Abstract
Cancers of the nervous system are clinically challenging tumors that present with varied histopathologies and genetic etiologies. While the prognosis for the most malignant of these tumors is essentially unchanged despite decades of basic and translational science research, the past few years have witnessed the identification of numerous targetable molecular alterations in these cancers. With the advent of advanced genomic sequencing methodologies and the development of accurate small-animal models of these nervous system cancers, we are now ideally positioned to develop personalized therapies that target the unique cellular and molecular changes that define their formation and drive their continued growth. Recently, the National Cancer Institute convened a workshop to advance our understanding of nervous system cancer mouse models and to inform clinical trials by reconsidering these neoplasms as complex biological systems characterized by heterogeneity at all levels.
Keywords: brain tumor, genetically engineered mice, glioma, medulloblastoma, xenograft
Tumors of the nervous system comprise a heterogeneous group of neoplasms that vary in location, age at onset, histologic features, tendency for progression and migration, and response to therapy.1 In this regard, these tumors exhibit a wide spectrum of histologic subtypes reflecting their potential cell of origin, causative molecular changes, local microenvironments, and clinical behavior (Table 1). Recent studies have underscored this heterogeneity, even within a histologically defined tumor subtype, demonstrating that histologically similar tumors represent several distinct molecular subtypes,2–4 each with a unique pattern of deregulated growth control pathways.5 Similarly, other CNS tumors (e.g., medulloblastoma, ependymoma) harbor distinct gene expression patterns that suggest that this molecular heterogeneity may be harnessed to develop more individualized therapies for these deadly cancers.6–8
Table 1.
Diversity of Nervous System Cancers
| Tumor typea | WHO gradeb | Agec | Location | Genetic Alterationsd |
|---|---|---|---|---|
| Astrocytic | ||||
| Pilocytic astrocytoma | I | 0–20 | Optic nerve, hypothalamus, thalamus, basal ganglia | NF1 loss |
| Astrocytoma | II | 30–40 | Frontal and temporal lobes, brain stem, spinal cord | p53 loss, PDGFRα, IDH1/2 mut (R132H) |
| Anaplastic astrocytoma | III | 30–60 | Cerebral hemispheres | p53, Rb, Cdkn2a, PTEN loss, CDK4 amp |
| Glioblastoma | IV | 45–70 | Subcortical white matter of cerebral hemispheres | PDGFRα and EGFR mut/overexpression, IDH1/2 mut, Cdkn2a, PTEN, NF1 loss |
| Oligodendroglial | II–III | 40–60 | Cortex and white matter of cerebral hemispheres | LOH chr 1p, 19q, EGFR, PDGFR +ligands overexpression, loss of CDKN2a |
| Oligoastrocytic | II–III | 35–45 | Cerebral hemispheres | LOH chr 1p, 19q, loss of p53 |
| Ependymal | I–III | 0–16 and 30–40 | Along the ventricular system and spinal canal | NF2 loss |
| Embryonal | ||||
| Medulloblastoma | IV | 0–20 | Cerebellum | c-myc amp, p53, ptch loss |
| PNET | IV | 0–10 | Supratentorial | n-myc amp, p53 loss |
| Cranial/peripheral nerves | ||||
| Schwannoma | I | 40–60 | Peripheral nerves of head and neck region | NF2 loss |
| MPNST | II–IV | 30–60 | Large and medium nerves | NF1 and p53 loss |
Abbreviations: PDGFR, platelet-derived growth factor receptor; PNET, primitive neuroectodermal tumor; CDK4, cyclin-dependent kinase 4; IDH1, isocitrate dehydrogenase 1; LOH, loss of heterozygosity.
aBased on World Health Organization (WHO) Classification of Tumours; Pathology and Genetics of Tumours of the Nervous System, P. Kleihues and W. Cavenee, eds.
bWHO grading system, from benign (grades I–II) to malignant (grade III–IV).
cPeak incidence range.
dMost common alterations listed.
To begin to address this issue of heterogeneity, the National Cancer Institute convened the fifth Mouse Models of Human Cancers Consortium (MMHCC) Nervous System Tumors Workshop, held in Montreal, Canada, on November 18, 2010 (Table 2). The workshop was divided into four topics, each moderated by an expert in the field. The meeting opened with presentations on the identification and characterization of the cell of origin of brain tumors in different mouse models, followed by talks that focused on the role of the microenvironment in tumor initiation and growth. The meeting concluded with sessions on genomics and systems biology as well as the use of mouse models for therapeutic target discovery and evaluation.
