Abstract
The human major histocompatibility complex (MHC) on chromosome 6p21 is a paradigm for genomics, showing remarkable polymorphism and striking association with immune and non-immune diseases. The complex genomic landscape of the MHC, notably strong linkage disequilibrium, has made resolving causal variants very challenging. A promising approach is to investigate gene expression levels considered as tractable intermediate phenotypes in mapping complex diseases. However, how transcription varies across the MHC, notably relative to specific haplotypes, remains unknown. Here, using an original hybrid tiling and splice junction microarray that includes alternate allele probes, we draw the first high-resolution strand-specific transcription map for three common MHC haplotypes (HLA-A1-B8-Cw7-DR3, HLA-A3-B7-Cw7-DR15, and HLA-A26-B18-Cw5-DR3-DQ2) strongly associated with autoimmune diseases including type 1 diabetes, systemic lupus erythematosus, and multiple sclerosis. We find that haplotype-specific differences in gene expression are common across the MHC, affecting 96 genes (46.4%), most significantly the zing finger protein gene ZFP57. Differentially expressed probes are correlated with polymorphisms between haplotypes, consistent with cis effects that we directly demonstrate for ZFP57 in a cohort of healthy volunteers (P = 1.2 × 10−14). We establish that alternative splicing is significantly more frequent in the MHC than genome-wide (72.5% vs. 62.1% of genes, P ≤ 1 × 10−4) and shows marked haplotypic differences. We also unmask novel and abundant intergenic transcription involving 31% of transcribed blocks identified. Our study reveals that the renowned MHC polymorphism also manifests as transcript diversity, and our novel haplotype-based approach marks a new step toward identification of regulatory variants involved in the control of MHC-associated phenotypes and diseases.
The human major histocompatibility complex (MHC), located on chromosome 6p21 in humans, previously referred to as the “human leukocyte antigen (HLA) complex,” plays a pivotal role in immune function (Dausset 1981). This region of 3.5 Mb is the most gene-dense of the genome, with 230 known genes and pseudogenes (Horton et al. 2004). It is classically divided into the class I region, which includes genes such as HLA-A, HLA-B, and HLA-C, and the class II region, including, for example, HLA-DP, HLA-DQ, and HLA-DR. These classical HLA genes encode molecules involved in antigen presentation and processing. The intervening MHC class III region notably includes genes encoding a variety of proteins involved in immunity including the Tumor Necrosis Factor (TNF) superfamily, components of the complement cascade, and molecular chaperones such as heat-shock proteins. The MHC is remarkable for its extensive polymorphism (de Bakker et al. 2006) and ranks first for the number of associations with immune and non-immune diseases (Shiina et al. 2004; Rioux et al. 2009). This has raised considerable interest across disciplines, from immunology and genetics, to medicine and evolutionary biology. Remarkable recent advances in our understanding of the genetic basis of common diseases have been achieved by genome-wide association studies (GWAS) (Wellcome Trust Case Control Consortium 2007; Manolio 2010), which have confirmed the preeminence of the MHC in terms of the magnitude of effect, statistical confidence, and the number of associations with autoimmune, infectious, and inflammatory diseases, together with cancer and adverse drug effects (Conde et al. 2010; Hamza et al. 2010; Hor et al. 2010; Singer et al. 2010).
The fine mapping of causal variants has proved challenging for the majority of complex traits, and we rarely understand the mechanisms through which DNA sequence polymorphisms operate (Knight et al. 2004). Their identification has been confounded by their multiplicity, their frequency in the general population, their modest effects, and linkage disequilibrium (LD). The latter is most remarkable in the MHC, where it may extend over several megabases (Ahmad et al. 2003; Yunis et al. 2003; Vandiedonck and Knight 2009). As a result, diseases are often found to be associated with common extended ancestral MHC haplotypes encompassing hundreds of genes, many of which are candidates.
Gene expression levels are considered as relevant intermediate phenotypes in complex diseases (Vafiadis et al. 1997; Giraud et al. 2007; Cookson et al. 2009; Nica et al. 2010; Teslovich et al. 2010). These expression phenotypes are heritable (Yan et al. 2002) and can be mapped as quantitative traits (Emilsson et al. 2008; Cheung and Spielman 2009). Such studies have already highlighted cis- and trans-acting SNPs within the MHC (Dixon et al. 2007; Vandiedonck and Knight 2009). However, these studies and more recent RNA-seq-based expression quantitative trait analyses (Montgomery et al. 2010; Pickrell et al. 2010) were focused primarily on single-point mapping of gene expression and did not account for the extended haplotypes relating the associated polymorphisms. A particular allele can indeed be found on more than one haplotype. Thus, the reciprocal question of which genes are differentially expressed between MHC extended haplotypes remains essential to resolving functionally important genetic variants as one might expect to find the disease-related genes among the genes whose expression is specifically modified on the risk haplotype.
Here we sought to draw for the first time a map of transcription for the human MHC at a haplotypic resolution in which the consequences of genetic variation in phase for a large contiguous chromosomal region can be established. We investigated three important haplotypes that are common in northern European populations, are highly conserved, and show evidence of selection and important associations with diseases: HLA-A1-B8-Cw7-DR3 (associated with type 1 diabetes, systemic lupus erythematosus and myasthenia gravis, together with other diseases including common variable immunodeficiency and infectious disease susceptibility) (Price et al. 1999); A3-B7-Cw7-DR15 (associated with protection from type 1 diabetes and susceptibility to multiple sclerosis and systemic lupus erythematosus) (Barcellos et al. 2003; Larsen and Alper 2004); and A26-B18-Cw5-DR3-DQ2 (associated with type 1 diabetes and Graves' disease) (Johansson et al. 2003). These haplotypes were fully resequenced as part of the MHC Project (Stewart et al. 2004; Traherne et al. 2006; Horton et al. 2008), but informative individuals carrying the specific haplotypes were not included in previous expression quantitative trait studies (Montgomery et al. 2010; Pickrell et al. 2010). In this study, we show how gene expression profiling of individuals homozygous for the region has allowed us to identify extensive haplotype-related transcriptional differences and highlight the importance of alternative splicing in this transcriptional diversity.
Results
The MHC array: design and validation
To understand more clearly the relationship between MHC sequence variation and gene expression, we aimed to investigate how transcription varies between commonly occurring haplotypes spanning the classical MHC, including resolution of strand-specific transcripts and alternative splicing. Conventional microarrays based on the human reference sequence are often confounded by sequence variation not accounted for in probe design (Walter et al. 2007), and, to date, the difficulties of mapping reads from high-throughput sequencing technologies to the highly polymorphic MHC have limited the application of RNA sequencing to this genomic region. Thus, we developed a hybrid microarray for the MHC (denoted “MHC array”) that included alternate allele probes to account for known sequence diversity (Supplemental Methods). Our array design also aimed to resolve genic and intergenic transcription in a strand-specific manner at high resolution by including a strand-specific tiling path probe set together with probes specific to known and predicted splice junctions. We sought to use the MHC array to analyze transcription at haplotypic resolution using lymphoblastoid cell lines (LCLs) established from individuals MHC-homozygous for the three autoimmune disease-associated haplotypes of interest—COX (HLA-A1-B8-Cw7-DR3), PGF (A3-B7-Cw7-DR15), and QBL (A26-B18-Cw5-DR3-DQ2) (Horton et al. 2008).
