Abstract
Freshwater planaria are a very attractive model system for stem cell biology, tissue homeostasis, and regeneration. The genome of the planarian Schmidtea mediterranea has recently been sequenced and is estimated to contain >20,000 protein-encoding genes. However, the characterization of its transcriptome is far from complete. Furthermore, not a single proteome of the entire phylum has been assayed on a genome-wide level. We devised an efficient sequencing strategy that allowed us to de novo assemble a major fraction of the S. mediterranea transcriptome. We then used independent assays and massive shotgun proteomics to validate the authenticity of transcripts. In total, our de novo assembly yielded 18,619 candidate transcripts with a mean length of 1118 nt after filtering. A total of 17,564 candidate transcripts could be mapped to 15,284 distinct loci on the current genome reference sequence. RACE confirmed complete or almost complete 5′ and 3′ ends for 22/24 transcripts. The frequencies of frame shifts, fusion, and fission events in the assembled transcripts were computationally estimated to be 4.2%–13%, 0%–3.7%, and 2.6%, respectively. Our shotgun proteomics produced 16,135 distinct peptides that validated 4200 transcripts (FDR ≤1%). The catalog of transcripts assembled in this study, together with the identified peptides, dramatically expands and refines planarian gene annotation, demonstrated by validation of several previously unknown transcripts with stem cell-dependent expression patterns. In addition, our robust transcriptome characterization pipeline could be applied to other organisms without genome assembly. All of our data, including homology annotation, are freely available at SmedGD, the S. mediterranea genome database.
Planarian flatworms are free-living members of the phylum Platyhelminthes and belong with annelids and molluscs to the Lophotrochozoa, a major animal phylum comprised of understudied taxa (Paps et al. 2009). The spectacular regenerative capabilities of planarians have been studied for more than 100 yr and are known to be mediated by a large population of pluripotent stem cells, which, by morphological criteria, represent ∼30% of all cells in the animal. With the development of new molecular and genetics approaches, planarians have recently reemerged as a model system for the study of regeneration, tissue homeostasis, and stem cell biology (Newmark and Sánchez Alvarado 2002; Agata 2003; Reddien and Sánchez Alvarado 2004; Sánchez Alvarado 2006; Handberg-Thorsager et al. 2008; Rossi et al. 2008; Friedländer et al. 2009). Moreover, the recent genome sequencing of the sexual strain of the species Schmidtea mediterranea (A Sánchez Alvarado, unpubl.) is opening planarian research to powerful genomics approaches and is expected to further boost the interest in planarian research.
For any kind of genomic approaches, a comprehensive description of the full complement of transcripts is one of the most important resources required. However, the current gene annotation in the S. mediterranea genome, which is largely based on computational predictions complemented with partial supporting evidence from EST libraries (Zayas et al. 2005b; Robb et al. 2008), is not complete. In the current version of SmedGD, the S. mediterranea genome database (http://smedgd.neuro.utah.edu/), 30,930 “MAKER” transcripts from 30,333 genome loci were predicted (Cantarel et al. 2008; Robb et al. 2008) and exonic nucleotides cover, in total, 2.8% of the genome (24.8 Mb). Many of these transcripts and gene models await further validation, and the number of missing transcripts in SmedGD is unknown.
Our motivation for this study was twofold. On the one hand, we wanted to improve the planarian transcriptome annotation in order to provide a much needed resource for the community. On the other hand, we were interested in investigating general strategies for sequencing and assembling a complex transcriptome without using the genome sequence (“de novo”). The latter is of great practical importance since the genomes of many organisms are known to be extremely difficult to assemble, even with the power of current and upcoming sequencing technologies. The major reasons for these difficulties are polyploidy, low complexity, and repetitive DNA. Very recently, first attempts have been made to de novo sequence and assemble the transcriptomes of animals such as the coral (Meyer et al. 2009), the whitefly (Wang et al. 2010), and butterflies (Vera et al. 2008). However, in all cases the mean length of assembled transcripts (197 nt for butterfly, 266 nt for whitefly, and 440 nt for coral) was substantially shorter than the estimated average mRNA length (>1000 nt). We reasoned that assembly performance would be improved with (1) the combination of complementary sequencing technologies that provide either long and relatively few sequencing reads (454 Life Sciences [Roche] technology), or many but relatively short reads (for example, Illumina technology); (2) the efficient normalization of cDNA libraries prior to sequencing, because the high dynamic range of mRNA expression (usually spanning five to seven orders of magnitude) is a problem for comprehensive de novo mRNA sequencing and assembly. Both strategies define the success of the mRNA sequencing and assembly pipeline that we devised and describe in this study.
