitor blocked the TPA-induced Cx43 degradation that occurred concomitantly with the inhibition of TPA-induced increase in Cx46 protein. This led us to speculate whether the degradation of Cx43 is necessary for the increase of Cx46 protein. To check this, we silenced Cx43 expression by using a selective anti-Cx43 siRNA and examined the effect of Cx43 down-regulation on Cx46 protein levels. As shown in Fig. 7, transfection of HLECs with anti-Cx43 siRNA decreased the Cx43 protein levels significantly after 48 h. Interestingly, the decrease in Cx43 protein level (Fig. 7B) was associated with a notable increase in Cx46 protein at 48 h of siRNA transfection (Fig. 7C). However, treatment with anti-Cx43 siRNA did not cause a change in the amount of Cx50 protein (Fig. 7A). These data indicate that endogenous Cx43 is involved in the regulation of Cx46 protein expression in HLECs and further confirmed the endogenous reciprocal relationship of Cx46 and Cx43 expression.
FIGURE 7.
siRNA-mediated knockdown of Cx43 results in increased Cx46 protein levels. A, 70% confluent HLECs were treated with 50 nm of either anti-Cx43 siRNA or 50 nm of AllStars Negative Control siRNA for the indicated time periods. Increase in Cx46 level was noticed at 48 h of anti-Cx43 siRNA treatment when Cx43 was maximally down-regulated. B and C, densitometric analyses show siRNA-mediated selective down-regulation of Cx43 causes an increase in Cx46 protein levels. Data are represented as mean ± S.E. *, significant statistical difference (p < 0.05) between indicated data and control (at 0 h); **, statistical insignificance between the data and control (at 0 h).
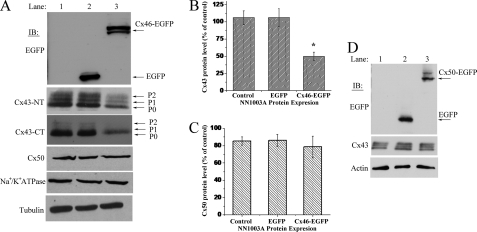
Overexpression of Cx46 Causes a Decrease in Cx43 Protein Levels in NN1003A Rabbit Lens Cells
Because the degradation of Cx43 was associated with an up-regulation of Cx46, we next investigated whether overexpression of Cx46 had an effect on Cx43. To examine this, we stably overexpressed the Cx46-EGFP fusion protein in rabbit lens epithelial NN1003A cells and analyzed the effect of Cx46 overexpression on the other lens connexins endogenously present in these cells, as well as other membrane proteins. The Cx46-EGFP-expressing cells showed the expression of fusion protein as two distinct bands around 75 kDa as determined by Western blot using an anti-EGFP antibody (Fig. 8A). The two bands in Fig. 8A are suspected to be phospho-Cx46-EGFP isoforms. Interestingly, stable overexpression of Cx46 decreased the Cx43 protein levels of the phosphorylated isoforms P0, P1, and P2. Stable overexpression of Cx46 reduced the Cx43 protein levels to 50% when compared with untransfected or EGFP-transfected cells (Fig. 8B). However, the Cx50 protein level was not changed due to the stable overexpression of Cx46 (Fig. 8, A and C). We also examined the effect of overexpression of Cx50 on Cx43 protein levels. Stable overexpression of Cx50-EGFP had no effect on Cx43 protein levels (Fig. 8D). Collectively, these data indicate that only Cx46 influences the decrease of Cx43 and that another closely related α-type connexin, Cx50, does not.
FIGURE 8.
Overexpression of Cx46 causes reduction in Cx43 protein levels in rabbit lens epithelial NN1003A cells. A, Western blot analyses of Cx46-EGFP, Cx43, Cx50, tubulin, and Na+/K+ ATPase in untransfected control cells and in cells stably overexpressing EGFP or Cx46-EGFP. Whole cell lysates were made, and equal amounts of total protein were run in SDS-PAGE followed by immunoblot. The blot was probed with anti-EGFP antibody to detect Cx46-EGFP expression. The blots were also probed with Cx43CT (against C terminus) and Cx43NT (against N terminus) antibodies to detect Cx43 to rule out any C-terminal truncation of Cx43. B, densitometric analyses showed significant reduction of Cx43 protein levels in Cx46-EGFP stably transfected NN1003A cells, whereas (C) the Cx50 protein level showed no change. The bands representing Cx43, Cx50, and tubulin were digitized by UN-SCAN-It gel software. The average pixel values were calculated for Cx43 bands, normalized against average pixel values for loading control (tubulin), and then plotted in percent of control. Data are represented as mean ± S.E. of three independent experiments. *, significant statistical difference (p < 0.05) between indicated data and control cells. D, overexpression of Cx50 does not cause a decrease in Cx43 protein levels. NN1003A cells were stably transfected with plasmid encoding Cx50-EGFP. Cell lysates were prepared, and equal amounts of total protein were run in to SDS-PAGE followed by Western blot. The Cx50-EGFP and Cx43 proteins were detected by probing the blots with EGFP and Cx43 antibodies, respectively.