The MHC array includes 505,686 probes of 25-mers interrogating 3.5 Mb of the classical MHC between coordinates chr6:29,748,239–33,231,091 (hg18), including 230 genes with a total of 2755 exons (Supplemental Fig. 1). One set of 398,626 overlapping probes (denoted the tiling path probe set) tiles both strands with a final resolution of 18 bases, allowing identification of any new transcript and its transcriptional orientation. A second set of 15,348 junction probes in four replicates aimed to monitor all known or predicted splice events, corresponding to 1043 junctions in the MHC class III region (12 overlapping probes on average per junction). For any junction or tiling probe, its reverse complement was also incorporated into the design. Importantly, alternate probes were specifically designed for all known SNPs or segmental duplications.
We first carried out experiments to assess the performance of the MHC array. Our design incorporated 10,572 shared probes with the Affymetrix Exon 1.0 ST array allowing comparison across platforms for these probes. We analyzed three biological replicate samples for each of three cell lines using the custom MHC array and the Affymetrix Exon 1.0 ST array. Intensity data from the shared probes were highly correlated for the nine samples hybridized to both platforms (Pearson correlation coefficients ranged from 0.83 to 0.91) (Supplemental Table 1), proving that our sample preparation and hybridization conditions were satisfactory. Interestingly, differences between cell lines were also correlated between platforms, suggestive of haplotypic differences (Supplemental Fig. 2). When all probes of the MHC array were considered, the correlation coefficient between culture replicates ranged between 0.96 and 0.98 (Supplemental Fig. 3). In addition to the usual standard quality controls for hybridization and sensitivity, we estimated the strand specificity as 84.7% (±4.1%) based on the observed ratios of expression between the two strands of known expressed housekeeping genes (Supplemental Methods). We also verified the coverage of full transcripts by comparing the signal intensities from probes tagging both ends of housekeeping genes (coefficient of variation, 13%).
To assess the signal specificity of alternate allele probes, we compared the signal intensity of the PGF samples in the transcribed regions (see below) measured on PGF-specific probes with that measured on COX- and QBL-specific probes. We found a significantly higher signal on PGF-specific probes (ANOVA, P = 2.4 × 10−5). We also compared the signal of the PGF samples on the 123 perfect match probes paired with probes carrying one mismatch corresponding to the COX path. The signal was consistently higher on perfect match probes (ANOVA with repeated measures, P = 2 × 10−4). We evaluated the junction probes' performance by using CD79A and CD79B genes that code for both main chains of the invariant component of the B-cell receptor complex and are expressed in LCLs. The comparison of array data and quantitative PCR data showed similar proportions between isoforms. For CD79A, we measured a ratio of 4.96 ± 0.17 between the long and the short isoforms using the array, and of 5.35 ± 0.46 by RT-PCR. For CD79B, we obtained a ratio of 2.71 ± 0.21 between the long and the short isoforms with the array, compared to 2.84 ± 0.48 by RT-PCR.
A high-resolution strand-specific MHC transcription map
Identification of transcriptionally active regions (TARs)
Using this validated platform, we initiated experiments in which we aimed to generate a high-resolution strand-specific transcriptomic map of the MHC. We first verified chromosome and MHC integrity of the selected homozygous cell lines by DNA-FISH and then analyzed RNA prepared from PGF, COX, and QBL cells grown in triplicate and hybridized to the MHC array. After preprocessing of all probes, we analyzed the tiling probes on each strand, in terms of the “shared paths” corresponding to probes shared and identical between the three haplotypic sequences, and the “alternate paths,” which also include haplotype-specific probes for each haplotype. Hence, a total of eight sequence paths were considered (one shared and three haplotypic sequence paths for each strand). After signal smoothing, we determined transcriptionally active regions (TARs) (Bertone et al. 2004) as any 51-base windows with median signal intensity exceeding a threshold determined by permutation (Supplemental Methods). An overview of the signal across the entire region with the “shared paths” relative to the PGF reference assembly sequence is provided in Supplemental Figure 4. Overlapping TARs at a false discovery rate (FDR) of 1% were merged to define transcribed blocks, whose size range was similar between strands and haplotypes (from 51 to 1380 bases, mean = 108.2 bases). On average, there was one transcribed block per 1.4 kb. These are listed in Supplemental Table 2 including location relative to path, strand, and each transcript as annotated in Vega, currently the most comprehensive annotation of the MHC locus. Overall, we found that 6% of the MHC sequence is transcribed, with an equal distribution of 2% for transcribed blocks on the forward, reverse, or both strands.
Genic and intergenic transcription
We then sought to determine the extent of genic and intergenic transcription based on Vega gene annotations. We defined Vega genes as being transcribed based on the inclusion of at least one TAR using a 5% FDR on each “alternate path.” Their proportion was similar between haplotypes and remarkably high, >92% for the genes and >70% for the pseudogenes, underscoring the accuracy of Vega annotations for the MHC (Supplemental Table 3). An overview of strand-specific gene transcription occurring across the MHC is provided in the associated Figure 1 (see foldout).
Figure 1.
The first transcriptional map of the human MHC: Haplotype-specific pattern of expression. Each cell line transcription level against the minimal level of the three cell lines is displayed; (pink) PGF, (orange) COX, and (purple) QBL. Bars corresponding to genes on the minus strand are displayed leftward and those for the plus strand rightward. Boxes in the class III and class II regions indicate haplotype-specific segmental duplications. Overall, the MHC class II region presents the highest minimal expression (dark blue). The most differentially expressed gene is ZFP57, located at the beginning of the class I region.
In terms of intergenic transcription, a remarkably high proportion (31%) of the transcribed blocks did not map to known genes. These intergenic blocks had an average size of 69 bases (range 51–367) in total, reaching 1.7% of the combined length of both strands, thus corresponding to 28.3% of overall transcribed genomic sequence length. When looking at the distribution of the distances of these TARs to known neighboring genes, the median was found to be 10 kb (Supplemental Fig. 5). One-half of the intergenic transcribed blocks thus mapping within 10 kb of annotated genes on either the 5′ or the 3′ side, could be new exons or regulatory elements as suggested by previous studies using either tiling arrays or RNA-seq (Bertone et al. 2004; Gaulton et al. 2010; van Bakel et al. 2010). The remaining 50% of intergenic transcribed blocks were more distant (>10 kb) and tended to cluster (>50% are <0.9 kb apart) in regions of lower gene density (65.1% in class I, 25.5% in class II, and 9.4% in class III). Most notably, 95% of them colocalized with repeat elements, 78% of which mapped to an Alu sequence. This is not simply a consequence of cross-hybridization with Alu sequences transcribed from elsewhere in the genome as only 37% of Alu repeats covered by the array design (necessitating probes to be of genome-wide unique sequence) overlapped a TAR. The same proportion was found when considering recent Alu subfamilies, AluY and AluSg. Similarly, this signal could not be attributed to edited RNAs from genome-wide Alu sequences that are widespread in human, as we found that only 0.1% of probes present in these distant intergenic TARs matched the A-to-I or C-to-U edited RNA sequences cataloged in the comprehensive DAtabase of RNa EDiting (Kiran and Baranov 2010). Altogether, our data support an abundant transcriptional activity from Alu sequences in the intergenic regions.