A crucial and nontrivial task, which is, however, often missing in previous similar studies, is to carefully assay the quality of the assembled transcripts. In this study we used different and complementary strategies to do so. Computer simulations and Illumina sequencing were used to assay the importance of our cDNA normalization. RACE was used to check the length of 24 randomly selected transcripts. Mapping to the genome allowed us to estimate the overall quality of our transcripts and the number of distinct genomic loci covered. Further computational exercises enabled us to flag transcripts with a clear homology match to proteins from other species and to infer the rates of mistakes in our assembly, e.g., frameshifts, fusion, and fission events. However, it is difficult to assess the number of false negatives (missing transcripts) and also, importantly, which transcripts are truly translated. We therefore used massive shotgun sequencing of planarian protein extracts by mass spectrometry and mapped sequenced peptides to our transcripts. We were able to flag thousands of transcripts that show strong evidence of being genuine protein-coding mRNAs. To roughly estimate how many proteins are not represented by our transcriptome assembly, we also tallied peptides that did not match our transcripts, but did match the existing gene annotation. We provide transcripts, mapping to the genome, homology information, and peptide sequences in flat files and in the S. mediterranea genome database SmedGD. Finally, we used whole-mount in situ hybridization to successfully validate the expression of several previously unknown transcripts. Using irradiated animals, we further demonstrate that the expression of these transcripts is stem cell dependent, showing that BIMSB transcripts are an important resource for investigating stem cell biology in planaria.
Results
Sequencing and assembly of planarian transcripts
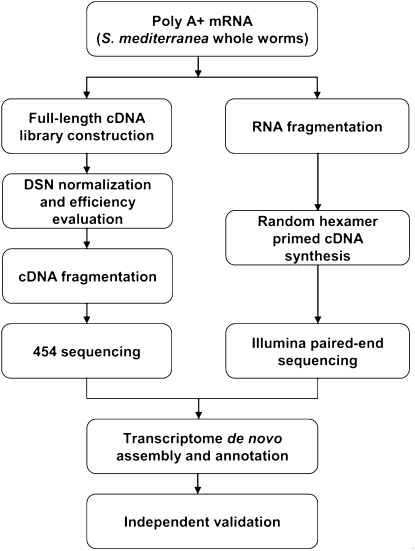
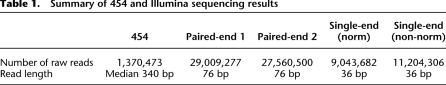
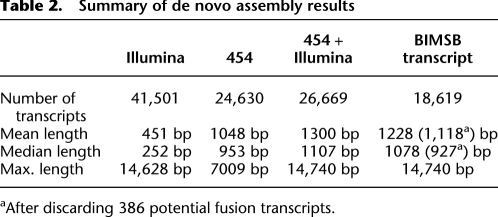
Our experimental scheme for transcriptome cloning, sequencing, and assembly is summarized in Figure 1. First, poly(A) RNA extracted from adult asexual worms was used to construct a full-length cDNA library, which was then normalized using a duplex-specific nuclease (Methods). After evaluating the normalization efficiency (Supplemental material), we proceeded with sequencing the normalized library on the 454 GS FLX platform (Methods) and obtained 1,370,473 reads with a median length of 340 nt that passed the 454 quality filter (Table 1). After trimming off the 454 sequencing adaptors and the adaptors used in the cDNA library construction, all 454 reads were used as input for the Newbler assembler, a 454 proprietary assembly software package distributed together with a 454 sequencing machine. The majority of reads (83.99%) was successfully assembled and formed 24,630 contigs with a median length of 953 nt and a maximum length of 7009 nt (Table 2). Probably due to the lower efficiency of the reverse transcriptase for long transcripts, transcripts longer than 6 kb were under-represented in our full-length cDNA library. To compensate for this, we constructed another cDNA library, in which random priming was used to reverse transcribe the fragmented poly(A) RNA (Fig. 1). Both ends of this library were then sequenced using Illumina paired-end sequencing technology. We obtained ∼28 million pairs, each pair consisting of two 76-nt-long reads (Table 1). Using the SOAPdenovo software (Methods), these reads were then assembled together with the ∼20 million single-end reads. In total, 41,501 contigs, each of length >100 nt, formed (median/maximum length 252/14,628 nt). All 454 sequencing reads and processed contigs assembled from Illumina sequencing runs were then used together for a final “Newbler” assembly (Methods). This assembly produced 26,669 contigs with a dramatically improved median length of 1107 nt. On the other hand, the maximum length at 14,740 nt still indicates that extremely long planaria transcripts are missing from the assembly (i.e., Smed-titin; A Sánchez Alvarado, unpubl.). However, it largely exceeds the expected length of the full-length cDNA library.
Figure 1.
Experimental design.
Table 1.
Summary of 454 and Illumina sequencing results
Table 2.