Cx46 Is Predominantly Localized to the Intracellular Compartments in Lens NN1003A Cells
We determined the intracellular localization of Cx46 in the stably transfected cells by intrinsic EGFP fluorescence and immunofluorescence microscopy. Green fluorescence of Cx46-EGFP expression was predominantly detected to an intracellular perinuclear compartment (Fig. 9A). The fluorescence signal of Cx46-EGFP was not observed at the plasma membrane region.
FIGURE 9.
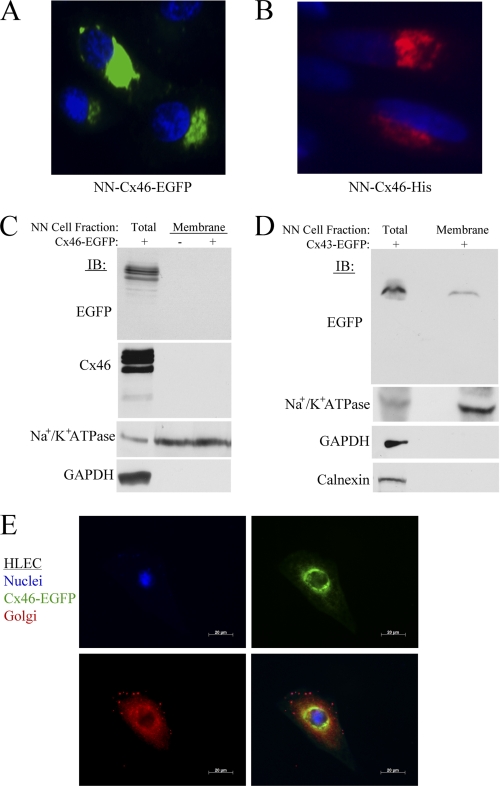
Cx46 protein is predominantly localized to the intercellular compartments in lens NN1003A cells. A, fluorescence microscopy images of NN1003A cells stably overexpressing Cx46-EGFP. All cells were fixed and stained with DAPI to visualize nuclei. B, the localization of Cx46-His protein (pentahistidine-tagged) in NN1003A cells. Cells were transfected with plasmid encoding Cx46-His and, after 24 h of transfection, cells were fixed, permeabilized, and labeled with Penta-His antibody (red) and DAPI (blue). C, Western blot analyses were performed for Cx46 (with anti-EGFP or anti-Cx46) on equal amounts of total protein from lysates of Cx46-EGFP-expressing cells (lane 1) and plasma membrane protein fractions isolated from untransfected control cells (lane 2) or Cx46-EGFP stable (lane 3) cells. The blots were also probed with antibodies against a plasma membrane marker Na+/K+ ATPase, and a cytosolic marker (GAPDH) to demonstrate the purity of plasma membrane protein extracts. D, Western blot analyses show the localization of Cx43-EGFP in the plasma membrane of Cx43-EGFP-expressing NN1003A cells. Cells were transiently transfected with plasmid encoding Cx43-EGFP for 24 h. Then, the cells were fractionated into plasma membrane protein fractions. Equal amounts of total protein of lysate and plasma membrane fraction of Cx43-EGFP-expressing cells were subjected to Western blot analyses with antibodies against EGFP, Na+/K+ ATPase, GAPDH, and calnexin (an ER marker). E, fluorescence microscopy images of HLEC cells expressing Cx46-EGFP (green) that were fixed, permeabilized, and labeled with 58K/FTCD antibody (red, a Golgi marker) and DAPI (blue). Cx46-EGFP did not co-localize well with the Golgi marker. NN, NN1003A cells.
The EGFP protein is a rather large fusion protein tag and has a molecular mass of 27 kDa. Therefore, we speculated whether EGFP tagging at the C terminus of Cx46 prevented it from reaching the cell surface. To investigate this, we transfected cells with a plasmid encoding Cx46-His, which encodes six histidines fused to the C terminus of Cx46. We then examined the localization of Cx46-His fusion protein via immunofluorescence to assess differences in the localization of Cx46. Immunofluorescence using the anti-His antibody also demonstrated that Cx46 localized predominantly to the intracellular perinuclear compartments and not to the plasma membrane (Fig. 9B, red channel).
To confirm the localization of Cx46, untransfected or stably transfected cells were fractionated to isolate plasma membrane proteins. Western blot analyses with antibodies against EGFP and Cx46 detected no immunoreactive bands that corresponded to endogenous or Cx46-EGFP or Cx46 protein in the plasma membrane fraction of untransfected or transfected cells (Fig. 9C). In all the preparations of plasma membrane fractions, a marker for plasma membrane (Na+/K+ ATPase) was enriched; meanwhile, the cytosolic marker GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was not, confirming the purity of the membrane fractions.
To further demonstrate that EGFP tagging does not affect the localization of connexin proteins in NN1003A cells, cells were transiently transfected to express Cx43-EGFP. Using Western blot analysis, Cx43-EGFP fusion protein (around 70 kDa) was detected in plasma membrane fractions isolated from Cx43-GFP-overexpressing NN1003A cells (Fig. 9D). These data clearly suggested that EGFP tagging had no influence on the Cx43 or Cx46 transport and localization within the lens cell. Our data are consistent with other literature confirming that C-terminal tagging of connexins does not inhibit their intracellular transport (41, 42).
