Abstract
Formalizing eligibility criteria in a computer-interpretable language would facilitate eligibility determination for study subjects and the identification of studies on similar patient populations. Because such formalization is extremely labor intensive, we transform the problem from one of fully capturing the semantics of criteria directly in a formal expression language to one of annotating free-text criteria in a format called ERGO Annotation. The annotation can be done manually, or it can be partially automated using natural-language processing techniques. We evaluated our approach in three ways. First, we assessed the extent to which ERGO Annotations capture the semantics of 1000 eligibility criteria randomly drawn from ClinicalTrials.gov. Second, we demonstrated the practicality of the annotation process in a feasibility study. Finally, we demonstrate the computability of ERGO Annotation by using it to (1) structure a library of eligibility criteria, (2) search for studies enrolling specified study populations, and (3) screen patients for potential eligibility for a study. We therefore demonstrate a new and practical method for incrementally capturing the semantics of free-text eligibility criteria into computable form.
Keywords: Eligibility criteria, clinical trials, natural-language processing, ontology, OWL, relational databases
I. Introduction
Human studies are the most important source of evidence for advancing our understanding of health, diseases, and treatment options. It is therefore crucial to optimize the design, execution, and use of human studies. Yet the current practices are fraught with problems. Clinical trials have difficulty recruiting study subjects. For example, a recent study in UK found that less than one third of trials in a cohort of studies recruited their original target within the time originally specified and 45% of the trials recruited below 80% of their original recruitment target [1]. At the point of care, clinicians are inundated with study results that relate only partially to their patients [2]. Both problems would be ameliorated by making computer-interpretable the eligibility criteria which define the target populations of studies. At the design stage, study investigators could query a library of computable criteria to help define their study population by comparing the content and selectivity of their criteria to those of related studies. At the execution stage, investigators could query electronic health records to find potentially eligible subjects. Finally, at the usage stage, providers at the point of care could query for studies that enrolled patients similar to theirs.
Formalizing eligibility criteria in a computer-interpretable language, however, is an extremely labor-intensive task that requires knowledge of the detailed syntax and semantics of the representational language [3]. In this paper, we briefly discuss current approaches to creating computer-interpretable languages for eligibility criteria (Section II), the use cases and research questions that motivate this work (Section III), and the insights that led to the new methodology described in this paper (Section IV). This methodology aims at incrementally capturing the semantics of eligibility criteria by defining a representation intermediate in expressiveness between domain-specific enumerative criteria and expression languages. This intermediate representation, called ERGO Annotation, is informed by both the complexity of natural language and the requirements for computability. (ERGO Annotation is based on the Eligibility Rule Grammar and Ontology (ERGO) previously defined in The Trial Bank Project.1) We define ERGO Annotations for classes of eligibility criteria based on their logical and comparative structure and on their noun phrases. This intermediate representation allows the encoding of free-text criteria to be partially automated through the application of natural-language processing (NLP) and other computational methods. From ERGO Annotations we generate computable expressions, such as OWL DL queries [4] or SQL queries, that can be used to satisfy valuable use cases (Section III). We validate the methodology through detailed examples (Section VI) and through two preliminary evaluations that (1) demonstrate the practicality of the encoding process using NLP methods, and (2) assess the extent to which ERGO Annotations capture the semantics of sample eligibility criteria (Section VII).
II. Background
Currently, eligibility criteria are available only in free text that are difficult to parse or process computationally. As a consequence, study repositories such as ClinicalTrials.gov generally ignore eligibility criteria and use instead some notion of “health condition studied” as the primary proxy for a study's target population. This approach is frequently inaccurate and does not allow for very specific searches. For example, searching ClinicalTrials.gov for studies on “small cell lung cancer” returns 443 trials2. If one were interested only in studies in “small cell lung cancer” patients with existing “brain metastases,” entering these terms into the basic search field returns 12 studies, of which only 3 require the presence of brain metastases, while five studies are on preventing brain metastases (i.e., patients with brain metastases are specifically excluded). The Advanced Search also has errors. Entering these terms into the Targeted Search fields for Condition and specifying NOT “brain metastases” in the Outcomes field returns 3 studies, but only 2 of the 3 that require brain metastases and erroneously including 1 study on preventing brain metastases in patients without brain metastases. None of the other public trial registers (e.g., WHO3, ISRCTN4) support targeted searches on eligibility criteria either, but even if they did, simple keyword searches of the criteria text would not be sufficient, for reasons we discuss extensively in this paper. If the eligibility criteria were computable, however, one could search for studies enrolling patients with specific clinical features such as prior chemotherapy or radiotherapy, or the extent and location of metastases. Such clinically specific searches are needed to more accurately find studies for patients to enroll in, or to find completed studies on patients similar to a given patient for whom clinical trial evidence is being sought.
Over the years, informatics researchers have developed representations of eligibility criteria. Some, like the ASPIRE project [5], seek to develop consensus on a core coded set of generic (e.g., age, gender) and disease-specific (e.g., breast cancer stage, estrogen and progesterone status) data elements for representing eligibility criteria. Each data element has an associated value set that defines its legal values (e.g., {Male, Female} for the Sex data element). Other computable representations of eligibility criteria, like Arden Syntax [6], GELLO [7], and other database or logic-based rule languages, employ domain-independent formal syntax for encoding computer-interpretable expressions that use external terminology systems. In Arden Syntax, the eligibility criteria of a study may be defined as a Medical Logic Module (MLM) that includes specifications for the events that trigger the MLM, the data needed to evaluate eligibility, the decision logic that computes an eligibility status, and actions for alerting providers or patients. In GELLO, formalizing an eligibility criterion involves (1) specifying a patient data model and codes from external terminologies, and (2) writing the criterion using a formal object-oriented expression grammar. In a rule language such as JESS,5 eligibility criteria can be encoded either as individual rules, or as declarative data structures that are evaluated using generic rules. Similarly, eligibility criteria can be written directly as SQL queries for some relational databases.