Table 2.
MMHCC Nervous System Tumors Workshop participants
| Participant | Institution |
|---|---|
| Suzanne Baker, Ph.D. | St. Jude Children's Research Hospital, Memphis, TN |
| Michael Berens, Ph.D | The Translational Genomics Research Institute, Phoenix, AZ |
| Gideon Bollag, Ph.D. | Plexxikon Inc., Berkeley, CA |
| Al Charest, Ph.D. | Tufts University School of Medicine, Boston, MA |
| Charles Eberhart, M.D., Ph.D. | Johns Hopkins University, Baltimore, MD |
| Frank Furnari, Ph.D. | Ludwig Institute, University of California–San Diego, La Jolla, CA |
| Marco Giovannini, M.D., Ph.D. | House Ear Institute, Los Angeles, CA |
| David Gutmann, M.D., Ph.D. | Washington University School of Medicine, St. Louis, MS |
| Eric Holland, M.D. | Memorial Sloan Kettering Cancer Center, New York, NY |
| C. David James, Ph.D. | University of California–San Francisco, San Francisco, CA |
| David Largaespada, Ph.D. | University of Minnesota, Minneapolis, MN |
| Scott Lowe, Ph.D. | Cold Spring Harbor Laboratories, Cold Spring Harbor, NY |
| Silvia Marino, M.D. | Barts and The London School of Medicine and Dentistry, London, England |
| John Ohlfest, Ph.D. | University of Minnesota, Minneapolis, MN |
| Karlyne Reilly, Ph.D. | NCI, Frederick, MD |
| Joshua Rubin, M.D., Ph.D. | Washington University School of Medicine, St. Louis, MS |
| Jann Sarkaria, M.D. | Mayo Clinic, Rochester, MN |
| Charles Stiles, Ph.D. | Dana Farber Cancer Institute, Boston, MA |
| Rob Wechsler-Reya, Ph.D. | Sanford-Burnham Medical Research Institute |
| William Weiss, M.D., Ph.D. | University of California–San Francisco, San Francisco, CA |
Modeling Nervous System Tumors in Mice
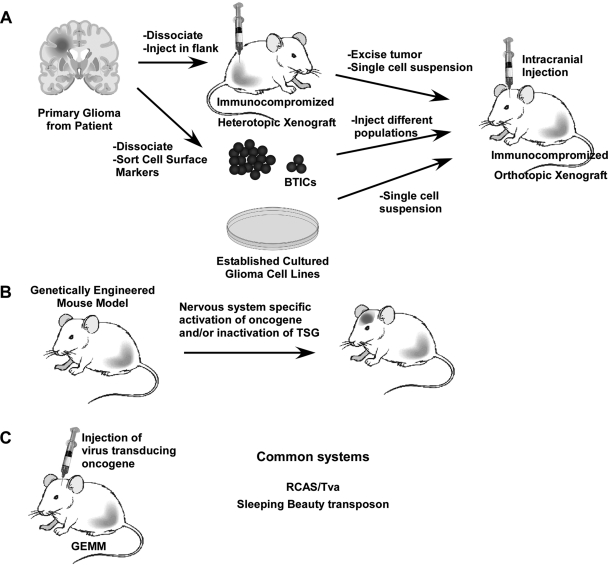
Brain tumor models in mice are being used to study many aspects of tumor biology and in preclinical settings to evaluate potential treatment modalities. In general, small-animal models can be divided into two basic categories: (1) those that implant tumor cells into recipient mice (xenograft) and (2) those that induce tumors in mice de novo (genetically engineered mouse [GEM] models) (Fig. 1). The first generation of nervous system tumor xenograft models employed tumor cell lines that had been maintained under artificial cell culture conditions for extended periods of time (often decades). Typically, the tumors generated from these cell lines fail to accurately reproduce the classical histopathologic appearances of their human counterparts,9 display no molecular resemblance to the original human tumor,3 and, more importantly, are not predictive of drug response in preclinical trials.10
Fig. 1.