Because Cx46 is localized to the perinuclear region, we further characterized which compartment Cx46 is localizing to by using three-color fluorescence microscopy. Fig. 9E shows that Cx46-EGFP localizes to the perinuclear region in HLECs but does not co-localize well with the Golgi marker 58K/FTCD protein. This suggests that Cx46-EGFP could reside in the endoplasmic reticulum compartment of HLECs.
Treatment with Proteasome Inhibitors Restore Cx43 Protein Levels, whereas Lysosome Inhibitors Have No Effect
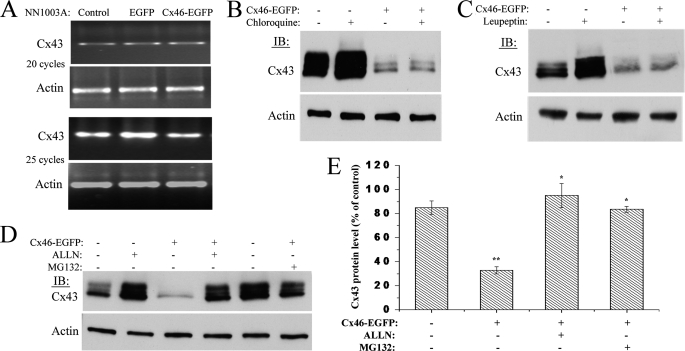
We then investigated whether Cx43 degradation was pre- or post-transcriptionally initiated in Cx46-overexpressing NN1003A cells. To check this, RT-PCR was carried out to detect the Cx43 transcript levels in cells stably or transiently overexpressing Cx46-EGFP. Fig. 10A shows that, with the overexpression of Cx46-EGFP, no change in Cx43 mRNA level was observed, indicating the occurrence of post-transcriptional regulation of Cx43. We then investigated the pathway that is involved in the Cx46-induced degradation of Cx43.
FIGURE 10.
Treatment with lysosome inhibitors has no effect, whereas proteasome inhibitors have a robust effect on Cx46-induced degradation of Cx43. A, RT-PCR analyses of Cx43 mRNA levels in NN1003A cells overexpressing Cx46-EGFP. Total RNA was isolated from stably transfected cells followed by RT-PCR using specific primers for Cx43 cDNA and β-actin cDNA (internal control). The numbers of cycles of PCR are given. Untransfected control and Cx46-EGFP stably transfected cells were treated with lysosome inhibitors chloroquine (200 μm) (B) or leupeptin (200 μm) (C) for 4 h. Cell lysates were prepared, and equal amounts of total protein were analyzed by Western blot using antibody against Cx43CT and β-actin (loading control). D, control or Cx46-EGFP stably transfected NN1003A cells were treated with proteasome inhibitors, ALLN (100 μm) or MG132 (10 μm), for 4 h. Cell lysates were prepared, and equal amounts of total cell protein in each cell lysate were analyzed by Western blot with antibodies against Cx43CT and β-actin (loading control). E, densitometric analyses show that the treatment with ALLN or MG132 restored Cx43 protein levels in Cx46-EGFP-expressing cells. Cx43 and β-actin bands were digitized by UN-SCAN-It gel software. The average pixel value was calculated for Cx43 bands, normalized, and plotted in percent of control (β-actin). *, significant statistical difference (p < 0.05) between indicated data and Cx46-EGFP-expressing untreated cells (**). Data are represented as mean ± S.E. of three independent experiments.
Cx43 degradation has been reported to be mediated by both lysosomal and proteasomal pathways (4, 43–45). To identify which proteolytic degradation pathway is involved in the reciprocal relationship of Cx43 and Cx46, we treated stably transfected NN1003A cells with Cx46-EGFP then treated the cultures with a series of inhibitors for both the lysosome and proteasome. Next, we analyzed their effects on Cx43 degradation by Western blot. Treatment with the lysosome inhibitor, chloroquine, did not cause a change in the Cx46-induced loss of Cx43 (Fig. 10B). Treatment with another lysosome inhibitor, leupeptin, showed the same result as no change in the reduced levels of Cx43 were observed (Fig. 10C). However, when stably transfected NN1003A cells were incubated with the proteasome inhibitor, ALLN, Cx43 protein levels were not reduced (Fig. 10, D (lane 4) and E). Treatment with ALLN restored all three phospho-isoforms of Cx43 protein (P0, P1, and P2) to the levels of Cx43 protein in untransfected control cells (Fig. 10, D and E).
The restoration of Cx43 protein levels, due to the addition of the proteasome inhibitor, was further confirmed by treating the cells with another proteasome inhibitor, MG132 (Fig. 10, D and E). We observed that the Cx46-induced loss of Cx43 was strongly counteracted in Cx46-EGFP-expressing NN1003A cells by the treatment of MG132 (Fig. 10D, lane 6). Together, these results suggested that the proteasome is the major site where Cx43 is degraded upon Cx46 overexpression in NN1003A lens cells.
Overexpression of Cx46 Increases Cx43 Ubiquitination
Proteasomal degradation may involve direct proteolysis or ubiquitin-mediated degradation of targeted proteins. Ligation of ubiquitin has been previously shown to serve as an important signal in the regulation of Cx43 localization and turnover (31, 46, 47). To determine if Cx46-induced Cx43 degradation is initiated by ubiquitin conjugation, Cx43 was immunoprecipitated from untransfected, EGFP-transfected, or Cx46-EGFP stably transfected cells using anti-Cx43CT antibody. Immunoprecipitated samples were then analyzed for the presence of ubiquitinated Cx43 by Western blot using an anti-ubiquitin antibody.