Encoding eligibility criteria into existing representations presents a number of problems. ASIPRE's domain-specific coded eligibility criteria have not been fully standardized, and it will take years to create eligibility codes for all disease areas. This enumerative approach does not make use of existing reference terminologies. Thus, for example, it would represent “Presence of asthma” and “Presence of severe asthma” as two criteria whose relationship to each other cannot be inferred from “Asthma” (SNOMEDCT 195967001) and “Severe Asthma” (SNOMEDCT 370221004) in a standard terminology. Furthermore, having no mapping of ASPIRE elements to patient data models and terminologies, ASPIRE criteria cannot be used directly for eligibility screening using existing EHR data.
Computable expression languages such as Arden Syntax, GELLO, SQL, and rule languages combine the use of domain-independent generative syntax with standard terminologies and data models. However, they are designed primarily for providing patient-specific decision support in clinical information systems. They provide no method for classifying or reasoning with formalized eligibility criteria. Like ASPIRE, Arden Syntax and GELLO offer no way to identify a relationship between the criterion expressions for “Presence of Asthma” and “Presence of severe asthma.” In Arden Syntax, queries for such data would be hidden in the institution-specific “curly braces” [8]. In GELLO and other rule and database languages, on the other hand, encoding eligibility criteria requires a commitment to a specific patient data model. For example, to encode in GELLO a simple criterion such as “Presence of azotemia within the last 3 months” would require that the criterion be written in terms of some data structure (e.g., Observation) that has a code for azotemia, and a time stamp for the observation. The criterion could be represented as shown in Figure 1.
Figure 1.
The GELLO expression language assumes a patient data model that may includes an Observation class that has properties `effectiveTime' (an interval with high and low limits), `code' consisting of a coded concept (e.g., terminology and code), and `value' that may be a coded concept or a physical quantity that has a value and unit.
Commitment to a patient data model is necessary for linking criteria with patient data for screening purposes, but it poses several problems. It limits the possibility of using the encoded criteria for multiple use cases, as is desired for eligibility criteria (Section III). Furthermore, it complicates the encoding process. Not only does the encoder need to know the syntax and semantics of a complex language like Arden Syntax or GELLO, she also needs to make assumptions about how patient data are represented. This complexity adds to the difficulty and labor demands of formalizing eligibility criteria.
Another weakness of expression languages like GELLO and Arden Syntax is that they do not support encoding of the noun phrases which contain much of the semantics of free-text criteria. Having been designed for matching criteria with patient data, those languages typically assume that potentially complex noun phrases in eligibility criteria (e.g., “Histologically or cytologically confirmed extensive stage small cell lung cancer”) can be represented using a single terminological code.
To overcome some of these shortcomings, we previously created ERGO, a template-based expression language that can capture the full expressivity of eligibility criteria from any clinical domain [9]. Like GELLO, ERGO is based on an object-oriented data model, and its expressions can be seen as a subset of GELLO, except that:
Instead of being a string-based language, ERGO expressions are largely (though not completely) defined by a set of frame-based templates;
ERGO allows not only Boolean combinations of statements, but also statements linked by semantic connectors such as defined by (e.g., “adult patients defined by age >= 18 years”) and by examples (e.g., “Planned coronary revascularization such as stent placement OR heart bypass”);
ERGO explicitly incorporates constraints on temporal relationships, such as “occurrence of a stroke within six months following a myocardial infarction”; and
ERGO explicitly models terminological expressions as part of the language.
In ERGO, clinical statements are built from other clinical statements and from expressions that can be data values (e.g., physical quantities, time points and time intervals as well as standard primitive data types), functions, queries, variables, and noun phrases. ERGO defines three subclasses of noun phrases:
Primitive noun phrases (e.g., “myocardial infarction”), which represent terms from vocabularies.
Logical combinations of noun phrases connected by and, or, or not (e.g., “ myocardial infarction or diabetes mellitus”). The and, or, and not operators are interpreted as intersection, union, and complement of the corresponding sets, respectively.
Noun phrases with modifiers that place restrictions on the root noun phrase. Modifiers follow the entity-attribute-value model (e.g., asthma induced by exercise). In cases where the attribute of the modifier is unclear, we use a default modified_by attribute (e.g. “asthma modified_by transient”).
We developed ERGO based on our past experiences in designing template-based expression languages for encoding guideline-based decision support knowledge [10, 11]. While encoding eligibility criteria using a template-based approach eliminates the need to remember and follow the strict syntax of a string-based language, it still requires a commitment to a patient data model and is still a labor-intensive process that scales poorly to encoding free-text criteria from tens of thousands of existing human studies. For example, using ERGO to encode the criterion “most recent white blood cell count > 4000/mm3” requires the instantiation of templates for comparing an expression with a value, for querying white blood cell count observations, and for selecting the most recent observation in the query result. A more practical method for encoding eligibility criteria is needed to optimize the design, execution, and use of human studies.
III. Research Question and Hypothesis
To address the problems inherent in existing methods for encoding eligibility criteria, we hypothesized that we could create a new method for encoding such criteria that uses an intermediate representation
which is midway in complexity between ASPIRE's domain-specific enumerative criteria and expression languages such as GELLO;
whose encoding requires few specialized skills and can be partially automated; and
which will nevertheless help us satisfy the use cases for eligibility criteria. These use cases include (1) constructing a library of eligibility criteria such that, for a given criterion, we can find more general or more specific criteria; (2) searching for studies whose target population satisfies certain criteria; and (3) screening an EHR database for patients potentially eligible for a study.
The method begins with the free-text eligibility criteria of clinical studies, incrementally classifies the criteria into well-defined statement types, and annotates the criteria using this intermediate representation. The representation allows us to support multiple different use cases of value across the study lifecycle from design to execution to application.