Nervous System Tumor Models. (A) Xenograft models are created from patients' tumors or from established glioma cell lines injected into immunocompromised mice. Brain tumor-initiating cells (BTICs) are isolated from freshly dissociated tumors and sorted based on cell surface markers. (B) GEM models are designed to produce tumors de novo by activating oncogenic mutations and/or inactivating tumor suppressor genes (TSGs) in a cell-type specific manner. (C) A variation of GEM utilizes intracranial injections of viruses to express oncogenes in the CNS. The RCAS/Tva system allows for cell-specific expression of oncogenes.
Over the past several years, a number of laboratories have developed orthotopic xenograft models using primary nervous system tumor cells from freshly isolated human brain tumors. These tumor models recapitulate certain features of the human tumors, including their invasive behaviors and tissue architecture. More recently, the isolation of brain tumor-initiating cell populations from dissociated patient tumors using cell surface markers has refined our understanding of glioblastoma multiforme (GBM) heterogeneity with respect to renewal and tumor-initiating capacities. Dr David James (University of California, San Francisco) provided a nice overview of the human glioma xenografts generated in his laboratory and outlined their use for preclinical therapeutic studies. Similarly, Dr Jann Sarkaria (Mayo Clinic) demonstrated that these human high-grade glioma xenografts retain most of the seminal genetic alterations observed in the original patient tumors, which were remarkably stable over time. While these brain tumor models have the advantage of deriving from actual human tumors, they are grown in mice lacking a functional immune system or a relevant microenvironment.
In contrast, GEM models are designed to induce brain tumors using relevant cancer-causing genetic changes in the context of an intact immune system and nervous system microenvironment. Their use for functionally validating the role of specific genetic changes to tumor formation and progression has been particularly instructive, and they have revealed important roles for local and genomic environments in tumorigenesis and continued tumor growth. In preceding studies in human tumors, accurate small-animal nervous system GEM strains have been used to test the efficacy of novel drugs and compounds in preclinical settings.
Cell of Origin and Developmental Neurobiology
The session on the interface between neuro-oncology and developmental neurobiology was moderated by Dr Charles Stiles (Dana Farber Cancer Institute) and focused on the various methodologies employed to identify the cell types that give rise to various brain tumors. A recurrent theme in this session was the concept of the varying degrees of permissiveness of stem and progenitor cells to specific cancer-causing genetic alterations. In this regard, tumorigenesis in the nervous system is dependent on a combination of specific cancer-associated genetic mutations occurring in receptive cell types during permissive periods of nervous system development.
Dr Rob Wechsler-Reya (Sanford-Burnham Medical Research Institute) presented data from mouse medulloblastoma modeling experiments suggesting that more personalized brain tumor treatments may come from a more complete understanding of the interplay between genetic mutations and the specific stem and progenitor cells in which these mutations occur. In these studies, he employed a combination of human xenograft and GEM models to re-create various genetic subtypes of medulloblastoma. He showed that forced expression of c-myc in cerebellar stem cells is mitogenic and results in transient hyperplasia, while simultaneous expression of c-myc and mutant p53 results in aggressive tumors that resemble human large-cell anaplastic medulloblastoma. Further characterization of these tumors indicated that they were molecularly distinct from those driven by Ptch mutation and exhibited different responses to therapy.
Dr Silvia Marino (Barts and The London School of Medicine and Dentistry) demonstrated that loss of p53 and Rb in two different populations of progenitor cells—cerebellar granule cell progenitors and cerebellar stem cells of ventricular zone (VZ) origin—gave rise to medulloblastomas in mouse models. In these studies, conditional inactivation of Rb and p53 was obtained in these cells either in vivo, through granule cell progenitors, or in vitro followed by orthotopical transplantation, through VZ-derived stem cells of nongranule cell lineage. Both populations gave rise to medulloblastoma tumors with identical histopathologic appearances; however, tumors originating from VZ progenitors preferentially expressed stem cell markers. This set of markers was shown to identify a subset of human medulloblastomas associated with a poorer clinical outcome.
Dr William Weiss (University of California, San Francisco) presented studies that focused on identifying the cell of origin in two glioma models. First, using advanced labeling techniques, he demonstrated that in an astrocytoma GEM model (GFAP-HaRas) developed by Dr Abhijit Guha,11 gliomas arise from SVZ-derived stem cells, whereas in the S100-vErb oligodendroglioma model,12 tumors originated from white matter NG2+ glial progenitor cells. He further demonstrated that NG2+, but not CD133+, cells isolated from human oligodendroglioma tumors were capable of forming tumors following implantation into immunocompromised mice. Collectively, these data support a model in which gliomas may develop from stem cells, whereas oligodendrogliomas derive from NG2+ progenitor cells.