Summary of de novo assembly results
Transcriptome annotation
Our 26,669 assembled contigs are likely to contain nearly identical ones. After merging these, 18,619 candidate transcripts remained (Table 2; Methods). We further removed 386 transcripts that likely contained erroneous fusions (see below). We will refer to this final set of 18,233 transcripts as BIMSB transcripts (“BIMSB”-Berlin Institute for Medical Systems Biology). The length distribution of the BIMSB transcripts (average/median length 1118/927 nt) and of the MAKER transcripts is shown in Supplemental Figure 5. Compared with our transcripts, MAKER transcripts are typically shorter. To estimate the number of distinct genes represented by BIMSB transcripts, we mapped them to the reference genome of S. mediterranea. All sequences transcribed from the same strand with strongly overlapping genomic alignments were clustered (Supplemental material). Each cluster was then flagged as a putative gene locus. With this procedure we could cluster a total of 17,546 transcripts (comprising 19.8 Mb) into 15,284 separate gene loci. The exons covered 1.9% of the genome reference sequences. We next compared BIMSB transcripts with MAKER transcripts. Using the exact same analysis as for BIMSB transcripts, we found that MAKER transcripts cover 30,333 gene loci and the exonic regions amount to 2.7% of genome sequences. Based on the genome alignments, 8365 gene loci overlapped between BIMSB transcripts and MAKER gene predictions, whereas 20,399 and 6533 loci appear to be specific to MAKER and BIMSB transcripts, respectively.
Based on the genome alignment of BIMSB transcripts, we searched for common splice-site motifs (GT/AG, GC/AG, AT/AC). Of the 17,546 mappable BIMSB transcripts, 13,256 (76%) were found to have such a motif for at least one splice junction. Of the remaining 4290 transcripts, 3345 consisted of a single exon and thus did not contain splice sites. For 5% of the BIMSB transcripts with multiple exons, we did not identify canonical motifs at splice junctions. However, since the genome alignments produced with BLAT are insensitive to these motifs and alignments are frequently non-unique at the boundaries of aligned blocks, the fraction of 5% of BIMSB transcripts without canonical motifs at splice junctions is very likely to be an overestimate. trans-Splicing, in which a spliced-leader (SL) RNA is appended to the most 5′ exon of independently transcribed pre-mRNAs has been described in S. mediterranea (Zayas et al. 2005a). We therefore searched for SL sequences at the ends of our BIMSB transcripts and found 245 and 252 containing SL-1, SL-2, respectively. The frequency of trans-splicing events observed in our transcripts (2.53%) is only slightly lower than that described in the previous study (3.02%), suggesting that we have obtained the complete 5′ end for most trans-spliced transcripts.
Of the 18,619 BIMSB transcripts, 2789 had little coding potential and are likely noncoding RNAs (Methods). For the remaining 15,830 candidate protein-coding transcripts, we wished to generate a high-confidence homology annotation. We therefore searched, using stringent parameter settings, for orthologs in C. elegans, Drosophila, mouse, and human, and found 6729 transcripts with orthologs in at least one of the species (Supplemental Table 4). Using the same parameter settings, we identified ∼4600 orthologs between C. elegans and humans, indicating that our ortholog calls are highly conservative.
Independent assays suggest high quality of BIMSB transcripts
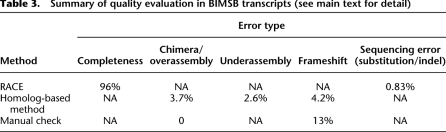
To determine whether the BIMSB transcripts contained full 5′ and 3′ ends, we performed 5′ and 3′ RACE (Rapid Amplification of cDNA Ends) for 24 transcript candidates covering high, moderate, and low expression levels (Supplemental material). Ten of these had no overlapping MAKER transcripts. Most (22 out of 24) RACE products were successfully amplified and Sanger sequenced (Supplemental Table 3). For the remaining two transcripts, the 3′ ends were validated by RACE, but 5′ RACE failed. In total 16,564 nt sequences were generated, out of which 16,520 nt can be aligned to our transcripts (Supplemental Methods). Based on these results, we confirmed that 22 transcripts have complete or nearly complete 5′ and 3′ ends (with a maximum of 50 nt shorter than RACE products). In addition to the completeness of 5′ and 3′ ends, we also estimated the sequence accuracy of our BIMSB transcripts based on the Sanger sequencing results of the RACE products. Out of 16,520 nt that can be aligned with our transcripts, we identified substitutions for 116 nt, and deletions/insertions affecting 9/24 nt. The overall error rate is therefore ∼0.90%.
Insertion/deletions introduced during the cloning, sequencing, or assembly process could potentially lead to frameshift mutations in the BIMSB transcripts. Additionally, out of 18,619 transcripts, 2745 contain a stretch of uncalled nucleotides. Thus, to estimate the frequency of such frameshift errors in the remaining 15,784 transcripts, we used Inparanoid (Remm et al. 2001) to infer orthology relations between the translated BIMSB transcripts and the human proteome (ENSembl release 57). We realigned the nucleotide sequence of each BIMSB transcript and the corresponding human protein sequence with the “protein2dna” model of Exonerate (Slater and Birney 2005). Based on the 3508 BIMSB transcript–human protein alignments, we detected 160 frameshifts. A total of 147 transcripts (4.2%) contained at least one frameshift. In total, 5,351,576 nt were scanned. After excluding the transcripts containing potential frameshift errors, we estimated the length distribution of open reading frames (ORFs) derived from the 3362 transcripts with homologs in human, as shown in Supplemental Figure 6 (Supplemental material), ORF lengths followed a logarithmic normal distribution, similar to ORFs in mouse transcripts.