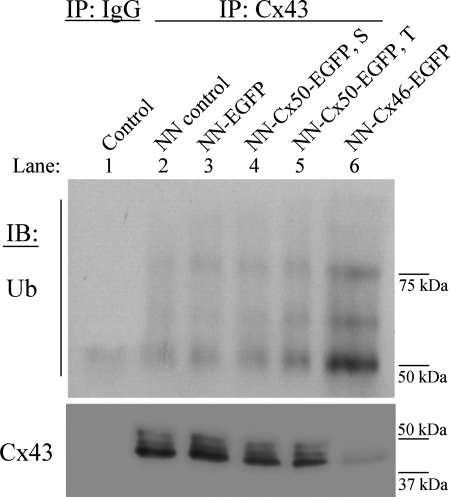
In untransfected and EGFP (empty vector)-transfected cells, a faint ladder of ubiquitin bands of high molecular mass (50- to 75-kDa region), characteristic of polyubiquitinated Cx43, was observed (Fig. 11, lanes 2 and 3). Interestingly, cells stably expressing Cx46-EGFP demonstrated a ladder of ubiquitin bands with strong intensities at the same region of the blot, corresponding to ubiquitinated Cx43 (Fig. 11, lane 6). However, in the negative control sample, where cell lysate made from Cx46-EGFP-expressing cells was immunoprecipitated with a rabbit nonspecific IgG, ubiquitin bands were not detected (Fig. 11, lane 1).
FIGURE 11.
Overexpression of Cx46 increases ubiquitination of Cx43. Lysates from untransfected NN1003A cells (lane 2) or EGFP (lane 3), Cx50-EGFP (lane 4), Cx46-EGFP (lane 6) stably transfected (S) or Cx50-EGFP transiently transfected (T) cells (36 h, lane 5) were prepared and subjected to immunoprecipitation by rabbit anti-Cx43CT antibody. Equal amounts of immunoprecipitated samples were loaded into 8% SDS-PAGE. Ubiquitinated Cx43 was detected by immunoblotting using anti-ubiquitin antibody. As a negative control, cell lysate, made from Cx46-EGFP-expressing cells, was immunoprecipitated with a rabbit nonspecific IgG. The same blot was stripped and reprobed with anti-Cx43 antibody (anti-Cx43CT) to show equal loading of immunoprecipitates. A band observed at the 50-kDa region in the negative control sample probably resulted from cross-reactivity of the secondary antibody with IgG. As a positive control of anti-ubiquitin antibody, a commercially available purified ubiquitin ladder (ubiquitin is polymerized through lysine 48) was subjected to 15% SDS-PAGE. Western blotting was performed and probed with anti-ubiquitin antibody (data not shown).
Additionally, we investigated whether Cx50 overexpression had any effect on Cx43 ubiquitination. Stable or transient overexpression of Cx50 showed the presence of a similar ubiquitin ladder to that of the controls with faint intensities (Fig. 11, lanes 4 and 5). These data indicated that Cx46 overexpression caused an increase in the ubiquitination of Cx43, which correlates well with our degradation pathway inhibitor study.
The C-terminal Tail Domain of Cx46 Is Required for Induction of Cx43 Degradation
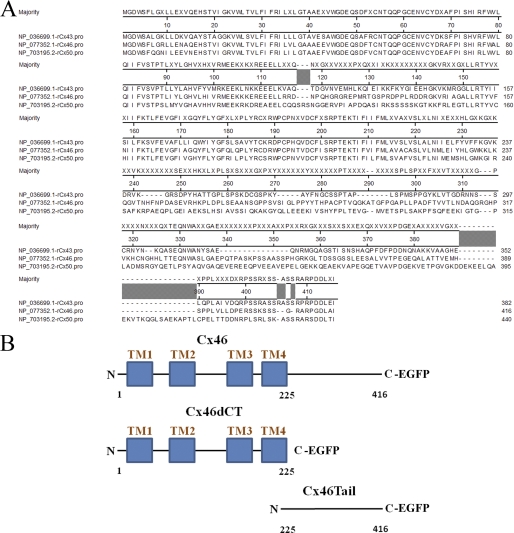
Primary amino acid sequence alignments of rat Cx43, Cx46, and Cx50 show significant sequence similarity at their N-terminal and transmembrane domains similar to that of all α-type connexins (48). The major variability in the amino acid sequences of Cx43, Cx46, and Cx50 is found at the cytoplasmic C-terminal tail domain (Fig. 12A). Because Cx50 overexpression was not found to mediate Cx43 degradation, we speculated whether the C-terminal tail domain (amino acids 225–416) is the active domain of Cx46 that induces Cx43 degradation. To test this hypothesis, we created two Cx46 mutants. One mutant contains the N terminus and all four transmembrane domains (amino acids 1–225, named Cx46dCT). The other Cx46 mutant contains amino acids 225–416, which encode only the C-terminal tail domain (named Cx46Tail). Each mutant is expressed as an EGFP fusion protein, similar to Cx46-EGFP (Fig. 12B).
FIGURE 12.