IV. Method Development Process
The methods we report on in this paper were informed by a separate study we conducted to analyze the types and range of complexity in eligibility criteria [12]. That study classified 1000 clinical trial eligibility criteria randomly selected from ClinicalTrials.gov according to their comprehensibility, semantic complexity, and content variation (Table 1). Comprehensibility refers to the degree to which a criterion has some discriminatory power – that is, logic as an inclusion or exclusion criterion – which is readily apparent to the average clinician. In that sample, 927/1000 criteria were judged “comprehensible.” We then classified those comprehensible criteria by their semantic complexity, as either “elementary” or “compound.” Elementary criteria consist of a single noun phrase (e.g. “uncontrolled hypertension”) or its negation (“not pregnant”), or a simple quantitative comparison (e.g., WBC > 5000 cells/mm3). All other criteria were deemed to be compound criteria. Table 1 shows various dimensions of content-based variation, whether semantic (negation, Boolean connectors, arithmetic comparison operators, temporal connectors and comparison operators, if-then statements) or not (requiring clinical judgment, dependent on metadata). Criteria that call for clinical judgment (e.g., “eligible for statin therapy”) or that implicitly reference other metadata about the study (e.g., “No evidence of metastases” where the type of primary carcinoma is specified elsewhere in the study protocol) are considered underspecified. Of the initial 1000 criteria analyzed, a total of 632 criteria were informative in that they were comprehensible and required no clinical judgments or additional metadata.
Table 1.
Semantic Complexity in Eligibility Criteria
| Criteria Type | Criteria Number | Proportion of Informative Criteria | |
|---|---|---|---|
| A | Total Criteria | 1000 | |
| B | Comprehensible Criteria | 932 | |
| C | Elementary Criteria EC | 139 | |
| D | EC Requiring Clinical Judgment | 14 | |
| E | EC Dependent on Study Metadata for Comprehensibility | 0 | |
| F | Compound Criteria: CC (B-C) | 793 | |
| G | CC Requiring Clinical Judgment | 152 | |
| H | CC Dependent on Study Metadata for Comprehensibility | 151 | |
| I | Overlap Between G & H | 22 | |
| J | Informative Criteria (B-D-E-G-H+I) | 637 | |
| K | Informative Elementary Criteria (C-D-E) | 125 | 0.20 |
| L | Informative Criteria With One or More Negations | 132 | 0.21 |
| M | Informative Criteria with One or More Arithmetic Comparison Operators | 112 | 0.18 |
| N | Informative Criteria with One or More Temporal Comparison Operators | 245 | 0.38 |
| O | Informative Criteria with One or More Boolean Connectors | 247 | 0.39 |
| P | Informative Criteria with If-Then Statements | 35 | 0.05 |
Based on the results of this study characterizing the complexity of eligibility criteria, and on our prior experiences encoding eligibility and decision criteria for guidelines and protocols, we observed that
Much of the operative semantics of eligibility criteria are captured in terminological expressions and comparison statements. These “surface” semantics can support the three use cases described in Section III without needing to capture 100% of the meaning that is in the criteria. For example, “heart failure” by itself captures much of the meaning of “severe heart failure.”
Natural language processing (NLP) techniques have been used to extract coded concepts from narrative text with high recall and precision [13, 14].
Computational methods such as description logic subsumption reasoning can organize terminological expressions into classification hierarchies that we can use to index eligibility criteria.
We first used the classification of clinical statements and noun phrases in ERGO to define ERGO Annotations as an intermediate representation that bridges the gap between natural language eligibility criteria and fully specified ERGO criteria. Instead of trying to capture all of the semantics of an eligibility criterion, an ERGO Annotation is essentially a terminological expression used to annotate and index the criterion. The process of encoding free-text criteria into ERGO Annotations entails extracting appropriate noun phrases and semantic connectors. This process can be partially automated using NLP techniques. Along the way, the noun phrases can be mapped to terms in standard terminologies and to UMLS semantic types whenever possible.
We recognized that it would be very hard to automatically recognize noun phrases in the types of complex sentence fragments that eligibility criteria are often written in. Therefore, in this proof-of-concept work, we added preprocessing steps to semi-structure the criteria. We evaluated our NLP encoding process to demonstrate the practicality of a computer-assisted process for annotating eligibility criteria. We also analyzed the extent to which ERGO Annotations could capture the range of complexity and semantics we identified in our eligibility criteria complexity study (Table 1). Finally, using concrete examples, we clarified how ERGO Annotations could be used with a description logic reasoner and a relational database to satisfy the three use cases enumerated in Section III. We defined algorithms for generating Web Ontology Language (OWL) [4] expressions and SQL queries, given specific assumptions about terminologies and patient data models when necessary.
V. Method Description
Our method for annotating eligibility criteria with ERGO Annotations has three components: (1) Classification of eligibility criteria statement types and the definition of ERGO Annotation for each statement type; (2) An encoding process that involves first rewriting eligibility criteria so that they fall into the criteria statement types, using NLP techniques to extract concepts, modifiers, Boolean connectives, other semantic connectors and comparison relationships, and then generating ERGO Annotations for each criterion; (3) For each use case, depending on the application's computational environment, generating computable expressions from ERGO Annotations so that they can be applied in the use cases.
V.A. Criteria Classification and ERGO Annotation
Our team has extensive prior experience encoding decision criteria in guidelines and protocols [11, 15], and analyzing the types and range of complexity in eligibility criteria [12]. Based on this experience, we categorize eligibility criteria into three classes of clinical statements that are mutually exclusive:
Simple statements making a single assertion (e.g., “asthma induced by exercise”). These may involve complex modifiers and constraints (e.g., “Tuberculosis of intrathoracic lymph nodes, confirmed bacteriologically and histologically within 6 months”) or may be expressed at a high level of abstraction (e.g., “treated for LDL-C”).
Comparison statements of the form Noun Phrase comparison operator (e.g., >, < =) Quantity.