Haplotype-specific transcription
Numerous genes are differentially expressed between haplotypes
Using this high-resolution, strand-specific transcriptional map of the MHC, we addressed the issue of haplotypic-specific gene expression. First, we considered the highest resolution using TARs generated at a conservative FDR of 1% on the “shared paths.” We found that 9%, 4.6%, and 11.1% of the TARs on PGF, COX, and QBL sequences, respectively, were identified in only one cell line, suggesting haplotype-specific expression.
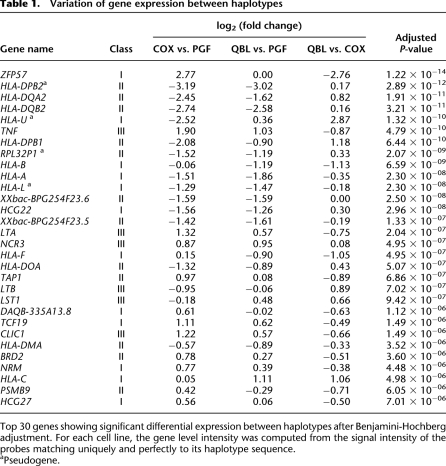
We next tested quantitative differences in expression levels. To this end, we used the probes from the “alternate paths” matching exactly the haplotypic sequence of the corresponding cell line, grouped into metaprobesets based on Vega annotations. Moreover, these metaprobesets contain 4.13 times more probes per gene than in the Affymetrix Exon 1.0 ST array (Supplemental Fig. 6). The MHC array thus provides “individualized” gene levels with a high level of accuracy. As shown in Supplemental Table 4, this resulted in a somewhat different list of differentially expressed genes. Overall, using the MHC array, we identified 96 differentially expressed genes between the three cell lines (Fig. 1; Table 1). These included a number of classical HLA class I (HLA-A, -B, -C, and -F) and class II genes (HLA-DQA2, -DQB2, -DPB1) as well as class III genes including TNF, LTA, NCR3, and LTB. We selected 12 genes showing haplotypic differences in expression for study by quantitative RT-PCR and found expression level differences between the cell lines reaching statistical significance in nine of them (Supplemental Fig. 7) (see below for ZFP57, HLA-DQB2, and HLA-C).
Table 1.
Variation of gene expression between haplotypes
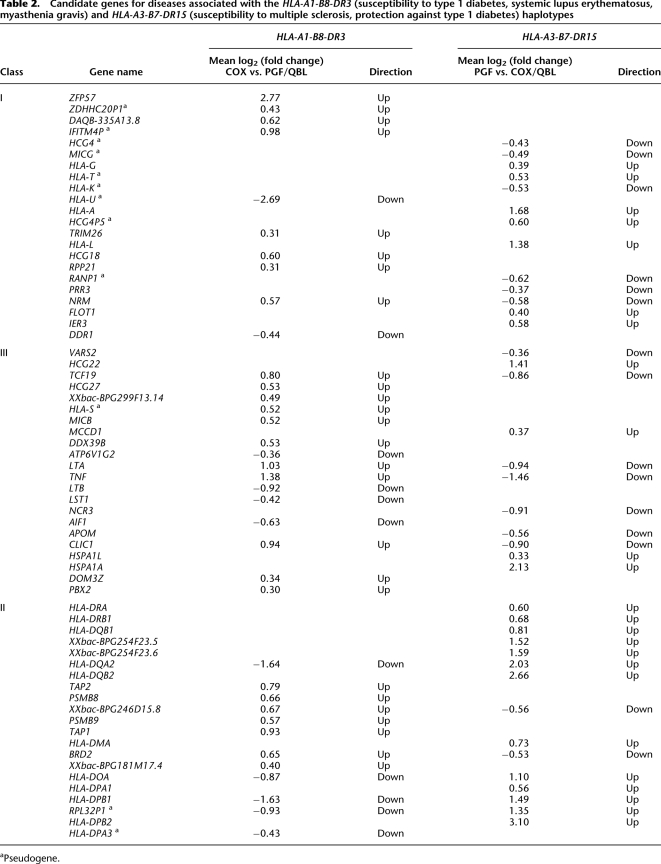
This analysis allows candidate genes to be defined for specific haplotypes. For example, we determined genes, ordered on the chromosome from telomere to centromere, that were significantly differentially expressed (adjusted P-value < 0.05) between either COX and PGF/QBL for the HLA-A1-B8-DR3 haplotype or between PGF and COX/QBL for the HLA-A3-B7-DR15 haplotype. Only genes up- or down-regulated in the same direction were selected (Table 2). This highlights, for example, ZFP57, LTA, TNF, HLA-DQA2, and HLA-DPB1 as showing greater than twofold differential expression with the HLA-A1-B8-DR3 haplotype and as being important candidate genes to investigate further for this important disease-associated haplotype.
Table 2.
Candidate genes for diseases associated with the HLA-A1-B8-DR3 (susceptibility to type 1 diabetes, systemic lupus erythematosus, myasthenia gravis) and HLA-A3-B7-DR15 (susceptibility to multiple sclerosis, protection against type 1 diabetes) haplotypes
Colocalization of differentially expressed probes and polymorphic SNPs
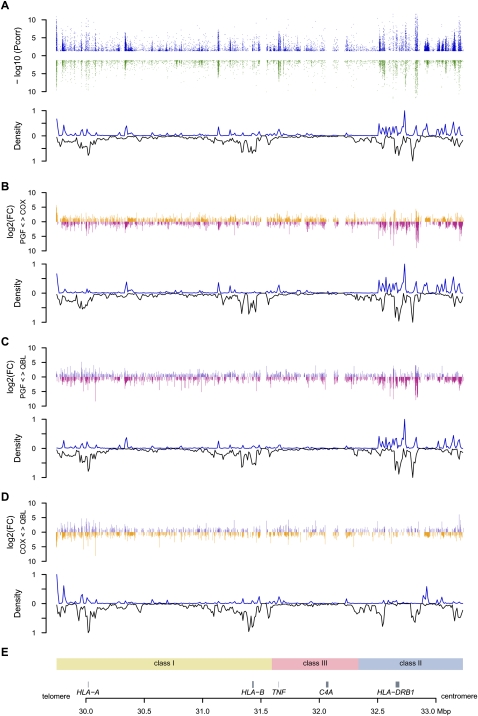
That these differences could result from haplotype-specific sequence variation was supported by the correlation we found between the location of differentially expressed probes and polymorphisms between haplotypes along the chromosome for two sets of interval series of 10-kb windows shifted by 5 kb across the MHC (Fig. 2). This was particularly significant (as low as P = 1.8 × 10−6 between PGF and QBL, Spearman test) when the analysis was restricted to windows including at least one gene (Supplemental Table 5). Conversely, no correlation was found in windows lacking genes or when testing genic windows against nonpolymorphic markers between the pairs of haplotypes. Altogether, these results are consistent with a role for cis-acting regulatory variants influencing levels of gene expression.