During the assembly process, separate transcripts might be erroneously joined together (overassembly/fusion) or a single transcript could be separated into different pieces (underassembly/fission). To estimate the frequency of such fusion and fission events in the protein-coding region, we again used the alignment of the BIMSB transcripts and the human proteome (ENSembl57) (Supplemental material). Of 10,369 transcripts, we found 386 cases (3.7%) of putative gene fusions, a significant proportion of which might be due to overassembly. Concerning fission events, we assessed 7081 transcripts and detected 186 fission events involving two or more transcripts.
In addition, we manually checked 50 randomly chosen transcripts for frameshift and fusion events (Supplemental material). Out of the 50 transcripts, we could align 31 transcripts to at least one protein sequence in the NCBI nonredundant protein database (nr Release May 11, 2010). None of the 31 transcripts could be aligned with different proteins from the same organism. Therefore, in these cases no fusion events were detected. In six out of the 31 transcripts, the peptides translated from different possible ORFs could be mapped to the same protein sequence, indicating frameshift errors. The frameshift in two transcripts could be explained by a stretch of uncalled nucleotides within the transcripts. Overall, the frameshift rate of 13% obtained manually was much higher than the estimate based on alignments to human proteins, The latter estimation was based on deeply conserved transcripts that are known to be expressed at higher levels, which are also, on average, shorter than poorly conserved transcripts. Thus, we believe that the true error rates fall somewhere between both estimates. The results of the quality evaluation are summarized in Table 3. BIMSB transcripts containing potential frameshifts, fusion, and fission errors are listed in Supplemental Table 5.
Table 3.
Summary of quality evaluation in BIMSB transcripts (see main text for detail)
Proteomic survey
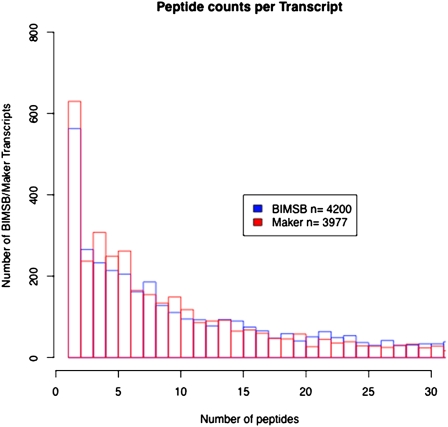
To further validate our BIMSB transcripts, we performed massive shotgun proteomic analysis using mass spectrometry (Methods). Using a protein database derived from BIMSB transcripts, at a false discovery rate of 1% (Methods), we could identify 105,688 (34.2%) MSMS spectra with a positive match on the 5017 ORFs derived from 4200 transcripts, corresponding to 16,135 different peptide sequences. In comparison, using the protein database derived from MAKER transcripts, 79,561 (25.7%) MSMS spectra corresponding to 11,985 peptides were identified with a positive match on 3977 MAKER transcripts derived proteins. In Figure 2, the distribution of transcripts with different numbers of peptide matches was plotted for BIMSB transcripts and MAKER transcripts. In comparison with MAKER transcripts, not only more BIMSB transcripts were supported with peptide matches, but also, on average, the number of peptide matches per BIMSB transcript (∼3.8) is higher than the MAKER transcript (∼3.0). This indicated that more BIMSB transcripts can be validated with higher confidence to be genuine protein-coding genes, which demonstrates the high quality of the BIMSB assembly. As discussed below, this effect might also reflect specific gene/protein expression profiles in our samples. The peptide sequences identified in this study are freely available in SmedGD, which can help the design of further proteomic research such as targeted proteomics analysis.
Figure 2.
Distribution of transcripts with different numbers of peptide matches. Distribution of distinct peptide matches to BIMSB ORFs and MAKER-predicted proteins. For each ORF generated from the BIMSB transcript sequences (see main text), the number of distinct peptide mappings is counted (x-axis) and the frequency of each count is plotted (y-axis). The same distribution is shown for the MAKER predicted proteins.