Complex statements -- that is, multiple statements joined by Boolean connectives AND, OR, NOT, IMPLIES or semantic connectors (e.g., evidenced by). The Boolean connectives are logically defined keywords, not linguistic terms that may be used imprecisely. Ideally, the semantic connectors should come from some controlled terminology.
For simple statements, we define a valid ERGO Annotation as the most specific noun phrase that can be extracted from the criterion, or noun phrases that are either semantically equivalent to or allowable generalizations of that most specific noun phrase. A noun phrase may be post-coordinated with modifiers. For example, the most specific ERGO Annotation of the criterion “asthma induced by exercise” is the UMLS code C00004096 (asthma) with the attribute induced by and the attribute value C0015259 (exercise). Other valid ERGO Annotations include C0004099 (asthma, exercise-induced), which is semantically equivalent to the most specific noun phrase, or C00004096 (asthma), which is a generalization. We categorically exclude several types of generalizations from being valid ERGO Annotations. One is noun phrases (e.g., “disease” or “syndrome”) that are equivalent to high-level UMLS Semantic Types and therefore are too general to be informative. We also exclude noun phrases that cannot be meaningfully asserted about a person (e.g., a person's “pressure” is not a meaningful generalization of a person's “high blood pressure”). Lastly, to be valid, generalizations must preserve the core meaning of the criterion. We operationalize this constraint by requiring ERGO Annotations to be no more general than the root of a noun phrase. For example, if the criterion is “severe asthma,” valid annotations include “severe asthma” and “asthma,” but not “pulmonary problem.”
If a simple statement's noun phrase is a logical combination of other noun phrases, we define valid ERGO Annotations for each component noun phrase as discussed above, join the annotations with the logical connectives, and then exclude any constructs that are self-contradictory. For example, the ERGO Annotations for “NOT severe asthma” include “NOT asthma” and “NOT severe asthma” because both “asthma” and “severe asthma” are valid ERGO Annotations of “severe asthma.” If the criterion were “asthma and NOT severe asthma,” however, “NOT asthma” cannot be a valid ERGO Annotation for “NOT severe asthma” even if “asthma” is valid at the component level because the overall joined criterion will then be “asthma AND (NOT asthma)” which is an unsatisfiable logical combination.
For comparison statements, a valid ERGO Annotation is the triplet {noun phrase, comparison operator, quantity}, where quantity may be a string when the quantity cannot be expressed as a value and unit. Finally, for complex statements, we define valid ERGO Annotations to be the valid ERGO Annotations of its component simple and comparison statements joined by the relevant Boolean and semantic connectors.
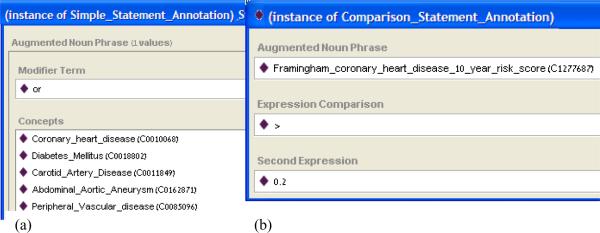
Figure 2 shows examples of ERGO Annotations for a simple statement and a comparison statement in the Protégé tool.
Figure 2.
ERGO Annotations. (a) A simple statement annotation for a noun phrase composed of five terms combined together using OR; (b) a comparison statement annotation for an assessment of Framingham risk score greater than 0.2.
V.B. Annotation Process
The process of encoding ERGO Annotation can be either completely manual or assisted by applying NLP techniques. Our current semi-automated annotation method includes three stages: manual pre-processing and rewriting, application of NLP to the rewritten criteria, and finally the generation of ERGO Annotations from the NLP output.
Free-text eligibility criteria are often sentence fragments whose meaning and complexity range from simple to highly complex. Because this work is only an initial feasibility study, we manually pre-processed and rewrote raw eligibility criteria to yield more tractable criteria for the automated process. Advances in NLP may help to automate these steps in the future. We:
Eliminated criteria or parts thereof that were too vague, were purely explanatory (e.g., parenthetical contents in “Low HDL cholesterol (`good cholesterol')),” or had no discriminating power (e.g., “Men or women”, or criteria describing allowable states, e.g., “Concurrent bisphosphonates allowed”).
Eliminated redundant words (e.g., “patients” in “adult patients”).
Separated “run-on” criteria into stand-alone criteria (e.g., “Stable CAD patients (men & postmenopausal women)” should be “Stable CAD patients” and “men & postmenopausal women”).
Eliminated or rewrote criteria or parts thereof that specified physician discretion for eligibility, resulting in either more or less stringent criteria than the original. E.g., “History of a psychological illness that interferes with the subject's ability to understand study requirements” becomes “History of a psychological illness”; or elimination of the entire exclusion criterion “Certain medications that may interfere with the study”.
After this pre-processing, the criteria were classified manually as Simple, Comparison, or Complex statements, and were then rewritten by:
Decomposing complex statements into simple and comparison statements, by making implied semantics explicit if needed (e.g., “adults, 18–75 years of age” is rewritten as “adults defined by (age >= 18 years AND age <= 75 years).
Regularizing comparison statements so that the variable and its value are on the left and right hand sides respectively (e.g., “at least 18 years old” becomes “age >= 18 years”; “WBC greater than 13,000 or lower than 3,000” becomes “WBC > 13,000 OR WBC < 3,000”)
Making Boolean connections explicit (e.g., “CHD, including patients with other CHD risk factors” is pre-processed to “CHD, including CHD risk factors” and then rewritten as “CHD OR CHD risk factors”).
Expanding acronyms (e.g., “CHD” becomes “coronary heart disease”).
Making diagnoses, conditions, and treatments explicit (e.g., “Severe asthma that is poorly controlled with medication” is rewritten as “Severe asthma AND poorly controlled asthma AND taking asthma medication”).