Protein sequence alignment of rat Cx43, Cx46, and Cx50 and schematic of Cx46 mutants. A, the amino acid residues of Cx43, Cx46, and Cx50 are given as a ClustalW alignment produced with Lasergene 9 MegAlign software (DNAStar, Inc., Madison, WI). Sequences are numbered according to the Cx46 sequence. The NCBI reference sequences used are NP_036699.1 for Cx43, NP_077352.1 for Cx46, and NP_703195.2 for Cx50. B, schematic diagram of wild-type Cx46 and Cx46 C-terminal deletion mutant (Cx46dCT, deletion of amino acids 225–416), as well as a schematic of the Cx46 tail domain mutant (Cx46Tail, deletion of amino acids 1–224).
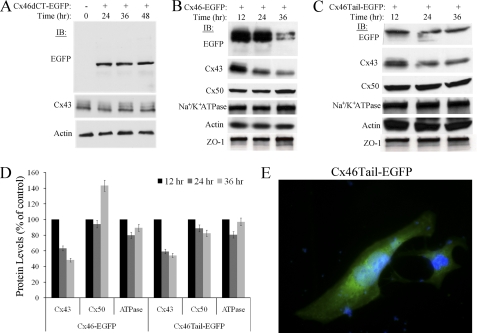
Next, we transfected NN1003A cells with either the plasmid-encoding wild-type Cx46-EGFP, Cx46dCT-EGFP, or Cx46Tail-EGFP, and then analyzed the effect of mutant Cx46 protein on the Cx43, Cx50, Na+/K+ ATPase, and ZO-1 protein levels (Fig. 13, A–D). Cx46dCT-EGFP was expressed at the predicted molecular mass of ∼50 kDa as determined by Western blot using anti-EGFP antibody (Fig. 13A). Notably, overexpression of Cx46dCT-EGFP did not induce Cx43 degradation at 24–48 h after transfection (Fig. 13A). The role of the C-terminal tail domain of Cx46 in inducing Cx43 degradation was confirmed by overexpressing Cx46Tail-EGFP. Overexpression of Cx46Tail-EGFP (Fig. 13C), like wild-type Cx46-EGFP (Fig. 13B), was able to cause a reduction of Cx43 protein at 24 and 36 h of transient transfection (Fig. 13D). Additionally, Cx46Tail-EGFP exhibited no specific localization within HLECs as shown by fluorescence microscopy (Fig. 13E). These data demonstrate that the C-terminal cytoplasmic tail domain of Cx46 is responsible for inducing the degradation of Cx43 in lens cells.
FIGURE 13.
The C-terminal tail domain of Cx46 is required for Cx43 degradation. A, overexpression of Cx46 deletion mutant (Cx46dCT-EGFP) does not induce Cx43 degradation. Western blot analyses of Cx46dCT-EGFP and Cx43 in untransfected cells (0 h) or in cells transiently transfected with plasmid encoding the Cx46 deletion mutant (Cx46dCT-EGFP). The expression of Cx46dCT-EGFP was confirmed by probing the blot with the EGFP antibody. The blot was also probed with Cx43CT and β-actin antibodies. B, NN1003A cells were transiently transfected with full-length Cx46-EGFP, and whole cell lysates were prepared after 12, 24, and 36 h post-transfection. Equal amounts of total protein were analyzed by 10% SDS-PAGE followed by immunoblot. The blots were probed with antibodies to detect EGFP, Cx43, Cx50, Na+/K+ ATPase, and β-actin. There is no change in any of the protein levels of membrane-associated proteins Cx50 or Na+/K+ ATPase, indicating that overexpression of Cx46-EGFP only affects Cx43 protein levels. C, overexpression of Cx46 C-terminal tail domain (Cx46Tail-EGFP, residues 225–416 of wild type) in NN1003A cells caused reduction in Cx43 protein levels after 24 and 36 h of transient transfection. This provides evidence that the CT tail domain of Cx46 induces the degradation of Cx43 through either direct or indirect interaction. D, densitometric analysis of Western blots used in B and C. Blots were digitized by UN-SCAN-It gel software. The average pixel value was calculated for all protein bands, normalized, and plotted in percent of control (β-actin). Data are represented as the mean of two independent experiments. E, fluorescence microscopy of HLECs expressing Cx46Tail-EGFP. The tail domain did not localize to any specific intracellular compartment, confirming cytoplasmic expression and not membrane association.
DISCUSSION
In the present study we investigated the reciprocal relationship between expression and turnover of two gap junction connexin proteins, Cx43 and Cx46, in lens epithelial cells. The use of vertebrate lens as a system to study gap junction communication and connexins function has been increasing in recent years. The developing lens also serves as a highly accessible paradigm for the universal study of cell proliferation, epithelial function, and cell differentiation, as well as studies of hypoxia. Here we show that TPA induces internalization and degradation of Cx43 in human lens epithelial cells. TPA-induced Cx43 degradation is mediated by the PKC pathway, whereas the MAPK pathway is apparently not involved, as determined by studies with specific inhibitors for these pathways. Previously, we have reported the association of PKCγ with Cx43 in response to TPA treatment in HLECs (28, 37). TPA-induced Cx43 hyperphosphorylation and internalization from membrane have been well established in the cultured rat liver epithelial cell line IAR20 (31). In HLECs, TPA-induced degradation of Cx43 was associated with an increase in phosphorylation of Ser-368, and treatment with PKC inhibitor prevented the phosphorylation, at this residue, and also prevented Cx43 degradation. Therefore, our study shows that the PKC-mediated phosphorylation of Cx43 on Ser-368 acts as a critical signal for internalization and degradation.