Dr Charles Eberhart (Johns Hopkins University) described the differences between various Notch isoforms in inducing glioma formation in the optic nerve and retina. In his studies, he showed that while Notch3 robustly induced optic nerve gliomas, tumors were not generated following expression of activated Notch1 or Notch2. These experiments clearly demonstrate differences in the susceptibility of tissues for oncogenic transformation by the Notch gene family. Using chimeric Notch constructs, the oncogenic portion of the Notch3 gene was found to reside in the carboxyl terminal domain of the protein.
Dr Marco Giovannini (House Ear Institute) described a new mouse model of schwannomatosis. In human schwannomatosis, NF2 mutations are common; however, mutations in the INI1 gene are observed in 30% of familial and 7% of sporadic cases. Whereas targeted deletion of Ini1 in mice is lethal, Schwann cell precursors with conditional Ini1 inactivation resulted in olfactory nerve, third cranial nerve, and trigeminal nerve tumors. Current studies are focused on developing mice with combined Ini1 and Nf2 inactivation in Schwann cell precursors.
Stromal Influences on Tumorigenesis
As has been reported for other cancers,13–15 it is becoming increasingly clear that the local microenvironment plays a critical role in brain tumor development and growth. This session was moderated by Dr Frank Furnari (Ludwig Institute, University of California, San Diego) and focused on the use of GEM strains to elucidate the complex relationship between neoplastic and nonneoplastic cells in the tumor microenvironment.
In particular, two presentations employed the inherited cancer predisposition syndrome, neurofibromatosis type 1 (NF1), to demonstrate that specific cell types and signals from the tumor microenvironment are important for gliomagenesis and continued glioma growth. The use of NF1 as a model system to study nervous system tumor-stroma interactions derives from studies first published by Dr Luis Parada (Southwestern University), in which targeted loss of Nf1 in Schwann cell precursors is insufficient for tumorigenesis unless coupled with heterozygosity for an inactivating Nf1 gene mutation in nonneoplastic cells.16 These initial observations have been extended to glioma17,18 and used to identify specific growth factors and cytokines that drive tumorigenesis and continued glioma growth.19–21
Dr David Gutmann (Washington University School of Medicine) described the critical role that microglia play in Nf1 GEM optic glioma growth. Using a combination of approaches, he demonstrated that pharmacologic and genetic microglia silencing inhibits optic glioma growth. Moreover, he described studies in which optic nerve microglia are uniquely sensitive to the effects of Nf1 heterozygosity during early glioma formation, leading to studies aimed at disrupting the interactions between microglia and preneoplastic/neoplastic cells during critical phases of gliomagenesis.
Dr Joshua Rubin (Washington University School of Medicine) next reported on his discovery of one key chemokine expressed in the nonneoplastic optic glioma microenvironment. He showed that CXCL12 (stroma-derived factor-1α) normally induces astrocyte apoptosis, whereas in Nf1-/- astrocytes, CXCL12 treatment leads to inappropriate astrocyte survival in vitro. This reduced apoptosis reflects decreased intracellular cyclic adenosine monophosphate (cAMP) production, prompting Dr Rubin to explore the possibility that ectopic suppression of cAMP in regions of the brains of Nf1 optic glioma might induce glioma formation. Indeed, cAMP reduction resulting from viral expression of phosphodiesterase-4–induced gliomas in the forebrain of these mice. Collectively, these studies highlight the critical interdependent relationship between neoplastic cells and signals from their nonneoplastic neighbors relevant to gliomagenesis and glioma maintenance.
Genomics and Systems Biology
With the recent explosion of comprehensive genomic studies on brain tumors, it is becoming increasingly clear that one has to view individual genetic mutations in the context of a global network. In the session chaired by Dr Scott Lowe (Cold Spring Harbor Laboratories), presentations focused on the various combinations of genetic mutations required for nervous system tumorigenesis.
Dr Suzanne Baker (St Jude Children's Research Hospital) presented data demonstrating the profound differences of Pten gene inactivation on gliomagenesis. Whereas postnatal, adult Pten ablation in astrocytes had no effect, combined deletion with other tumor suppressors induced astrocytomas with high penetrance. Co-deletion of Pten and Rb failed to induce astrocytomas, but co-deletion of Pten and p53, p53 and Rb, or Pten, p53, and Rb all induced astrocytomas. Secondary mutations within the phosphoinositide-3 kinase and retinoblastoma signaling pathways were found in tumors that were induced by inactivation of tumor suppressors in the same pathways. Tumors formed within and outside of proliferative niches in adult brain.