Rewriting partial lists (e.g., “treatment with drugs raising HDL (e.g., niacin, fibrates)” becomes “treatment with drugs raising HDL OR treatment with niacin OR treatment with fibrates”).
Using implication as a logical connector where needed (e.g., “Women must be post-menopausal or using effective birth control” is rewritten as “women IMPLIES (post-menopausal OR using effective birth control)”).
Rewriting terminological negation as EXCLUDING, to avoid confusion with Boolean negation (e.g., “Any life-threatening disease expected to result in death within 2 years (other than heart disease)” becomes “life-threatening disease EXCLUDING heart disease AND life expectancy<=2 years”).
At this point in the encoding process, the raw eligibility criteria have been pre-processed, rewritten, and structured into Simple, Comparison, and Complex statements whose contents are still in free text. Any number of biomedical concept extraction techniques [13, 14, 16] can then be applied to parse or chunk the free text into parse trees or segments that isolate the noun phrases and comparisons and that annotate the noun phrases with UMLS concept unique identifiers (CUIs) when such annotations are available. To convert this material into ERGO Annotations, we defined heuristic algorithms that incorporate both linguistic and clinical heuristics (details can be found online in Appendix 1). An example of a linguistically based heuristic is that, in English, the root noun phrase is usually the right-most noun phrase that is modified by adjectival phrases occurring to its left. An example of a clinically based heuristic is the following. If there is more than one noun phrase in a criterion, the algorithm first checks to see if they can be connected by a terminological AND, OR, or Excluding. If not, the algorithm gives preference to noun phrases that have semantic types Disease or Syndrome, Clinical Drug, Procedure, and Finding in that order. If the noun phrases are not of the above semantic types or if the noun phrases cannot be mapped to UMLS CUIs, we arbitrarily select the left-most noun phrase as the ERGO Annotation for the criterion. The overall result of this pre-processing, rewriting, NLP processing, and heuristic noun phrase identification process can now be instantiated as ERGO Annotations either automatically or by a human coder.
V.C. Use of ERGO Annotations
Once the encoding process produces ERGO annotations for eligibility criteria, they can be applied to the use cases described in Section III. The first use case is constructing a library of eligibility criteria such that a researcher can search for any given criterion, and can find more general or more specific criteria.
1) Library of Eligibility Criteria
An information resource indexes its entries to facilitate search. A library of eligibility criteria can use words in the criteria (with appropriate filtering of stop words) or terms derived from automated term-recognition for indexing purposes. If the recognized terms come from a reference terminology, searching for criteria in the library can make use of hierarchical relationships in the terminology. However, the structure of ERGO Annotations allows us to create a classification hierarchy of eligibility criteria that is much more precise. We formulate ERGO Annotations as fully defined description logic (DL) concepts that extend an existing reference terminology (e.g., SNOMED [17]), from which a DL reasoner will be able to automatically construct the needed hierarchies for indexing eligibility criteria. Individual criteria and their ERGO Annotations will therefore be placed in the proper hierarchical relationship to other more general or more specialized criteria in the library.
To construct the eligibility criteria classification hierarchy as a hierarchy of DL expressions, we need to pick a preferred ERGO Annotation to index each criterion. For simple or comparison statements, we can simply take the intersection of all valid ERGO Annotations as the preferred one (e.g., intersecting “Severe anemia” and “Anemia” yields “Severe anemia”, and intersecting “Severe anemia” and “Chronic anemia” yields (“Severe anemia and Chronic anemia”). In this case, the preferred annotation is the most specific annotation supported by the criterion.
For complex statements, the preferred ERGO Annotation is constructed recursively by applying Boolean and semantic connectors to the preferred ERGO Annotations of the component statements. For example, the preferred ERGO Annotation of the criterion “Elevated blood pressure defined by systolic blood pressure > 140 mm Hg and diastolic blood pressure > 80 mm Hg” is “Elevated blood pressure” defined_by {systolic blood pressure, `>', 140 mm Hg} AND {diastolic blood pressure, `>', 80 mm Hg}.
Using Web Ontology Language (OWL) [4] as the DL language, we show how to construct DL expressions from ERGO Annotations. For a simple statement, an ERGO Annotation is essentially a noun phrase that may have certain modifiers. A modifier may have an explicit attribute (e.g., “severity”) and an attribute value (e.g., “severe”) or it may be an adjective with no explicit attribute, in which case we use “modified_by” as the default attribute. A noun phrase N with modifier M can be written as the OWL expression (using the Manchester syntax) (N and (modifier some M)), where modifier is an OWL object property. A comparison statement can be defined in OWL 2.0 as restrictions on a noun phrase and a quantity. “White blood cell count > 5000 /mm3,” for example, can be written as (WhiteBloodCellCount and has_value some (Physical_quantity and has_unit value `/mm3' and has_realvalue some real[>5000]), where WhiteBloodCellCount is a class representing white blood cell count measurement and Physical_quantity is a class that has unit and real number components. For complex statements, we treat the Boolean connectors as OWL intersection, union, and set complement operations and the semantic connector defined_by as an OWL equivalence relation. Other semantic connectors, such as evidenced_by or caused_by, become OWL object properties so that, for example, “coronary heart disease evidenced by angiography” becomes the OWL restriction “coronary_heart_disease AND (evidenced_by some angiography).” This example shows that if, for example, the all eligibility criteria in ClinicalTrials.gov were annotated with preferred ERGO Annotations, the criteria could be searched for hierarchically related criteria, to facilitate standardization and reuse of criteria.
2) Searching for Studies Enrolling Specific Patient Populations
The second use case—searching for studies whose target population satisfies certain criteria—can also be formulated as a classification problem. For any study, the conjunction of all inclusion criteria and the negations of all exclusion criteria define the target population. For each criterion, we can find the preferred ERGO Annotation and formulate the associated OWL expression as described above. The conjunction of the OWL expressions associated with the inclusion criteria and the negation of the exclusion criteria approximates the characteristics of the study's target population. A query for studies based on certain criteria (e.g., all studies that include female subjects who are HIV positive and have viral load below a certain threshold) can be resolved by finding all studies whose associated OWL expressions are more specific than the query for all of the inclusion and negated exclusion criteria.