The up-regulation of Cx46 in response to TPA treatment is a novel finding of our study. TPA indeed has been previously reported to cause transcriptional up-regulation of Cx26 in a PKC-mediated manner (49). The up-regulation of Cx46 message level in response to TPA treatment suggests a similar possibility. Therefore, further studies are required to characterize the promoter region of the Cx46 gene for the presence of a TPA response element or xenobiotic response element that will give insight into the mechanism of TPA-induced up-regulation of Cx46. Current work in our laboratory has identified possible regulatory regions in the Cx46 promoter.
The reciprocal expression of Cx43 and Cx46 was further observed in rabbit lens epithelial cells, overexpressing Cx46-EGFP. These cells exhibited a reduction of endogenous Cx43 protein levels. We confirmed the reduction of all three phospho-isoforms of Cx43 protein (P0, P1, and P2) by Western blotting with antibodies against C- and N-terminal domains. This precaution aids in ruling out the possibility that proteolytic cleavage of Cx43 epitopes may be contributing to the reduction of Cx43 observed via Western blotting.
Several studies have previously reported the involvement of both proteasomal and lysosomal pathways in the degradation of connexins (4, 43–45). The lysosomal pathway was shown not to be involved in the degradation of Cx43 when it is internalized from cell membranes. Treatment with lysosome inhibitors leupeptin and chloroquine showed no effect on the degradation of Cx43 in Cx46-EGFP-expressing cells. But, the treatment with proteasome inhibitors ALLN and MG132 strongly counteracted Cx46-induced Cx43 degradation. In addition, co-immunoprecipitation analyses showed an increase in Cx43 ubiquitination associated only with Cx46-EGFP-expressing cells. Together, these results suggest that Cx46-induced degradation of Cx43 is mediated by the ubiquitin-proteasome pathway, whereas the lysosome degradation pathway does not appear to be involved.
In this study, we observed that Cx46 is mainly localized to the intracellular compartments of lens epithelial cells. Even upon overexpression of Cx46 or after TPA treatment, the localization of Cx46 was not detected in the plasma membrane, as determined by immunofluorescence confocal and EGFP-intrinsic fluorescence microscopy. This was further confirmed by Western blot analyses of cellular fractionation. The intracellular localization of Cx46 was also shown in bone osteoblast (39) and lung alveolar cells (40). Conventional techniques such as fluorescence microscopy and Western blot do not completely rule out the possibility of membrane localization of Cx46 in our lens cultures, because Cx46 has a major membrane-localized function in vivo in the lens. Connexins have rapid turnover rates with half-lives of a few hours (50–52). Therefore, if Cx46 is rapidly internalized after being delivered to the plasma membrane, it would not be detectable at the cell surface by conventional techniques. Nonetheless, because membrane localization was not observed, it is unlikely that overexpression of Cx46 induced Cx43 degradation by facilitating loss from the plasma membrane.
Due to its observed intracellular localization, Cx46 is likely to induce Cx43 degradation by a gap junction-independent mechanism. This leads us to speculate two possibilities for the degradation of Cx43. One, that endoplasmic reticulum (ER)-associated degradation mediates the Cx46-induced degradation of ubiquitinated Cx43 species observed in our studies (either folded or unfolded Cx43). Or, two, Cx46 associates with Cx43 either directly or indirectly in the ER prior to transit to the cellular membrane and induces the degradation of Cx43 outside of the ER. Nonetheless, these hypotheses need to be thoroughly tested.
Additionally, the Cx46dCT-EGFP mutant failed to induce Cx43 degradation. However, overexpression of the Cx46Tail-EGFP mutant, which localizes to the cytoplasm, caused a reduction of Cx43 protein levels. This is interesting because the Cx46Tail mutant does not contain the membrane-anchoring signals found in the N terminus of Cx46 but yet can still cause degradation of endogenous Cx43. It is possible that Cx46Tail stimulates ER-associated degradation of Cx43 during translation, but this does not rule out potential Cx46Tail interaction with Cx43 at the plasma membrane. Therefore, it is likely that Cx46 mediates its action through putative binding partners at the C-terminal tail domain. The cytoplasmic tail domain of Cx46 has predicted binding sites for various kinases, such as PKC, CK-1, Akt/PKB, and MAPK, among others. Further studies with mutant Cx46 proteins that are defective in phosphorylation sites or kinase-binding sites are required to unravel the mechanism of Cx43 degradation upon Cx46 overexpression. Also, studies elucidating the roles of various cellular compartments aiding in the interaction between Cx43 and Cx46 would provide insight into which kinases or other binding proteins are involved in the reciprocal relationship.