Dr David Largaespada (University of Minnesota) described work on the use of the sleeping beauty (SB) transposon system for mutagenesis screens in mice conditionally deleted for Pten and p53. Using this approach, he was able to generate cerebellar tumors with different complements of genetic alterations. For example, one of the genes inactivated by SB in this genetic screen was Slit3, which his laboratory demonstrated was also inactivated by mutation or promoter methylation in human medulloblastoma.
Dr Karlyne Reilly (NCI Frederick) presented her work on the identification of genetic modifiers of Nf1:p53-Cis–driven malignant peripheral nerve sheath tumor (MPNST). In these studies, she leveraged the differential susceptibility to MPNST in A/J compared with C57Bl/6J mice. One candidate gene was found to be an imprinted gene. Using a targeting strategy, she discovered that this modifier gene acts in a tumor suppressive manner when inherited from the mother. These findings support a model in which the severity of MPNSTs depends on whether maternal or paternal copies of chromosomes are altered during tumorigenesis.
Therapeutic Targets
In the final session of the meeting, approaches to discovering and exploiting therapeutic targets were discussed. This session was moderated by Dr Gideon Bollag (Plexxikon) and emphasized the complexities associated with performing preclinical trials in mice and the adaptability of tumors to therapeutic interventions.
Dr Al Charest (Tufts University School of Medicine) presented work using a GBM model driven by wild-type epidermal growth factor receptor (EGFR). Drawing upon the observation that human GBMs overexpressing wild-type EGFR also express EGFR ligands, he described a model by which somatic expression of EGFR and of transforming growth factor-α (an EGFR ligand) in the context of loss of cdkn2a and/or Pten tumor suppressor gene function yields tumors with molecular and histopathologic features of “classical” GBM tumors. He also described differences in the sensitivity of cdkn2a-null and cdkn2a;Pten-null tumor cells to EGFR inhibitors. His laboratory found that this difference arises from tumor cells switching their dependence for mitogenic signaling from one receptor tyrosine kinase to another. These data illustrate one molecular mechanism for the primary resistance of GBMs to EGFR tyrosine kinase inhibition.
Dr Michael Berens (The Translational Genomics Research Institute) described the Ivy Genomics-based Medicine Project, which is a 9-institute preclinical study relating chemovulnerability to molecular profiling in human primary GBM orthotopic xenografts. Funded by the Ben and Catherine Ivy Foundation, the update reported survival outcomes of 21 GBM models tested with 4 treatment regimens; genomic data (expression profiling, array comparative genomic hybridization, cytosine-phosphate-guanine methylation, and selected DNA sequencing of the models) are being produced. Engagement of pharmaceutical companies to provide a larger spectrum of targeted therapeutic agents remains in motion. Using extensive genomic profiling of 40 GBM xenograft lines, one initial objective was to establish a proband set of xenograft tumors with genomic signatures that represent the spectrum of patients with GBM as portrayed in The Cancer Genome Atlas. A follow-up study proposes to use the molecular profiling data to inform treatment planning (clinical trial) by matching the therapeutic responses of the various xenograft lines to their genetic signatures and aligning these against patient tumor signatures.
Dr Eric Holland (Memorial Sloan Kettering) described a procedure to generate a recurrent model of GBM using the replication-competent ALV splice acceptor (RCAS) virus/Tva model system. Tumor-bearing animals were given fractionated ionizing radiation or temozolomide therapy. Gene expression profiling was performed before and after treatment to identify genetic signatures most predictive of recurrence-free survival. In parallel, Dr Holland also presented data on the relative sensitivity of the various cell types within the GBM tumor to therapeutic intervention in vivo. Using differential cell purification methods, he was able to identify specific genes within the Olig2+ population of cells that might mediate resistance to radiation.