Figure 2.
Distribution of differentially expressed (DE) probes versus polymorphic SNPs. Only probes shared by the three haplotypes were included. (A) Three-haplotype comparison. (Upper panel) Significance level of DE probes for either unstimulated (blue) or stimulated (green) cells. The −log10 of significant adjusted P-values are plotted against the genomic coordinates. (Lower panel) Density curve of DE probes normalized using the number of probes designed (upward) mirroring the density curve of polymorphic SNPs between the three cell lines (downward) for 350 10-kb windows spanning the MHC. Densities have been normalized. (B–D) Pairwise comparisons of COX versus PGF, QBL versus PGF, and QBL versus COX. For each pair, the log2 of the intensity fold change (FC) is represented in the upper panel. For example, when expression is higher in COX than in PGF, the FC is set positive and an orange bar is represented above the x-axis. Conversely, when expression is higher in PGF, the FC is negative and represented by a pink bar below the x-axis. The density curves of DE probes and of SNPs polymorphic between both cells are plotted in the lower panel. (E) Genomic context.
Cis control of MHC gene expression in LCLs and primary cells
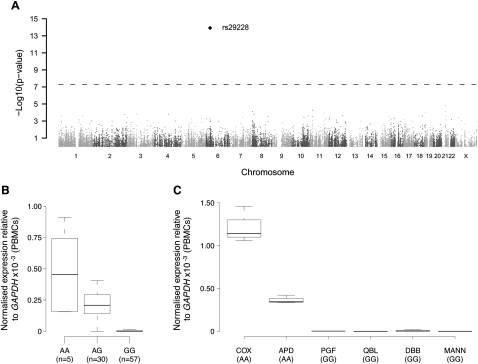
To further test whether variation in expression could be attributed to haplotypic effects, we investigated the three most significant differentially expressed genes—ZFP57, HLA-DQA2, and HLA-DQB2 (Table 1). ZFP57 encodes a zinc finger protein involved in transcriptional regulation and DNA methylation (Li et al. 2008) and is located at the telomeric end of the MHC class I region. We mapped its quantitative expression in peripheral blood mononuclear cells (PBMCs) of 93 healthy volunteers using 45,237 SNPs genotyped on the Illumina HumanCVDv1 BeadChip (Keating et al. 2008). Strikingly, this showed a highly significant association between expression of ZFP57 and the rs29228 SNP located 16.8 kb downstream from ZFP57 (P = 1.2 × 10−14) (Fig. 3A,B). The COX cell line is homozygous for the minor allele of the SNP associated with expression and when we tested three additional LCLs, only those homozygous for the rare allele showed evidence of ZFP57 expression (Fig. 3C). In addition, rs29228 is in complete linkage disequilibrium with rs3129073, which is also significantly associated with ZFP57 expression (effect = −1.088; P = 5.4 × 10−30; rank = fourth) in LCLs from an independent familial asthma cohort (Dixon et al. 2007). There is evidence of association of the COX haplotype with type 1 diabetes, while mutations of ZFP57 itself have been associated with transient neonatal diabetes (Mackay et al. 2008).
Figure 3.
Expression quantitative trait mapping for ZFP57. Expression of ZFP57 was determined by quantitative real-time RT-PCR in peripheral blood mononuclear cells of 93 healthy volunteers and analyzed for association using 45,237 SNPs enriched for immune and inflammatory genes. (A) Manhattan plot showing a highly significant association for an SNP, rs29228, 16.8 kb centromeric to ZFP57. The horizontal dashed line indicates the genome-wide threshold significance. The absence of other association with neighbor SNPs on chromosome 6 is not unexpected due to moderate SNP coverage in the region and low level of linkage disequilibrium. (B,C) Boxplots of ZFP57 gene expression relative to GAPDH depending on rs29228 genotype in 92 successfully genotyped individuals (Kruskal-Wallis test on genotypes, P = 6.7 × 10−11) (B) or for MHC-homozygous lymphoblastoid cell lines (C).
We also performed genome-wide eQTL mapping for HLA-DQA2 and HLA-DQB2 using the same cohort of healthy volunteers. For both genes, we found significantly associated SNP markers in the MHC. For HLA-DQA2, rs2269423 was the most significantly associated SNP in the MHC, located 653 kb away from the gene, and the sixth genome-wide (P = 2.13 × 10−4). Individuals possessing a copy of the A allele showed higher levels of expression with consistent results seen in the panel of six MHC-homozygous LCLs for this SNP (Supplemental Fig. 8A). Similarly, for HLA-DQB2, rs9469220 located 65 kb downstream from the gene is the best associated SNP in the MHC and the seventh genome-wide (P = 1.01 × 10−4) (Supplemental Fig. 8B).
We specifically investigated the SNP rs9264942 located 35 kb upstream of HLA-C, which was previously reported to be associated with expression of HLA-C in PBMCs (Thomas et al. 2009). Using the MHC array, we find that higher expression of HLA-C is seen in QBL, which is homozygous CC for this SNP compared to COX and PGF, which are homozygous TT, consistent with the previous report of higher expression associated with possession of the C allele. Moreover, when we genotyped our 96 healthy volunteers by Sanger sequencing and looked at expression of HLA-C at the transcript level in PBMCs, we found that possession of a copy of the C allele is associated with 22.6% higher expression of HLA-C (Mann Whitney, P = 0.023, two-tailed) (Supplemental Fig. 8C).
These results validate the use of homozygous LCLs to identify haplotype-specific expression patterns. Although our study does not rule out the involvement of trans-acting variants, the correlation of differential expression with adjacent polymorphisms and our findings from expression quantitative trait mapping are consistent with several studies reporting a majority of cis eQTLs (Cheung and Spielman 2009).