Several novel BIMSB transcripts are specifically expressed in stem cells
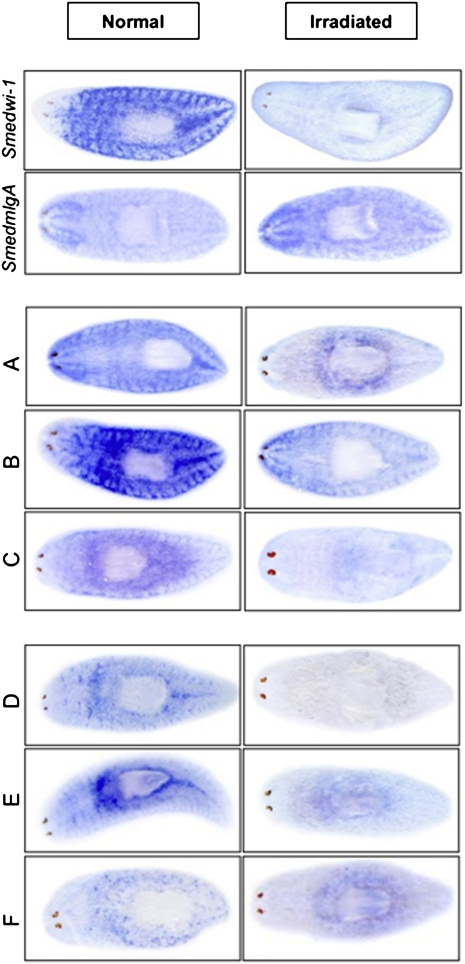
To demonstrate the value of our novel BIMSB transcripts for planarian biology, we compiled a list of transcripts that are likely to be expressed specifically in planarian stem cells (P Oenal, D Gruen, C Adamidi, A Rybak, G Mastrobuoni, Y Wang, U Ziebold, and N Rajewski, unpubl.). We then randomly selected six transcripts that did not overlap with MAKER predictions and assayed their expression via whole-mount in situ hybridization in normal as well as irradiated animals that are specifically depleted in stem cells (Methods). In addition, smedwi-1 (Reddien et al. 2005) and smedmlgA (Higuchi et al. 2008) were chosen as positive and negative controls for stem cell-specific expression, respectively. As summarized in Figure 3, all of the six novel BIMSB transcripts were expressed in a stem cell–dependent way. Interestingly, we detected transcripts with a strictly smedwi-1 like expression (transcripts A and C), transcripts with expression overlapping stem cells and the central nervous system (B), and transcripts with expression in a subpopulation of smedwi-1-positive cells (D–F). A further characterization of all stem cell-specific transcripts including, as we have shown here, novel BIMSB transcripts, will provide a much needed resource to study the heterogeneity as well as the differentiation capacity of planarian stem cells.
Figure 3.
Enriched expression of novel BISMB transcripts in planarian stem cells. Whole-mount in situ hybridization was performed on normal and irradiated asexual planarians using either smedwi-1, smedmlgA, or novel BIMBSB transcripts (human homologs) labeled: A (Misu or NSUN2), B (MOV10L1), C (LRRK2), D (HES5), E (RNMTL1), and F (CWF19L2). See Supplemental Table 7 for details.
Discussion
Traditionally, genome annotation has been based on sequencing cDNA libraries. Using Sanger sequencing, the procedure is very laborious and cost prohibitive. This situation has been dramatically improved with the recent introduction of massive parallel sequencing technology, which can sequence DNA orders of magnitude faster and at a much lower cost. So far, the sequencing of cDNA using these new technologies, so called RNA-seq, has mostly been applied to quantify the expression level of already annotated loci and to identify differentially expressed genes. RNA-seq has also been used to refine annotated gene structures such as alternative splicing, alternative 5′ and 3′ ends, or, most recently, to even build gene models de novo (Trapnell et al. 2010). However, all of these attempts require knowledge of genome sequences, which, in many situations, is not available or very difficult to obtain. In this study, we show that it is possible to obtain a high-quality representation (18,619 BIMSB nonredundant transcripts with an average length of 1228 nt and 1118 nt after filtering) of a complex animal transcriptome without using genomic sequences. Our data and analyses strongly argue that the two key ingredients of this success are (1) the simultaneous usage of complementary sequencing technologies and (2) careful normalization of the cDNA library. However, our BIMSB transcripts do contain errors (frameshifts or other assembly errors). We were able to estimate the average frequencies of these errors by using independent strategies. Overall, error frequencies were low. However, we would like to caution that our analyses also suggest that there might be substantial biases in these error rates when studying particular classes of transcripts; for example, very long and lowly expressed transcripts.
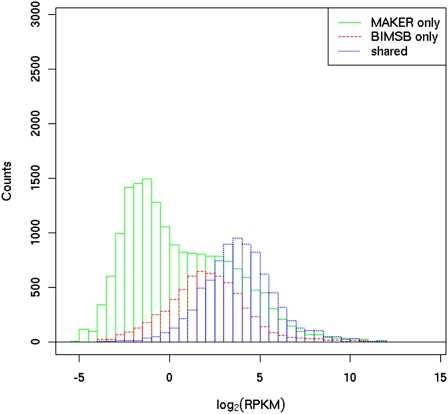
The comparison of our BIMSB transcripts to the current MAKER gene annotations showed only a limited overlap between both datasets, which could, in principle, be due to a low quality of either of the two sets. However, we believe that the observed differences, at least in part, arise from the specific expression profiles of the transcripts that we have sequenced. To confirm this hypothesis, all reads from one of the two Illumina paired-end sequencing data sets that we had used for the transcriptome assembly were mapped both to MAKER and BIMSB transcripts and used to estimate the respective expression levels in RPKM units (reads per kilobase of exon per million mapped sequence reads) (Mortazavi et al. 2008). As shown in Figure 4, BIMSB and MAKER transcripts that are transcribed from the same gene loci are expressed at an average/median abundance of 77.4/16.4 RPKM. However, transcripts that derive from gene loci specific to the MAKER annotation have lower average/median RPKM values (19.3/0.7). In comparison, the average/median RPKM specific to BIMSB transcripts (34.0/4.0) was substantially higher. These results confirm that a substantial fraction of MAKER annotations missed by our efforts were not detected because no or only a few sequencing reads have been generated from the corresponding loci. Two possible reasons for this effect are obvious: (1) Even after normalization, the abundance of these transcripts is very low; or (2) these transcripts are not expressed under the conditions at which we extracted RNA from the animals. Nevertheless, the relatively low abundance of many BIMSB transcripts missed by the MAKER annotation demonstrates that the MAKER annotation also misses many lowly expressed genes. These genes are presumably under-represented in the EST libraries used for MAKER annotations. The highest overlap (80% of the corresponding gene loci) between BIMSB and MAKER transcripts was obtained for the 6729 BIMSB transcripts for which an ortholog could be identified in either C. elegans, D. melanogaster, mouse, or human. This high overlap is probably due to the fact that highly conserved genes tend to be more highly expressed, and because the MAKER annotation uses homology analyses. Interestingly, 8% of the 6729 conserved BIMSB transcripts could not be aligned to the genome. Several possibilities could account for this: (1) missing/misassembled genomic sequences; (2) the fact that the genome of sexual animals was sequenced while we worked with asexual animals; (3) cryptic/complex splicing events. Nevertheless, it seems unlikely that these transcripts could have been predicted by using traditional genome annotation methods.