Our results can be extended to understand the molecular mechanism of connexin-influenced regulation of oncogenesis. Cx43 has anti-tumor properties, and expression of Cx43 can be down-regulated in tumors (22–25). Moreover, in previous studies, we have shown that Cx46 is up-regulated in breast tumor samples, MCF-7 xenografts, and Y79 retinoblastoma xenografts and acts as an oncogene to favor early tumor growth (24, 25). Therefore, we speculate that up-regulation of Cx46 promotes tumor growth by inducing the degradation of the tumor suppressor protein Cx43. Indeed, in a recent study we have found that the down-regulation of Cx46 by siRNA inhibited Y79 retinoblastoma xenograft tumor growth, which was associated, interestingly, with an increase in Cx43 protein levels in vivo (25).
In conclusion, our data indicate that Cx43 and Cx46 are expressed and regulated in a reciprocal manner in lens epithelial cells. Depletion of Cx43 is associated with an up-regulation of Cx46, and both processes are mediated by PKC. We also provide evidence that endogenous Cx43 regulates endogenous Cx46 at the protein level. Exogenous expression of Cx46, on the other hand, induces degradation of Cx43. The C-terminal tail domain of Cx46, which has less sequence similarity with other connexins, is required to induce degradation of Cx43. The ubiquitination of Cx43 is strongly increased in Cx46-expressing cells, and proteasome inhibitors counteract Cx43 degradation. Therefore, Cx46-induced Cx43 degradation is likely to be mediated via the ubiquitin-proteasome pathway in the perinuclear region of lens cells, although the exact cellular mechanism warrants further characterization.
This work was supported, in whole or in part, by National Institutes of Health Grant EY13421 (to D. J. T.) and NIH K-INBRE Grant P20RR016475 (to D. M.). This work was also supported by Terry C. Johnson Cancer Center Innovative Research Grant (to D. J. T.) and Terry C. Johnson Cancer Center Summer Stipend (to D. B. and S. D.).
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- MG132
- N-[(phenylmethoxy)carbonyl]-l-leucy-l-N-[(1S)-1-formyl-3-methylbutyl]-l-leucinamide
- ALLN
- N-acetyl-Leu-Leu-Nle-CHO
- HLEC
- human lens epithelial cell
- PDD
- 4α-phorbol-12,13-didecanoate
- CT
- C-terminal
- NT
- N-terminal
- qRT-PCR
- reat-time quantitative PCR
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Goodenough D. A., Goliger J. A., Paul D. L. (1996) Annu. Rev. Biochem. 65, 475–502 [DOI] [PubMed] [Google Scholar]
- 2. Loewenstein W. R. (1980) Ann. N.Y. Acad. Sci. 339, 39–45 [DOI] [PubMed] [Google Scholar]
- 3. Yamasaki H., Naus C. C. (1996) Carcinogenesis 17, 1199–1213 [DOI] [PubMed] [Google Scholar]
- 4. Laird D. W. (2006) Biochem. J. 394, 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salameh A. (2006) Adv. Cardiol. 42, 57–70 [DOI] [PubMed] [Google Scholar]
- 6. Brissette J. L., Kumar N. M., Gilula N. B., Dotto G. P. (1991) Mol. Cell. Biol. 11, 5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lampe P. D. (1994) J. Cell Biol. 127, 1895–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bevans C. G., Harris A. L. (1999) J. Biol. Chem. 274, 3711–3719 [DOI] [PubMed] [Google Scholar]
- 9. Trexler E. B., Bukauskas F. F., Bennett M. V., Bargiello T. A., Verselis V. K. (1999) J. Gen Physiol. 113, 721–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lampe P. D., Lau A. F. (2004) Int. J. Biochem. Cell Biol. 36, 1171–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Musil L. S., Beyer E. C., Goodenough D. A. (1990) J. Membr. Biol. 116, 163–175 [DOI] [PubMed] [Google Scholar]
- 12. Dahm R., van Marle J., Prescott A. R., Quinlan R. A. (1999) Exp. Eye Res. 69, 45–56 [DOI] [PubMed] [Google Scholar]
- 13. Rong P., Wang X., Niesman I., Wu Y., Benedetti L. E., Dunia I., Levy E., Gong X. (2002) Development 129, 167–174 [DOI] [PubMed] [Google Scholar]
- 14. TenBroek E. M., Johnson R., Louis C. F. (1994) Invest. Ophthalmol. Vis. Sci. 35, 215–228 [PubMed] [Google Scholar]
- 15. Paul D. L., Ebihara L., Takemoto L. J., Swenson K. I., Goodenough D. A. (1991) J. Cell Biol. 115, 1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White T. W., Bruzzone R., Goodenough D. A., Paul D. L. (1992) Mol. Biol. Cell 3, 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mathias R. T., White T. W., Gong X. (2010) Physiol. Rev. 90, 179–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beyer E. C., Kistler J., Paul D. L., Goodenough D. A. (1989) J. Cell Biol. 108, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong X., Li E., Klier G., Huang Q., Wu Y., Lei H., Kumar N. M., Horwitz J., Gilula N. B. (1997) Cell 91, 833–843 [DOI] [PubMed] [Google Scholar]