The extent of alternative splicing in the MHC
Alternative splicing is increased in the MHC compared to non-MHC genes
Alternative splicing (AS) is critical to the generation of transcriptomic diversity and is known to be modulated by sequence variation with important implications for disease (Wang and Cooper 2007; Keren et al. 2010). Here we sought to investigate the extent of haplotype-specific alternative splicing within the MHC. First, we used the Affymetrix Exon 1.0 ST array hybridized with the PGF samples (whose MHC sequence is the human reference) to establish the extent of AS in this region in comparison with the rest of the genome. Absolute exon normalized intensities (NI) were determined by subtracting the log2 exon intensity from the log2 gene intensity, positive and negative values indicating exon inclusion and exclusion, respectively, with an NI value >1 indicating that the exon is expressed at least twice more or less than the overall gene level. The proportion of exons with NI values different from zero was determined for MHC and other gene sets. This analysis revealed that AS events were strikingly enriched in MHC genes compared to non-MHC genes generally, or specifically to non-MHC genes with an immune function (Fig. 4). This was true whether we considered the number of spliced exons or the number of genes with at least one splice event. Overall, 72.5% of the MHC genes underwent AS of at least a twofold magnitude compared to 62.1% of the non-MHC genes (Fig. 4A). To avoid potential bias due to the number of annotated exons per gene, we determined the significance of these observations by permutations on genes with at least four annotated exons (median number in the genome) in either Vega (Fig. 4B) or Ensembl databases (P ≤ 1 × 10−4 for MHC vs. non-MHC and MHC vs. non-MHC immune; not significant for non-MHC immune vs. non-MHC non-immune) (Supplemental Table 6). The extent of AS in the MHC could therefore be considered as a further means to increase diversity of gene expression in this genomic region already characterized by its extreme polymorphism.
Figure 4.
Extent of alternative splicing in the MHC. Absolute values of exon level intensities normalized against gene intensities [NI = log2(exon/gene)] were computed from the median signal of the three PGF sample replicates hybridized to the Affymetrix Exon 1.0 ST array. Thus, absolute NI > 1 indicates that the exon is expressed at least twice more or less than the overall gene level. Mean percentage of exons (A) and of genes with at least one exon (B) with NI value(s) exceeding the indicated thresholds for MHC (gray bars) and non-MHC genes (white bars). Error bars depict standard errors of the means of the three replicates (C–E) Comparisons of the median NIs (dashed vertical line) in the 131 MHC genes (C,E) or in 733 non-MHC immune genes (D) having at least four annotated exons in Vega with the density distribution of median NIs obtained in 10,000 random sets of similar numbers of non-MHC (C), non-MHC non-immune (D), and non-MHC immune (E) genes.
The MHC haplo-spliceo-transcriptome
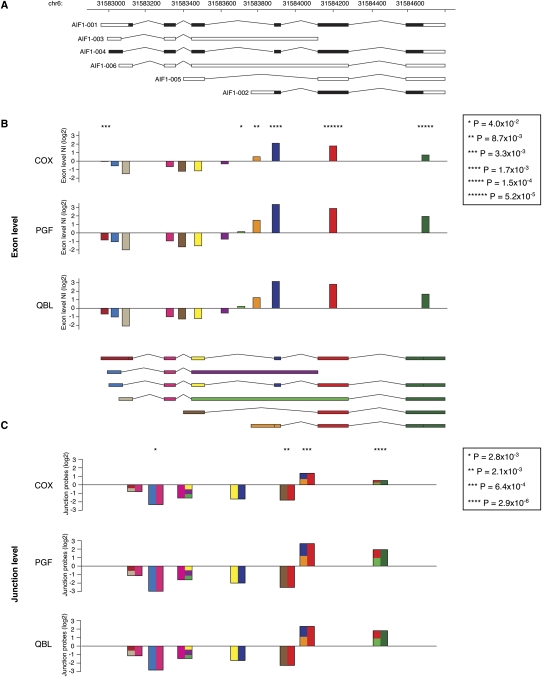
We next considered the extent to which alternative splicing varies between haplotypes. To do this, gene and exon level expression of genes were determined from the probes matching uniquely and perfectly to each haplotype. This analysis showed that these AS events also demonstrate haplotype-specific differences (Supplemental Table 7). In total, we found that 526 (23.9%) exons in the MHC showed haplotypic differences, notably affecting classical HLA genes such as HLA-DPB2, HLA-DQB2, HLA-C, or HLA-G. In the latter, AS has been described as playing a critical role in immunomodulation, susceptibility to preeclampsia, and sensitivity to tumor lysis by natural killer cells (Yao et al. 2005; Carosella et al. 2008). We complemented our analysis at the exon level with splice junction resolution in the class III region (where we designed junction probes). We computed junction level intensity values, which were then normalized against the gene intensity. We identified 27 out of 58 genes (46.6%) as showing haplotypic differences in AS (Supplemental Table 8). A number of genes in this region are known to undergo AS such as AIF1 (Hara et al. 1999). We validated the array results for AIF1 by RT-PCR, both in terms of exon normalized intensities and junction normalized intensities (P < 0.02 for all junctions, ANOVA) with evidence of haplotypic differences (Fig. 5).
Figure 5.
Variation of splicing events in AIF1 between haplotypes. (A) Gene transcripts as they are annotated in Vega. (B) Barplots of all exon normalized intensities (NI) for each cell line. The color code for each exon is indicated in the transcript scheme underneath. (C) Barplots for the junction normalized intensities (JNI). Donor and acceptor exons are represented on each half of the junction with the same color code as in B. If the junction is shared between different transcripts, the corresponding site is depicted as a composite of all possible exons. (B,C) Asterisks above barplots indicate the level of significance, as listed in the caption, for differential expression between the three cell lines. For example, the isoform AIF1-002 tagged by the exon in orange is proportionally more represented in QBL and PGF than in COX. Conversely, the isoform AIF1-005 characterized by the junction between the brown and the red exons is better represented in COX than in PGF and QBL.
Discussion
Our results provide the first high-resolution, strand-specific transcriptional map of the MHC. We find that both intergenic transcription and alternative splicing are abundant in the MHC and that the transcript diversity mirrors the unusually high level of polymorphism found in this region. Specifically, for common disease-associated haplotypes, we have been able to define transcription at haplotype-specific resolution using MHC-homozygous LCLs. This has highlighted the extent of variation in gene expression that exists between haplotypes when a large contiguous homozygous sequence is analyzed, in this case, a 3.5-Mb region spanning the classical MHC. Haplotype-specific analysis is critical to advance our understanding of the nature and consequences of genetic variation within the diploid human genome. Given its biological significance and wealth of disease associations, the full haplotype-specific sequence for eight common haplotypes spanning the MHC were defined by the MHC Project (Stewart et al. 2004; Traherne et al. 2006; Horton et al. 2008). Here we complement this data with a haplotype-specific map of transcription in which differentially expressed genes were quantified in the context of phased sequence variants, allowing any regulatory variants to exert allele-specific effects in the naturally occurring genomic context. The use of MHC-homozygous LCLs avoids the confounding effects normally encountered in analysis of the diploid genome and provides a route map for further fine mapping and functional analysis of observed MHC disease associations.