Figure 4.
Estimated expression level (represented by RPKM) for transcripts predicted by both MAKER and BIMSB annotation and for transcripts predicted only by MAKER or BIMSB annotation. For the transcripts covered by both annotations, only the expression level, estimated based on BIMSB annotation, is depicted (the expression level estimated based on the MAKER annotation is highly correlated with a correlation coefficient of 0.938).
In summary, we believe that we have developed a powerful method that can be successfully used to obtain a high-quality, complex transcriptome without the need to sequence and assemble genomic DNA. Future improvements of this method would use strand-specific sequencing protocols and cDNA derived from specific cells or conditions. However, with our current data we were already able to substantially improve the current annotation of S. mediterranea. Our massive shotgun proteomics approach allowed us to validate thousands of novel transcripts. This data set can also be used to validate transcripts predicted in future studies or to guide transcript/protein discovery.
Methods
RNA preparation
Total RNA from asexual planaria was isolated using the standard TRIzol protocol (Invitrogen) with an additional 70% ethanol precipitation step in the end. Poly(A) RNA was then purified by two rounds of selection using the Dynabeads mRNA Purification Kit (Invitrogen) according to the manufacturer's instructions and quantified by Nanodrop 7500 spectrophotometer. Sample quality was assessed by Bioanalyzer (Agilent Technologies).
Full-length enriched cDNA library construction and normalization
The construction of normalized full-length-enriched cDNA libraries for 454 sequencing requires three steps: (1) the synthesis of double strand cDNA using a modified RACE technique, (2) the subsequent removal of poly(A):T tails using the methylation sensitive type II restriction enzyme GsuI, followed by the ligation of a new DNA adaptor, and (3) the normalization of the resulting cDNA library using duplex-specific nuclease (DSN). The DSN normalization method is based on the denaturation–reassociation of double-stranded (ds) cDNA coupled with the degradation of the ds cDNA fraction formed by abundant transcripts (Shagin et al. 2002; Zhulidov et al. 2004) and requires the presence of adaptor sequences at each terminus of the cDNA to prime PCR amplification. A more detailed protocol can be found in the Supplemental material.
454 FLX titanium sequencing
Normalized cDNA library was quantified with Quant-iT dsDNA HS Assay Kit (Invitrogen). A total of 5 μg was used to prepare the sequencing library with the 454 GS FLX Titanium General Library Preparation Kit according to the manufacturer's manual. The 454 sequencing library was then sequenced 200 cycles on a 454 GS FLX sequencer according to the manufacturer's manuals.
Single-end cDNA sequencing using Illumina GAIIX
Five micrograms of full-length cDNA was used to construct a single-end sequencing library using Illumina Genomic DNA Single End Sample Prep kit according the manufacturer's manuals. The DNA concentration was measured with a Nanodrop 7500 spectrophotometer, and a 1-μL aliquot was diluted to 10 nM. Adaptor-ligated DNA was hybridized to the surface of flow cells, and DNA clusters were generated using the Illumina cluster station, followed by 36 cycles of sequencing on the GAIIX, in accordance with the manufacturer's protocols.
Paired-end RNA-seq using Illumina GAIIX
A total of 300 ng of poly(A) RNA was fragmented at 94°C for 3.5 min using 5 × fragmentation buffer (200 mM Tris-Acetate at pH 8.1, 500 mM KOAc, 150 mM MgOA) in a total volume of 20 μL. The fragmented RNA was precipitated and converted to first-strand cDNA using Supescript II (Invitrogen), followed by second-strand cDNA synthesis with E.coli DNA pol I (Invitrogen) and RNase H (Invitrogen). Then the paired-end sequencing library was prepared using the Illumina Genomic DNA Paired End Sample Prep kit according the manufacturer's manuals. The DNA concentration was measured with a Nanodrop 7500 spectrophotometer, and a 1-μL aliquot was diluted to 10 nM. Adaptor-ligated DNA was hybridized to the surface of flow cells, and DNA clusters were generated using the Illumina cluster station, followed by 2×76 cycles of sequencing on the GAIIX, in accordance with the manufacturer's protocols.