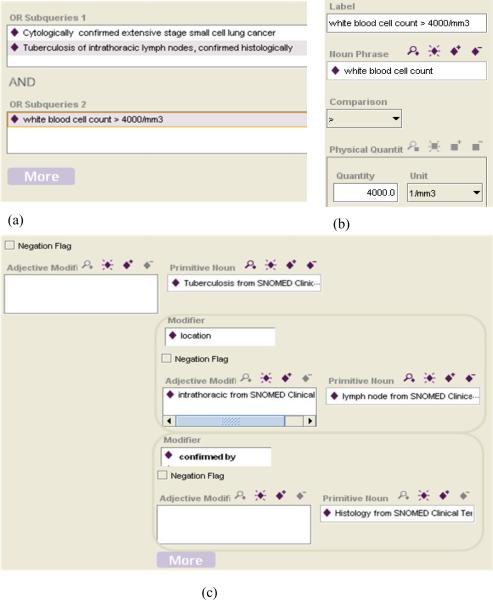
To perform a search for studies based on their eligibility criteria, a user will construct query expressions consisting of a Boolean combination of noun phrases and comparisons. Figure 3 (a)–(c) shows a possible interface that allows a user to construct such queries without having any knowledge of description logic or OWL.
Figure 3.
(a) A possible user interface for specifying a conjunction of disjunctive queries. Clicking on the “More” button refreshes the screen and creates a new set of OR subqueries. The  button allows the creation of a new comparison ERGO Annotation as shown in Figure 3 (b) or a new noun phrase ERGO Annotation as shown in Figure 3 (c).
button allows the creation of a new comparison ERGO Annotation as shown in Figure 3 (b) or a new noun phrase ERGO Annotation as shown in Figure 3 (c).
Figure 3 (c). A possible user-interface for specifying a noun phrase such as “Tuberculosis of intrathoracic lymph nodes, confirmed histologically,” initially consisting of a primitive noun and its adjectival modifiers. By clicking on the “More” button, a user can specify additional modifier attributes and noun phrases. Tool support can easily facilitate the task of searching terminological references such as primitive nouns and adjectival modifiers.
3) Screening for Potentially Eligible Participants
We could apply the same approach to the third use case—screening an EHR database for potentially eligible patients—by matching the OWL expressions as in the second use case to a description logic characterization of the state of a patient. This approach, however, is unappealing for several reasons. First, when the exclusion criteria are formulated as negated expressions, it is difficult to prove, with the open-world assumption of OWL, that a patient satisfies those criteria without first specifying explicit closure axioms. Second, and more important, it is unrealistic to expect EHR data to be widely available as OWL expressions.
In this use case, translating ERGO annotations into SQL queries for use in relational database technology is preferred over using OWL. Each institution will customize such translation of standardized ERGO Annotations according to the requirements of their EHR. As we demonstrate in the following example, such translation can be specified using mapping tables and generic mapping rules. Unlike a variable in a Medical Logic Module that is implemented using an idiosyncratic curly brace, ERGO Annotations can be translated systematically. The expectation is that translation can be automated so that, as new studies are added to the library, their eligibility criteria can receive standard annotation once, and multiple institutions can translate them to use their institution-specific EHR data. Informaticians will be involved in designing and implementing the translation mechanism for the institution as a whole. End users such as study coordinators should be able to download the ERGO Annotations and apply the automated translation to generate institution-specific queries.
To illustrate this through an example, we make certain assumptions to duplicate the features of a real example. First, we assume an implementation of SQL and a certain patient data model. For illustrative purposes, we chose Microsoft Access and a simple data model for patient data keyed to the patient ID:
DemographicsData table containing patient demographic data (e.g., date of birth, gender) whose key is the patient ID;
ProblemList table containing the problem name (e.g., disease name), a code for the problem, and its start and end times;
Medication table containing drug names and codes, the prescribed dose and frequency for each drug, and prescription start and end times;
LaboratoryTestResult table containing test names and codes, the values and units of test results, and the test dates;
Assessment table containing computed or assessed results rather than laboratory test results (e.g., the Framingham coronary heart disease score), their codes, and the assessment dates.
We must also make assumptions regarding terminology. We assumed that problems are specified as ICD-9 codes. In order to map ICD-9 codes and drug codes to the UMLS terms used in the ERGO Annotations, we created a TerminologyMapping table which, for each UMLS code, specified the possible EHR ICD-9 and drug codes corresponding to a patient's problems and medications.
A third assumption concerns the assignment of terms used in the eligibility criteria to the appropriate tables in the patient data model. We employed several rules for this task. For example, criteria terms whose UMLS semantic type was Disease or Syndrome were mapped to the ProblemList table, terms whose semantic type was Pharmacologic Substance or Clinical Drug were mapped to the Medication table, and terms whose semantic type was Laboratory or Test Result were mapped to the LaboratoryTestResult table. For specific noun phrases, such as age, gender, or sex, we used special rules that mapped them to the DemographicData table. For example, “age” had to be treated in a special way, because the information in the patient data model was date of birth, which had to be converted to age using SQL functions. We did not develop rules for mapping terms into the Assessment table.
We illustrate the process of creating SQL queries from the ERGO Annotations of a simple statement. For simple statements, the ERGO Annotation consists of a noun phrase built up from a primitive noun phrase, its modifiers, and/or the conjunction, disjunction, and complement of other noun phrases. A primitive noun phrase has the properties preferred_name, code_system, and code. For primitive noun phrases whose semantic types are Disease or Syndrome, Pharmacologic Substance, or Clinical Drug, we created an SQL query of the form:
SELECT * FROM ProblemList, TerminologyMapping
WHERE problem_code=EHR_code and and UMLS_code=code);
where the TerminologyMapping table was used to map eligibility criteria terms to patient data model terms that appear in the ProblemList table.