The transcriptional landscape we have described is likely to vary in a context-specific manner, and it will be important to extend our approach to relevant cell/tissue types and conditions for specific MHC-associated diseases. Nevertheless, the three LCLs we used were our material of choice for these first studies of the MHC transcription at a haplotype-specific resolution. Firstly, these cell lines are MHC-homozygous for the disease-associated haplotypes of interest, and we benefited from having available the full MHC sequence for each, facilitating our approach as if we had been studying mouse strains, thus avoiding the issue of recombination. Secondly, linkage of expression phenotypes was first demonstrated in LCLs, proving these phenotypes are not artifactual. Since then, most eQTL mapping studies have investigated that material with reproducible findings (Stranger et al. 2007; Cheung and Spielman 2009; Montgomery et al. 2010; Pickrell et al. 2010). Thirdly, in the MHC we found that 87% of the genes in the region are expressed in that cell type, making it a relevant choice for investigating differential expression between individuals. Finally, we proved with ZFP57, HLA-DQA2, HLA-DQB2, and HLA-C that our findings could be replicated in primary peripheral blood cells.
We have shown how a high-density tiling path array design incorporating sequence diversity and splice junctions is a powerful tool to help dissect the haplo-spliceo-transcriptome (Graveley 2008) of a large genomic region of interest. Like RNA-seq, our array overcomes some major issues associated with commercial expression arrays; notably, it accounts for underlying sequence polymorphism, allows for identification of new transcribed regions, and monitors splice events. Moreover, these microarrays currently provide a much less costly tool than RNA-seq to assess the transcription status of a chromosomal region the size of the MHC, although we recognize the greater dynamic range and allele-specific resolution of this technology (Wang et al. 2009). We have applied the MHC array to MHC-homozygous individuals, but the custom array should also be informative when applied to heterozygous samples. Here it will be necessary to know the DNA sequence or relevant genotypic information to select the correct probes for analysis and interpret the data correctly. The tiling path probe set can then be defined across the MHC by individual. At positions where the individual is heterozygous, the average of the two informative allele-specific probes can be taken.
Our findings that only 6% of the region is transcribed might appear as a low figure. This, however, is the same order of magnitude as, for example, 4.6% of the total length of ENCODE regions screened on tiling arrays (ENCODE Project Consortium 2007). That 31% of the TARs, representing 28.3% of total transcribed MHC sequence, were found in intergenic regions is also in line with the 25% recent estimate of “dark matter transcripts” obtained by RNA-seq (Ponting and Belgard 2010) and is, however, of considerable interest, given that the MHC region is the most gene-dense region of the human genome. Their expression was overall low, which might explain why we failed to detect haplotype-specific differences and correlation with SNPs localization, unlike for TARs in genic areas. The biological significance of these TARs is unclear. We have not investigated their processing, but the fact that of the 50% localizing distantly (>10 kb) from annotated genes, the majority colocalize with Alu sequences is particularly intriguing. It is known that Alu sequences can be actively transcribed and may contribute to the emergence of alternative splicing or even new genes and pseudogenes (Deininger et al. 2003). Some classical HLA genes have been postulated to derive from such repeat elements. The intergenic TARs we detect might therefore reflect an ongoing process of exonization of transposed elements leading to the emergence of new MHC genes, also important for the regulation of existing genes and therefore eventually for MHC-associated pathology.
It has been suggested that alternative splicing is a key modulator of immune gene expression (Lynch 2004), possibly leading to antagonist effects as seen for MYD88 or CD44. A previous study has revealed that up to 94% of human genes are alternatively spliced across 15 tissues tested from different individuals (Wang et al. 2008). We found that in a single cell type from a single individual, 72.5% of the MHC genes are alternatively spliced. Moreover, alternative splicing is enriched among MHC genes compared to non-MHC immune genes. We also demonstrate that alternative splicing is related to the haplotypic structure. In the context of common ancestral MHC haplotypes, one can thus imagine that alternative splicing is used by evolution to generate more transcript diversity in the MHC while preserving some of the haplotypic structure. Consequences of such splicing patterns can lead to dramatic consequences as already highlighted by mutations in the BTLN2 gene associated with sarcoidosis (Valentonyte et al. 2005).
Our study presents a proof of principle that, beyond standard SNP-based eQTL mapping studies, it is possible to directly study haplotype-specific gene expression at a high resolution for a 3.5-Mb region and find striking differences. Our approach will be of value in a generic sense for characterizing other genomic intervals identified by GWAS or other approaches. Risk haplotypes are ultimately associated to the phenotypes, and identifying genes differentially expressed can reduce the number of genes to study at the disease locus region.
For the MHC, this is of particular interest given the remarkable number of associations with common diseases reported, while the fine mapping of functionally important regulatory variants remains a challenge. That such important differences in gene expression could be detected by investigating only three haplotypes supports the hypothesis that they play a role in the autoimmune diseases associated with these haplotypes. Our data suggest a number of candidate variants and gene transcripts for further characterization. For example, Figure 1 presents a graphical overview of the locus showing differentially expressed genes by haplotype, while Table 2 lists 37 candidate genes potentially accounting for the association of the HLA-A1-B8-DR3 haplotype with numerous diseases.
For the MHC, both structural and regulatory genetic variants are important in determining disease susceptibility, and our approach to this region needs to consider such variants if causal relationships are to be established. Our analysis has provided new insights into how transcription differs between individuals across the classical MHC, and our custom array can be used to quantify haplotype-specific differences in related contexts such as DNA methylation or chromatin accessibility based on DNase hypersensitivity (Sabo et al. 2006; Weber et al. 2007). As our knowledge of the complexities of gene regulation continues to grow, it is important to acknowledge how much remains to be understood and the need for a more complete picture of gene expression beyond transcript level analysis. At a mechanistic level, much attention has focused on modulation of transcriptional initiation, but sequence diversity will impact in multifaceted ways on the whole process of transcription and translation, as well as how chromatin is packaged and gene expression coordinated at a local and global level within the nucleus. It will be critical to establish the nature and basis of individual epigenetic variation, defining how this may be modulated by underlying DNA sequence variation as well as environmental factors relevant to disease. We believe our analysis opens the door to such studies and provides an important further step in our quest to define the functional basis of the remarkable disease associations found for this region of the genome.
Methods
Full methods and any associated references are available in the Supplemental Material.