- 20. Lee S. W., Tomasetto C., Paul D., Keyomarsi K., Sager R. (1992) J. Cell Biol. 118, 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirschi K. K., Xu C. E., Tsukamoto T., Sager R. (1996) Cell Growth Differ. 7, 861–870 [PubMed] [Google Scholar]
- 22. Kanczuga-Koda L., Sulkowski S., Tomaszewski J., Koda M., Sulkowska M., Przystupa W., Golaszewska J., Baltaziak M. (2005) Oncol. Rep. 14, 325–329 [PubMed] [Google Scholar]
- 23. Shao Q., Wang H., McLachlan E., Veitch G. I., Laird D. W. (2005) Cancer Res. 65, 2705–2711 [DOI] [PubMed] [Google Scholar]
- 24. Banerjee D., Gakhar G., Madgwick D., Hurt A., Takemoto D., Nguyen T. A. (2010) Int. J. Cancer 127, 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burr D. B., Molina S. A., Banerjee D., Low D. M., Takemoto D. J. (2011) Exp. Eye Res. 92, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hombach S., Janssen-Bienhold U., Söhl G., Schubert T., Büssow H., Ott T., Weiler R., Willecke K. (2004) Eur. J. Neurosci. 19, 2633–2640 [DOI] [PubMed] [Google Scholar]
- 27. Kelsell D. P., Dunlop J., Stevens H. P., Lench N. J., Liang J. N., Parry G., Mueller R. F., Leigh I. M. (1997) Nature 387, 80–83 [DOI] [PubMed] [Google Scholar]
- 28. Akoyev V., Takemoto D. J. (2007) Cell Signal. 19, 958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Solan J. L., Lampe P. D. (2009) Biochem. J. 419, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zampighi G. A., Planells A. M., Lin D., Takemoto D. (2005) Invest. Ophthalmol. Vis. Sci. 46, 3247–3255 [DOI] [PubMed] [Google Scholar]
- 31. Leithe E., Rivedal E. (2004) J. Biol. Chem. 279, 50089–50096 [DOI] [PubMed] [Google Scholar]
- 32. Rivedal E., Yamasaki H., Sanner T. (1994) Carcinogenesis 15, 689–694 [DOI] [PubMed] [Google Scholar]
- 33. Matesic D. F., Rupp H. L., Bonney W. J., Ruch R. J., Trosko J. E. (1994) Mol. Carcinog. 10, 226–236 [DOI] [PubMed] [Google Scholar]
- 34. Musil L. S., Cunningham B. A., Edelman G. M., Goodenough D. A. (1990) J. Cell Biol. 111, 2077–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Musil L. S., Goodenough D. A. (1991) J. Cell Biol. 115, 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lampe P. D., Lau A. F. (2000) Arch. Biochem. Biophys. 384, 205–215 [DOI] [PubMed] [Google Scholar]
- 37. Akoyev V., Das S., Jena S., Grauer L., Takemoto D. J. (2009) Invest. Ophthalmol. Vis. Sci. 50, 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Das S., Wang H., Molina S. A., Martinez-Wittinghan F. J., Jena S., Bossmann L. K., Miller K. A., Mathias R. T., Takemoto D. J. (2011) Cur. Eye Res. 10.3109/02713683.2011.573899 [DOI] [PubMed] [Google Scholar]
- 39. Koval M., Harley J. E., Hick E., Steinberg T. H. (1997) J. Cell Biol. 137, 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Avanzo J. L., Mesnil M., Hernandez-Blazquez F. J., da Silva T. C., Fukumasu H., Mori C. M., Yamasaki H., Dagli M. L. (2006) Life Sci. 79, 2202–2208 [DOI] [PubMed] [Google Scholar]
- 41. Paemeleire K., Martin P. E., Coleman S. L., Fogarty K. E., Carrington W. A., Leybaert L., Tuft R. A., Evans W. H., Sanderson M. J. (2000) Mol. Biol. Cell 11, 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomas T., Jordan K., Simek J., Shao Q., Jedeszko C., Walton P., Laird D. W. (2005) J. Cell Sci. 118, 4451–4462 [DOI] [PubMed] [Google Scholar]
- 43. Laing J. G., Tadros P. N., Westphale E. M., Beyer E. C. (1997) Exp. Cell Res. 236, 482–492 [DOI] [PubMed] [Google Scholar]
- 44. Laing J. G., Beyer E. C. (1995) J. Biol. Chem. 270, 26399–26403 [DOI] [PubMed] [Google Scholar]
- 45. Qin H., Shao Q., Igdoura S. A., Alaoui-Jamali M. A., Laird D. W. (2003) J. Biol. Chem. 278, 30005–30014 [DOI] [PubMed] [Google Scholar]
- 46. Leithe E., Rivedal E. (2007) J. Membr. Biol. 217, 43–51 [DOI] [PubMed] [Google Scholar]
- 47. Leithe E., Rivedal E. (2004) J. Cell Sci. 117, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 48. White T. W., Bruzzone R., Wolfram S., Paul D. L., Goodenough D. A. (1994) J. Cell Biol. 125, 879–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li G. Y., Lin H. H., Tu Z. J., Kiang D. T. (1998) Gene 209, 139–147 [DOI] [PubMed] [Google Scholar]
- 50. Fallon R. F., Goodenough D. A. (1981) J. Cell Biol. 90, 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beardslee M. A., Laing J. G., Beyer E. C., Saffitz J. E. (1998) Circ. Res. 83, 629–635 [DOI] [PubMed] [Google Scholar]
- 52. Hervé J. C., Derangeon M., Bahbouhi B., Mesnil M., Sarrouilhe D. (2007) J. Membr. Biol. 217, 21–33 [DOI] [PubMed] [Google Scholar]