For noun phrases made up of the conjunction and/or disjunction of other noun phrases (AndOr_Noun_Phrases) whose semantic types are Disease or Syndrome, Pharmacologic Substance, or Clinical Drug, the WHERE part of the query repeats for all concepts and modifier terms of the AndOr_Noun_Phrases. (And or Or) is translated into the corresponding SQL operator AND or OR. This resulted in queries of the form:
SELECT * FROM ProblemList, TerminologyMapping
WHERE problem_code=EHR_code and UMLS_code=concept1.code)…
(problem_code=EHR_code and UMLS_code =conceptn.code);
The online Appendix 2 contains details for generating SQL queries from other types of ERGO Annotations.
VI. Validation through Examples
We tested the feasibility and utility of capturing eligibility criteria in ERGO Annotation by rewriting a set of eligibility criteria and applying alternative NLP methods to generate ERGO Annotations. In the following sections, we illustrate this process. We then demonstrate the possibility of using ERGO Annotations for the use cases described in Section III.
VI.A. Examples of Annotation Process
1. Preprocessing the eligibility criteria
We entered the eligibility criteria from the selected trials (Section VIIB) in an Excel spreadsheet and used it to record all applied transformations: pre-processing, rewriting and breaking down of complex statements into their constituent parts. To demonstrate this, we use the trial with ClinicalTrials.gov id NCT00655473, which has inclusion criteria such as “adult patients, 18–75 years of age” and “CHD, including patients with other CHD risk factors,” and exclusion criteria such as “previous exposure to any CETP inhibitor or vaccine” and “poorly controlled diabetes.”
We first pre-processed and classified the criteria into Simple, Comparison, and Complex according to the steps described in Section V.B. Table 2 shows the results of the pre-processing and classification. Table 3 shows the results of further rewriting.6
Table 2.
Selected eligibility criteria after pre-processing and categorization as described in Section Error! Reference source not found.. “I” denotes inclusion criteria and “E” denotes exclusion criteria
| I/E | Original text | After pre-processing | Classification |
|---|---|---|---|
| I | adult patients, 18–75 years of age | adult, 18–75 years of age | complex |
| I | CHD, including patients with other CHD risk factors | CHD, including CHD risk factors | complex |
| E | previous exposure to any CETP inhibitor or vaccine | previous exposure to any CETP inhibitor or vaccine [no change] | complex |
| E | poorly controlled diabetes | poorly controlled diabetes [no change] | simple |
Table 3.
Selected eligibility criteria after re-writing and splitting into atomic elements
| After re-writing | Atom1 | Atom2 | Atom3 |
|---|---|---|---|
| adult DEFINED_BY (age >= 18 years AND age <= 75 years) | adult | age >= 18 years | age <= 75 years |
| coronary heart disease OR coronary heart disease risk factors | coronary heart disease | coronary heart disease risk factors | |
| previous exposure to any cholesteryl-ester transfer protein inhibitor OR previous exposure to any cholesteryl-ester transfer protein vaccine | previous exposure to any cholesteryl-ester transfer protein inhibitor | previous exposure to any cholesteryl-ester transfer protein vaccine | |
| poorly controlled diabetes [no change] | poorly controlled diabetes |
2. Automated generation of ERGO Annotations
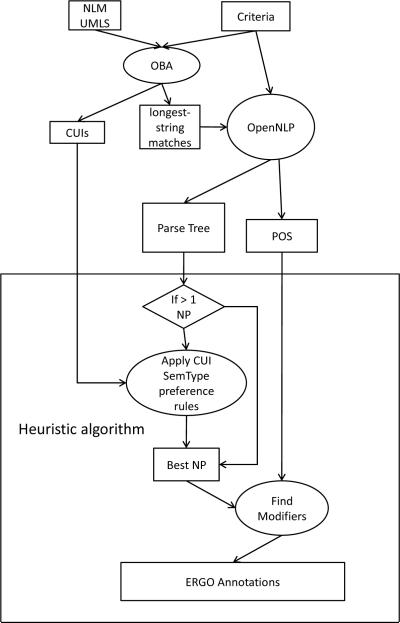
Multiple approaches to applying NLP to the atomic eligibility criteria are possible. We tried three different NLP parsers7 8 9, and we used the Open-Biomedical Annotator (OBA) of the National Center for Biomedical Informatics [8] and NLM's MMTx to obtain two sets of results. We describe below the NLP procedures we used in this feasibility study. They serve as examples of how to use NLP techniques to generate ERGO Annotations.
To obtain the first set of results, we took the following steps:
Apply OBA to the preprocessed criteria to get the longest coded string and their UMLS CUIs and semantic types.
Words from the phrases corresponding to CUIs are formed into single compound words (e.g., if “postauricular scar” is a phrase that correspond to some CUI, we will use “postauricularscar”).
Run preprocessed criteria through the OpenNLP Parser to find noun phrases in parse trees.
Use the heuristic algorithm described in Section V.B to extract noun phrases and their modifiers for simple criteria and noun phrases, comparison operators, and quantities for comparison criteria, discarding everything else.
The steps are illustrated in Figure 4.
Figure 4.
Steps in automated generation of ERGO Annotations.
To obtain the second set of results, we applied the following procedure:
Apply MMTx to the preprocessed criteria to get the best phrase chunking and CUI matches.
Apply NpParser (part of the MMTx toolkit) to obtain the parts of speech of the words in the criterion.
Use the same heuristic algorithm to generate the candidate ERGO Annotations.
The outputs of the semi-automated annotation process are criteria with their acquired ERGO Annotations. For example, OBA recognized “heart failure” in “Severe heart failure” as a UMLS term, and the OpenNLP Parser generated the parse tree [NP Severe/JJ heart-failure/NN]. Our heuristic algorithm, in this case, used the noun (NN) as the root noun phrase and the adjective (JJ) as the modifier in the acquired ERGO Annotation for the criterion.