Samples
Lymphoblastoid cell lines (LCL)
COX was obtained both from The International Histocompatibility Working Group (IHW, ref 0922) and by the generosity of S. Marsh and N. Mayor (Anthony Nolan Research Institute, UK). PGF and QBL were purchased from the European Cell Culture Collection (Salisbury, UK; ref 94050342 and 94070713). Chromosome integrity was checked by FISH. In addition to chromosome 6 painting, the AF129756 BAC (The Sanger Institute) encompassing most of the class III region was used as a second probe to verify MHC integrity. Genotypes of HLA classical molecules (HLA-A, B, C, DR, and DQ) were verified by the Tissue Typing Laboratory in Oxford (Dr. Barnardo Martin), while the homozygosity and genotypes of microsatellites along the class III region (D6S272, D6S2800, MICA, TNFb, and D6S2789) were checked as described before (Vandiedonck et al. 2004). Apart from D6S272, which showed heterozygosity for PGF, all other markers showed the expected genotypes. Genotypes for 410 SNPs in the MHC region were also verified for COX, PGF, and QBL using a cardiovascular gene-centric 50 K SNP array (humanCVD bead array; Illumina) (Keating et al. 2008). With one exception (rs562047 found G/C in QBL), all genotypes were those expected. To follow up results on ZFP57, HLA-DQA2, HLA-DQB2, and HLA-C expression, three additional MHC-homozygous cell lines—MANN/MOU, DBB, and APD (IHW9050, 9052, 9291)—were studied, and their MHC genotypes were also checked with the cardiovascular array (Keating et al. 2008).
PBMCs from healthy volunteers
PBMCs from healthy volunteers were recruited with cDNA prepared as described in Fairfax et al. (2010). Their genomic DNA was extracted using Puregene kits (Gentra Systems, Inc.). Genotyping on the humanCVD bead array was performed using genomic DNA from the volunteers and homozygous LCLs DNA, as previously described (Fairfax et al. 2010). For two specific SNPs not included on the array—rs2269423 and rs9264942—genotyping was performed by direct Sanger sequencing (primer sequences available on request). For some genes subsequently interrogated by expression quantitative trait mapping, genotyping and/or gene expression data were not available for all volunteers. The total numbers of volunteers included for each gene analyzed are shown in the associated figure legends.
Design of the MHC array
The MHC array was designed for the Affymetrix platform (Affymetrix) using ad hoc algorithms as described in detail in the Supplemental Methods. Criteria of uniqueness against the genome and transcriptome and of structural conformation were considered. Known polymorphisms and segmental duplications have been incorporated into the design.
Experimental procedures
Cell culture
Lymphoblastoid cells were grown in triplicate at a minimum density of 6 × 105 cells/mL in RPMI 1640 (Sigma, lot 16K2379) supplemented with 10% Fetal Calf Serum Gold (PAA, lot A64095-0537) and 2 mM L-glutamine (PAA, lot M00406-0102) at 37°C in a 5% CO2 wet environment. Cultures were stimulated or not for 6 h with 200 nM phorbol 12-myristate 13 acetate (PMA; Sigma) and 125 nM ionomycin (Sigma) and harvested at 8 × 105 to 1 × 106 cells/mL in log growth phase. Volunteers' peripheral blood mononuclear cells (PBMC) were prepared as previously described (Fairfax et al. 2010).
RNA extraction
Total RNA was isolated using RNeasy midiprep kits (QIAGEN) including on column DNase I digestion. Quantifications were done by Nanodrop (ThermoScientific), and integrity was verified using a 2100 Bioanalyzer (Agilent). All samples had a RNA integrity number >9. Genomic DNA contamination was checked by real-time PCR and was <0.1%.
Array experimental design
We hybridized samples from the unstimulated and stimulated triplicate cultures of COX, PGF, and QBL LCLs to custom MHC arrays, while only unstimulated samples were hybridized to commercial Affymetrix Exon 1.0 ST arrays.
Sample labeling and array hybridization
We used the GeneChip Whole Transcript (WT) Sense Target Labeling kit (Affymetrix), following the manufacturer's instructions, starting with 1.5 μg of total RNA and including the ribosomal RNA depletion step (Ribominus kit; Invitrogen). Then, cDNA was synthesized using random hexamers tailed with a T7 promoter to avoid 3′ bias, and the complementary strand of RNA was generated by an in vitro transcription reaction. Subsequently, a new first strand of cDNA was synthesized, complementary to the initial cDNA, in the same orientation as the mRNA and denoted “ccDNA.” It was fragmented (range 40–70 bases), end-labeled, and hybridized to the MHC and GeneChip Exon 1.0 ST arrays (Affymetrix) for 16 h at 45°C following the manufacturer's instructions. Reduced RNA, cRNA following IVT, and fragmented ccDNA were verified on a 2100 Bioanalyzer. Hybridized arrays were then washed and stained on a GeneChip Fluidics 450 workstation (Affymetrix) using the FS450_0001 protocol. The arrays were scanned on a GCS3000 Scanner (Affymetrix).
cDNA synthesis and RT-PCR for validation
cDNA was synthesized using random hexamers and Superscript III Reverse Transcriptase (RT) (Invitrogen) as per the manufacturer's instructions including control reactions without reverse transcriptase for each sample. Quantitative PCR was performed on three technical replicates using SYBR Green Supermix (Bio-Rad) on an iQ Cycler (Bio-Rad). PCR efficiency was determined using serial dilutions of pooled cDNAs from COX, PGF, and QBL cells. Melt curve analysis was performed for gene-specific primer sets. Relative gene transcript levels were determined by the ΔΔCt method. Primer sequences are available upon request.
Array signal processing
Custom MHC array signals
Custom MHC array signals were processed using an in-house pipeline under R and Bioconductor environment and using Perl scripts as detailed in the Supplemental Methods. Briefly, after preprocessing all probes, tiling and junction probes were analyzed independently. Tiling path analysis was conducted to determine the extent of transcription on the shared path and on each of the alternate paths. Transcription within a gene was assessed by the inclusion of at least one TAR at a FDR of 5%. Alternative splicing was evaluated on each of the alternate paths both at the exon level and, for the MHC class III region, at the splice junction level. Exon and junction intensities were normalized against the gene level intensities.
Exon array signals
Exon array signals were processed using Affymetrix Power Tools, and R scripts (see Supplemental Methods). Briefly, probe-level analysis was carried out for cross-platform validation, while gene level and alternative splicing analyses were performed using custom CDF files from the Microarray Lab (http://brainarray.mbni.med.umich.edu/).
Quality controls
Quality controls are given in the Supplemental Methods.
Statistical analyses
All statistical analyses, including distribution of the TARs; comparison of expression between haplotypes; correlation of differentially expressed probes with SNP distribution; eQTL mapping for ZFP57, HLA-DQA2, and HLA-DQB2; and analysis of the extent of the MHC splicing were performed using R, Perl, and PLINK as provided in detail in the Supplemental Methods.
Acknowledgments
This study was funded by the Wellcome Trust (grant number 074318/075491/Z/04) and the Multiple Sclerosis Society (grant number 875/07). C.V. has been supported by the Fondation pour la Recherche Médicale since July 2009. We are indebted to T. Watt and C. Blancher for technical assistance, N. Wilson and E. Volpi for performing the FISH analysis, B. Martin and H.J. Garchon labs for genotyping classical MHC genes and microsatellites, and S. Marsh and N. Major for generously providing the COX cell line. We are grateful to the volunteers who agreed to participate. We are grateful for critical discussions to P. Donnelly, G. McVean, D.R. Campbell, R. Mott, C. Julier, and H.J. Garchon. We thank all other members of the Knight laboratory for technical assistance.