Dr John Ohlfest (University of Minnesota) described the importance of the multidrug resistance system in the treatment of brain cancer. There are two dominant mechanisms responsible for poor blood-brain barrier (BBB) penetration of certain molecularly targeted drugs: the efflux systems coded for by the Bcrp and Pgp genes. Ohlfest advocated that although the BBB in the tumor core is leaky, allowing for systemic drug delivery, the tumor-infiltrated normal brain (the site of recurrence) has an intact BBB, which prevents drug delivery. Using a combination of knockout mice and specific pharmacologic inhibitors, he demonstrated that BCRP and PGP cooperate by synergistically effluxing gefitinib, sorafenib, and dasatinib from the brain. However, in the case of sorafenib, Bcrp was dominant, while for gefitinib and dasatinib, PgP was dominant. Using a mouse glioma model based on SB-delivered oncogenes, he discovered that loss of function of both the Bcrp and Pgp genes more than doubled survival after treatment with dasatinib relative to wild-type mice. In addition, western blot data revealed that dasatinib markedly inhibited phosphorylation of Src only in the Bcrp Pgp compound knockout mice. Collectively, these results suggest that optimal penetration of these drugs into tumor-infiltrated normal brain where the BBB is intact is dependent on Bcrp and PgP function, such that administration of single inhibitors of PgP or Bcrp would have minimal clinical advantage over chemotherapeutic agent alone. More importantly, these data stress the need to consider penetration of molecularly targeted agents in the tumor-infiltrated normal brain where the BBB is intact.
Summary—Leveraging Heterogeneity
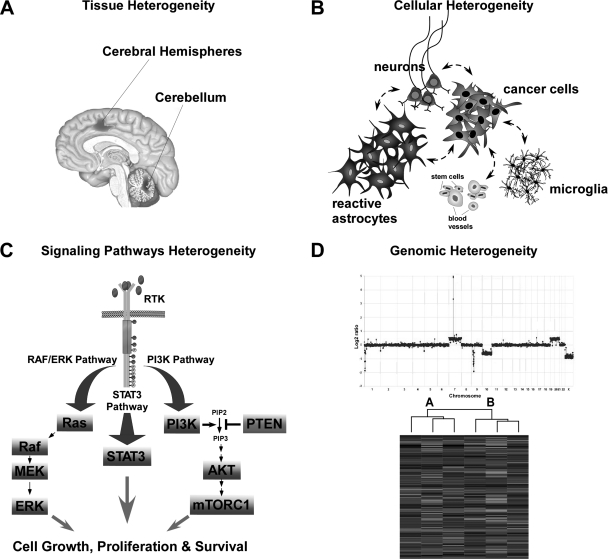
One of the common themes of this meeting was heterogeneity. Heterogeneity affects nervous system tumor formation and treatment in many ways (Figure 2). First, tumor susceptibility is influenced by genomic heterogeneity, such that both tumor formation and response to therapy are dictated in part by modifier genes in our individual genomes. Subtle polymorphisms in specific genes may change the local microenvironment, expression of specific tumor suppressor genes, or drug metabolizing enzymes.22–27 Second, progenitor cells and stem cells in distinct regions of the brain and during different times of development are unique, and may be differentially affected by cancer-causing genetic changes. For example, Nf1 inactivation in astrocytes or neural stem cells from the cortex has little effect on astrocyte proliferation or astrogliogenesis in vitro and in vivo, whereas Nf1 loss in brainstem neural stem cells or astrocytes results in increased proliferation and gliomagenesis.28 Third, the local microenvironment harbors specialized cells and signals capable of initiating and maintaining tumors in the nervous system. Fourth, the signaling pathways and transcriptional factor networks are highly adaptable and dynamic. In considering future therapies for brain tumors, we will need to employ a systems-based approach that integrates this heterogeneity at all levels to effect a more personalized treatment for these deadly cancers.
Fig. 2.
Nervous System Tumor Heterogeneity. (A) Tumors arise in different parts of the CNS with unique histopathologic presentations. (B) Within a tumor, there are multiple cell types that interact. (C) Within the tumor cells, there are multiple signaling pathways that integrate and create a physiological output. (D) Tumors categorized histologically as one entity differ considerably in their genomic makeup.
This workshop also provided a glimpse into future directions for glioma model research. Several laboratories are focusing on expanding the complexities of their models to better mimic human tumors for preclinical studies. As such, a great deal of resources and efforts are invested in studying therapeutic responses in models that are genetically designed to mirror patients’ tumors. In addition, more sophisticated studies on basic mechanisms of gliomagenesis are arising. Specifically, the concept of permissibility and resistance of glia and neuro stem vs non stem cell to oncogenic assault will no doubt reveal basic themes for CNS cancers. In the coming years, we will witness an unprecedented level of sophistication in these models and their use that will translate into major advances for both clinical and basic research.