De novo transcriptome assembly
The 454 reads were assembled using Newbler 2.3 (Roche) with default parameters. The Illumina assembly comprised of Illumina paired-end and single-end reads was obtained by using SOAPdenovo software (Li et al. 2010a,b) (http://soap.genomics.org.cn) with default parameters. The contigs longer than 100 bp in the Illumina assembly were combined together with 454 reads for the final assembly using Newbler with default parameters.
Redundancy filtering of transcripts
In order to remove redundant transcripts and retain a set of putatively unique isoforms, the following procedure was applied: The mutual overlap of candidate transcripts was determined by running BLAT (Kent 2002) with default parameters on all possible pairs of transcripts drawn from the full ensemble. From each pair, the shorter of the two transcripts was discarded whenever the number of nonaligned nucleotides fell below a threshold of 35 nt and the longer one had not been discarded previously. The threshold corresponds to the 5%-quantile of the cumulative exon length distribution as determined from the transcript alignments to the genome (see Supplemental material). By this filtering step, the set of 26,669 candidate transcripts was condensed to 18,619 unique transcripts.
Inference of ORFs from the BIMSB transcripts
We translated all 18,619 BIMSB transcripts and chose the three longest ORFs for each contig. A start codon (ATG) at the beginning of an ORF was not mandatory, but all ORFs had to differ in their end position. We retained only ORFs that were at least 60 nt long.
Delineation of homology relations on protein sequence level
We used the Inparanoid software (Remm et al. 2001) to infer pairwise homology relations between the translated BIMSB transcripts and the proteomes of Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans (ENSembl release 57). All pairwise relations were combined into groups of orthologous sequences by MultiParanoid (Alexeyenko et al. 2006). We transferred the orthology relations from the protein level to either the gene level (for H. sapiens, M. musculus, D. melanogaster, and C. elegans) or the nonredundant BIMSB transcript level (see Redundancy filtering of transcripts). This step established isoform-independent homology relations. To this end, we assigned each set of isoforms (e.g., proteins from the same gene) to the orthology group to which the longest isoform was mapped and collapsed all isoforms to a single representative sequence.
Assessment of coding potential
We used simple codon usage statistics to assess the coding potential of all 18,619 BIMSB transcripts. The reference codon usage table was determined from the set of human-planaria orthologs. The model for coding sequences (model 1, 60 free parameters) is a 0-order Markov model, which generates 61 triplets based on the observed triplet frequencies in the ortholog data set. The model for noncoding sequences (models 2, 3 free parameters) generates the 61 triplets from independent single-base frequencies, which were counted on the same data set. In essence, both models have the same single-base probabilities, yet model 1 generates the observed triplet probabilities. We computed for each ORF the log likelihood ratio that it originates from model 1 vs. model 2. We consider a transcript as noncoding if all of its ORFs fail to pass the log likelihood ratio test (P < 2.5×10−6).
Detection of trans-splicing leader sequences
We obtained the two trans-splice leader sequences SL1 (GCCGTTAGACGGTCTTATCGAAATCTATATAAATCTCTTATATG) and SL2 (GCCGTTAGACGGTCTTATCGAAATCTATATAAAAATCTTATATG) from a previous publication (Zayas et al. 2005a) and used BLAT (with default values except -minIdentity=90 -stepSize=5 -tileSize=11 -fine) to scan all BIMSB transcripts for any occurrence of trans-splicing leaders.
Proteomics
Proteins were extracted from eight whole worms in Urea buffer (8 M Urea, 100 mM TrisHCl at pH 8.5). After reduction and alkylation of disulfide bridges, an aliquot of the sample was enzymatically digested with LysC and trypsin (16 h and 4 h, respectively). Desalted peptide mixture was then fractionated in 10 fractions by isoelectrofocusing and analyzed in duplicate by LC-MS/MS analysis. Proteomics raw data were analyzed using the MaxQuant proteomics pipeline v1.1.25 and the built-in Andromeda search engine (Perkins et al. 1999; Cox and Mann 2008) using (1) a sequence database of MAKER protein predictions (MAKER search), and (2) a sequence database consisting of translated ORFs from each BIMSB transcript obtained in this study (the three longest ORFs were chosen for each transcript—Transcript search). A more detailed protocol can be found in the Supplemental material. The mass spectrometry data can be downloaded from the Proteome Commons Tranche repository (https://proteomecommons.org/tranche/) using the following hash: 5nrCDrDiY116ZM+FOaerTHlmLAh5UxNA0OG/nQbJOle5NamlwC3wDJx86ccFQ0TUFsY3PDk0V3V3AFB/qfBxm8aWgegAAAAAAAAM4A==
Whole-mount in situ hybridization (WISH)
Planaria were starved for at least 1 wk before harvesting for WISH. Irradiated planaria were exposed to 60 Gy and collected 7 d after irradiation. Digoxigenin-labeled RNA probes were prepared by using an in vitro transcription kit (Roche). Whole-mount in situ hybridization was carried out as described previously (Pineda and Saló 2002) with some modifications. A more detailed protocol can be found in the Supplemental material.