Authors' contributions: C.V. and J.K. designed the study; M.S.T. and C.V. designed the array; C.V. prepared samples and performed the array experiments and validations; K.P. performed the volunteers' experiment; B.F. collected the volunteers; C.V., H.L., C.D., J.M.T., J.B., and J.K. analyzed the array data; K.P. and B.F. analyzed the volunteers' data; all authors edited and proofread the manuscript; C.V. and J.K. wrote the manuscript.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE22455.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116681.110.
References
- Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling KL, Barnardo M, Goldthorpe S, et al. 2003. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet 12: 647–656 [PubMed] [Google Scholar]
- Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, et al. 2003. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet 72: 710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Carosella ED, Favier B, Rouas-Freiss N, Moreau P, Lemaoult J 2008. Beyond the increasing complexity of the immunomodulatory HLA-G molecule. Blood 111: 4862–4870 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS 2009. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet 10: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, Nieters A, Slager SL, Brooks-Wilson A, Agana L, et al. 2010. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet 42: 661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M 2009. Mapping complex disease traits with global gene expression. Nat Rev Genet 10: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dausset J 1981. The major histocompatibility complex in man. Science 213: 1469–1474 [DOI] [PubMed] [Google Scholar]
- de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, et al. 2006. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38: 1166–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr 2003. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13: 651–658 [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. 2007. A genome-wide association study of global gene expression. Nat Genet 39: 1202–1207 [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. 2008. Genetics of gene expression and its effect on disease. Nature 452: 423–428 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax BP, Vannberg FO, Radhakrishnan J, Hakonarson H, Keating BJ, Hill AV, Knight JC 2010. An integrated expression phenotype mapping approach defines common variants in LEP, ALOX15 and CAPNS1 associated with induction of IL-6. Hum Mol Genet 19: 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. 2010. A map of open chromatin in human pancreatic islets. Nat Genet 42: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud M, Taubert R, Vandiedonck C, Ke X, Levi-Strauss M, Pagani F, Baralle FE, Eymard B, Tranchant C, Gajdos P, et al. 2007. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature 448: 934–937 [DOI] [PubMed] [Google Scholar]
- Graveley BR 2008. The haplo-spliceo-transcriptome: common variations in alternative splicing in the human population. Trends Genet 24: 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, et al. 2010. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet 42: 781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Ohta M, Ohta K, Nishimura M, Obayashi H, Adachi T 1999. Isolation of two novel alternative splicing variants of allograft inflammatory factor-1. Biol Chem 380: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Hor H, Kutalik Z, Dauvilliers Y, Valsesia A, Lammers GJ, Donjacour CE, Iranzo A, Santamaria J, Peraita Adrados R, Vicario JL, et al. 2010. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat Genet 42: 786–789 [DOI] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, et al. 2004. Gene map of the extended human MHC. Nat Rev Genet 5: 889–899 [DOI] [PubMed] [Google Scholar]
- Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, et al. 2008. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics 60: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson S, Lie BA, Todd JA, Pociot F, Nerup J, Cambon-Thomsen A, Kockum I, Akselsen HE, Thorsby E, Undlien DE 2003. Evidence of at least two type 1 diabetes susceptibility genes in the HLA complex distinct from HLA-DQB1, -DQA1 and -DRB1. Genes Immun 4: 46–53 [DOI] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, et al. 2008. Concept, design and implementation of a cardiovascular gene-centric 50 K SNP array for large-scale genomic association studies. PLoS ONE 3: e3583 doi: 10.1371/journal.pone.0003583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Kiran A, Baranov PV 2010. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics 26: 1772–1776 [DOI] [PubMed] [Google Scholar]
- Knight JC, Keating BJ, Kwiatkowski DP 2004. Allele-specific repression of lymphotoxin-alpha by activated B cell factor-1. Nat Genet 36: 394–399 [DOI] [PubMed] [Google Scholar]
- Larsen CE, Alper CA 2004. The genetics of HLA-associated disease. Curr Opin Immunol 16: 660–667 [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 4: 931–940 [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40: 949–951 [DOI] [PubMed] [Google Scholar]
- Manolio TA 2010. Genomewide association studies and assessment of the risk of disease. N Engl J Med 363: 166–176 [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6: e1000895 doi: 10.1371/journal.pgen.1000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Belgard TG 2010. Transcribed dark matter: meaning or myth? Hum Mol Genet 19: R162–R168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P, Witt C, Allcock R, Sayer D, Garlepp M, Kok CC, French M, Mallal S, Christiansen F 1999. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev 167: 257–274 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, De Jager PL, Foisy S, Wang J, de Bakker PI, et al. 2009. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci 106: 18680–18685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. 2006. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3: 511–518 [DOI] [PubMed] [Google Scholar]
- Shiina T, Inoko H, Kulski JK 2004. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens 64: 631–649 [DOI] [PubMed] [Google Scholar]
- Singer JB, Lewitzky S, Leroy E, Yang F, Zhao X, Klickstein L, Wright TM, Meyer J, Paulding CA 2010. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat Genet 42: 711–714 [DOI] [PubMed] [Google Scholar]
- Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, et al. 2004. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14: 1176–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. 2007. Population genomics of human gene expression. Nat Genet 39: 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Apps R, Qi Y, Gao X, Male V, O'Huigin C, O'Connor G, Ge D, Fellay J, Martin JN, et al. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 41: 1290–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, Stewart CA, Ashurst JL, Atrazhev AM, Coggill P, Palmer S, et al. 2006. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet 2: e9 doi: 10.1371/journal.pgen.0020009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C 1997. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 15: 289–292 [DOI] [PubMed] [Google Scholar]
- Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, et al. 2005. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet 37: 357–364 [DOI] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiedonck C, Knight JC 2009. The human Major Histocompatibility Complex as a paradigm in genomics research. Brief Funct Genomics Proteomics 8: 379–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiedonck C, Beaurain G, Giraud M, Hue-Beauvais C, Eymard B, Tranchant C, Gajdos P, Dausset J, Garchon HJ 2004. Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci 101: 15464–15469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NA, McWeeney SK, Peters ST, Belknap JK, Hitzemann R, Buck KJ 2007. SNPs matter: impact on detection of differential expression. Nat Methods 4: 679–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA 2007. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW 2002. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
- Yao YQ, Barlow DH, Sargent IL 2005. Differential expression of alternatively spliced transcripts of HLA-G in human preimplantation embryos and inner cell masses. J Immunol 175: 8379–8385 [DOI] [PubMed] [Google Scholar]
- Yunis EJ, Larsen CE, Fernandez-Vina M, Awdeh ZL, Romero T, Hansen JA, Alper CA 2003. Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue Antigens 62: 1–20 [DOI] [PubMed] [Google Scholar]