Acknowledgments
We thank Dr Cheryl Marks, Janice Embry, Cynthia Graddy, and Jan Esenwein for their assistance in organizing this workshop.
Funding
Funding that supported this meeting was provided by the National Cancer Institute.
Conflict of interest statement. The authors declare no conflict of interest.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma MK, Mansur DB, Reifenberger G, et al. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67:890–900. doi: 10.1158/0008-5472.CAN-06-0973. doi:10.1158/0008-5472.CAN-06-0973. [DOI] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan C, Momota H, Hambardzumyan D, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. doi:10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbertson RJ, Gajjar A. Molecular biology of medulloblastoma: will it ever make a difference to clinical management? J Neurooncol. 2005;75:273–278. doi: 10.1007/s11060-005-6750-z. doi:10.1007/s11060-005-6750-z. [DOI] [PubMed] [Google Scholar]
- 7.Cho YJ, Tsherniak A, Tamayo P, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. doi:10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. doi:10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–4239. [PubMed] [Google Scholar]
- 11.Ding H, Roncari L, Shannon P, et al. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61:3826–3836. [PubMed] [Google Scholar]
- 12.Weiss WA, Burns MJ, Hackett C, et al. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–1595. [PubMed] [Google Scholar]
- 13.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. doi:10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 14.Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol. 2008;18:311–321. doi: 10.1016/j.semcancer.2008.03.013. doi:10.1016/j.semcancer.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. doi:10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y, Ghosh P, Charnay P, et al. Neurofibromas in NF1:Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. doi:10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajenaru ML, Hernandez MR, Perry A, et al. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63:8573–8577. [PubMed] [Google Scholar]
- 18.Zhu Y, Harada T, Liu L, et al. Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005;132:5577–5588. doi: 10.1242/dev.02162. doi:10.1242/dev.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang FC, Ingram DA, Chen S, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/– and c-kit-dependent bone marrow. Cell. 2008;135:437–448. doi: 10.1016/j.cell.2008.08.041. doi:10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daginakatte GC, Gutmann DH. Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth. Hum Mol Genet. 2007;16:1098–1112. doi: 10.1093/hmg/ddm059. doi:10.1093/hmg/ddm059. [DOI] [PubMed] [Google Scholar]
- 21.Warrington NM, Woerner BM, Daginakatte GC, et al. Spatiotemporal differences in CXCL12 expression and cyclic AMP underlie the unique pattern of optic glioma growth in neurofibromatosis type 1. Cancer Res. 2007;67:8588–8595. doi: 10.1158/0008-5472.CAN-06-2220. doi:10.1158/0008-5472.CAN-06-2220. [DOI] [PubMed] [Google Scholar]
- 22.Relling MV, Dervieux T. Pharmacogenetics and cancer therapy. Nat Rev Cancer. 2001;1:99–108. doi: 10.1038/35101056. doi:10.1038/35101056. [DOI] [PubMed] [Google Scholar]
- 23.Hunter K. Host genetics influence tumour metastasis. Nat Rev Cancer. 2006;6:141–146. doi: 10.1038/nrc1803. doi:10.1038/nrc1803. [DOI] [PubMed] [Google Scholar]
- 24.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. doi:10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosch TM. Pharmacogenomics of drug-metabolizing enzymes and drug transporters in chemotherapy. Methods Mol Biol. 2008;448:63–76. doi: 10.1007/978-1-59745-205-2_5. doi:10.1007/978-1-59745-205-2_5. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Hagemann A, DeMichele A. Immuno-modulatory gene polymorphisms and outcome in breast and ovarian cancer. Immunol Invest. 2009;38:324–340. doi: 10.1080/08820130902910567. doi:10.1080/08820130902910567. [DOI] [PubMed] [Google Scholar]
- 27.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. doi:10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 28.Lee da Y, Yeh TH, Emnett RJ, et al. Neurofibromatosis-1 regulates neuroglial progenitor proliferation and glial differentiation in a brain region-specific manner. Genes Dev. 2010;24:2317–2329. doi: 10.1101/gad.1957110. doi:10.1101/gad.1957110. [DOI] [PMC free article] [PubMed] [Google Scholar]