Acknowledgments
We thank Mirjam Feldkamp and Salah Ayoub for their excellent technical assistance and Jochen Rink for helpful discussions. As part of the Berlin Institute for Medical Systems Biology at the MDC, the research groups of Wei Chen, Stefan Kempa, and Christoph Dieterich are funded by the Federal Ministry for Education and Research (BMBF) and the Senate of Berlin, Berlin, Germany. Yongbo Wang is supported by a scholarship under the State Scholarship Fund from the Chinese government. Dominic Gruen received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement No. HEALTH-F4-2010-241504 (EURATRANS). The 454 sequencing reagents were kindly provided by Roche.
Authors' contributions: C.A., S.K., C.D., N.R., and W.C. conceived and designed the project. C.A. and Y.W. constructed and normalized the cDNA library. Y.W. performed 454 and Solexa sequencing, and RACE validation. D.G. and C.D. analyzed and annotated the assembled transcripts. G.M. and S.K. performed the proteomic experiments that were analyzed by D.T. and G.M. X.Y. assembled 454 and Illumina reads. M.D., S.M., and X.Y. simulated 454 sequencing experiments. A.G.D. calculated normalization efficiency. A.R. and P.O. performed the in situ experiments. E.R. and A.S.A. maintained SmedGD, the S. mediterranea genome database via which the data is released. A.S.A. contributed to the writing of the manuscript. Parts of the manuscript were prepared by C.A., D.G., G.M., S.K., and C.D. N.R. and W.C. wrote the paper.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA030605. Mass spectrometry data have been uploaded to the Proteome Commons Tranche repository (https://proteomecommons.org/tranche/).]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113779.110.
References
- Agata K 2003. Regeneration and gene regulation in planarians. Curr Opin Genet Dev 13: 492–496 [DOI] [PubMed] [Google Scholar]
- Alexeyenko A, Tamas I, Liu G, Sonnhammer ELL 2006. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics 22: e9–e15 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, et al. 2009. High-resolution profiling and discovery of planarian small RNAs. PNAS 106: 11546–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg-Thorsager M, Fernandez E, Salo E 2008. Stem cells and regeneration in planarians. Front Biosci 13: 6374–6394 [DOI] [PubMed] [Google Scholar]
- Higuchi S, Hayashi T, Tarui H, Nishimura O, Nishimura K, Shibata N, Sakamoto H, Agata K 2008. Expression and functional analysis of musashi-like genes in planarian CNS regeneration. Mech Dev 125: 631–645 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT–The BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. 2010a. The sequence and de novo assembly of the giant panda genome. Nature 463: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. 2010b. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, Colbourne JK, Willis BL, Matz MV 2009. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics 10: 219 doi: 10.1186/1471-2164-10-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A 2002. Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet 3: 210–219 [DOI] [PubMed] [Google Scholar]
- Paps J, Baguna J, Riutort M 2009. Bilaterian phylogeny: A broad sampling of 13 nuclear genes provides a new lophotrochozoa phylogeny and supports a paraphyletic basal acoelomorpha. Mol Biol Evol 26: 2397–2406 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pineda D, Saló E 2002. Planarian Gtsix3, a member of the Six/so gene family, is expressed in brain branches but not in eye cells. Mech Dev 119: S167–S171 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A 2004. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20: 725–757 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A 2005. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330 [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CEV, Sonnhammer ELL 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Robb SMC, Ross E, Sánchez Alvarado A 2008. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res 36: D599–D606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Batistoni R, Deri P, Gremigni V 2008. Molecular and cellular basis of regeneration and tissue repair. Cell Mol Life Sci 65: 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A 2006. Planarian regeneration: Its end is its beginning. Cell 124: 241–245 [DOI] [PubMed] [Google Scholar]
- Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S 2002. A novel method for snp detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res 12: 1935–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G, Birney E 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH 2008. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17: 1636–1647 [DOI] [PubMed] [Google Scholar]
- Wang X, Luan J, Li J, Bao Y, Zhang C, Liu S 2010. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics 11: 400 doi: 10.1186/1471-2164-11-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas RM, Bold TD, Newmark PA 2005a. Spliced-leader trans-splicing in freshwaterp planarians. Mol Biol Evol 22: 2048–2054 [DOI] [PubMed] [Google Scholar]
- Zayas RM, Hernández A, Habermann B, Wang Y, Stary JM, Newmark PA 2005b. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci 102: 18491–18496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA, et al. 2004. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res 32: e37 doi: 10.1092/nar/gnh031 [DOI] [PMC free article] [PubMed] [Google Scholar]