VI.B. Examples of Use
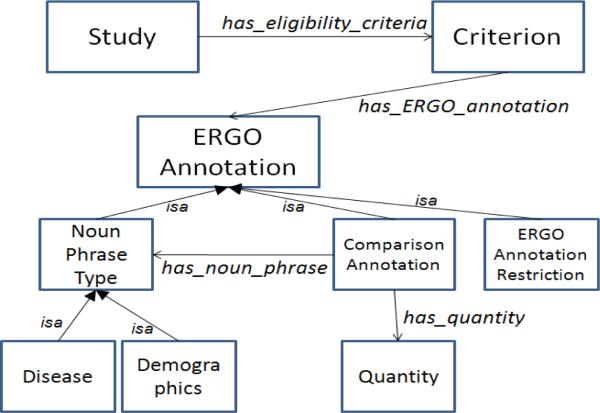
To demonstrate applicability of OWL-based ERGO Annotations to our use cases, we defined an OWL ontology (Figure 5) that consists of a Study class with a property called has_eligibility_criteria that is specialized into has_exclusion_criteria and has_inclusion_criteria subproperties. The Criterion class has a property called has_ERGO_annotation. ERGO Annotation may be a Noun_phrase_type or a Comparison_Annotation. Noun_phrase_type has subclasses: Demographic, Disease, and other UMLS semantic types. Comparison_Annotation has object properties has_noun_ phrase and has_quantity, and the Quantity class has a float value and unit. When the quantity part of a comparison annotation is a string, it plays no part in the OWL formalization of ERGO Annotation. ERGO Annotations for complex statements are written as OWL expressions involving Noun Phrase Type and Comparison Annotation.
Figure 5.
Predefined OWL ontology to illustrate how ERGO Annotations may be used to classify criteria and to search for.
We illustrate the application of ERGO Annotations to our uses cases by annotating three studies from ClinicalTrials.gov: NCT00655538, NCT00655473, and NCT00799903. The first two have the inclusion criterion “adult patients, 18–75 years of age” and the exclusion criterion “poorly controlled diabetes.” The third has the inclusion criterion “age >= 18 years.”
For “poorly controlled diabetes,” the valid ERGO Annotations are a) C0011849 (Diabetes_Mellitus), b) C0743131 (Uncontrolled_Diabetes), and c) C0011849 (Diabetes_Mellitus) modified by C0205318 (Uncontrolled). We can write a necessary and sufficient definition of Uncontrolled_Diabetes as Diabetes_Mellitus and (some attribute Uncontrolled). The annotation for “age > 18” is formalized as a Comparison Annotation “has_noun_phrase some Age and has_quantity some (Quantity and has_value some float[> 18] and has_unit value year), where year is an individual of the unit class.
Examining our first use case, investigators interested in using standard eligibility criteria for their study protocols may search for “Diabetes Mellitus” and retrieve all three ERGO Annotations and their associated eligibility criteria. They will see that “Diabetes Mellitus” is more general than the others.
A query such as (Study and exclusion_criteria some (Criterion and has_ergo_annotation some Diabetes_Mellitus)) will return studies with an exclusion criterion that includes any subclass of Diabetes_Mellitus (e.g, NCT00655538, NCT00655473). A query Study and inclusion_criteria some (Criterion and has_ergo_annotation some (Comparison_Annotation and has_ quantity some (has_value some float [> 16.0])and has_noun_phrase some Age))will return studies with inclusion criteria more restrictive than age > 16 (including all three studies discussed here).
Use case 3 involves querying a database of patient information to screen for potentially eligible patients. We exemplify our approach of translating an ERGO Annotation into an SQL query using the criterion “CHD OR CHD risk equivalent.” In order to match patient data that correspond to “CHD risk equivalent,” this criterion was first rewritten as two statements: “CHD or Diabetes mellitus or Carotid artery disease or Peripheral vascular disease or Abdominal aortic aneurysm” and “Framingham coronary heart disease 10 year risk score > 0.2.” This rewrite illustrates the fact that high-level terms in eligibility criteria (e.g., CHD risk equivalent) often need to be expressed in more concrete terms in order to match them against patient data. The corresponding ERGO annotations are shown in Figure 2.
Following the methods for converting an ERGO Annotation for a simple statement with an AndOr_Noun_Phrase into an SQL query, we obtain the following query corresponding to Figure 2(a):
SELECT * FROM ProblemList, TerminologyMapping
WHERE (problem_code=EHR_code and UMLS_code =“C0010068”) or
(problem_code=EHR_code and UMLS_code =“C0162871”) or
(problem_code= EHR_code and UMLS_code=“C0011849”) or
(problem_code= EHR_code and UMLS_code=“C0018802”) or
(problem_code= EHR_code and UMLS_code =“C0085096”);
VII. Preliminary Evaluations
VII.A. Ability of ERGO Annotations to capture the semantics of common eligibility criteria
We assessed the extent to which ERGO Annotations can capture the semantics of real-life eligibility criteria. For this evaluation, we used the study on eligibility criteria complexity described above [12], and specifically the 637 criteria in this study that were informative. Of these, 125 (20%) are elementary criteria whose semantics can be fully captured by ERGO Annotation. ERGO Annotation is unable to fully express temporal connectors and temporal comparison operators (present in 38% of the informative criteria in this sample), but can nevertheless capture some useful information. For example, annotating the criterion “chemotherapy treatment after surgery” with a conjunction of “chemotherapy” and “surgery” still captures some of the requirements specified in the criterion. Instead of omitting criteria that include temporal connectors and comparison operators, annotating them with ERGO Annotations that partially capture their semantics will help increase the overall accuracy of applying the criteria in the use cases. This analysis gives us a rough lower and upper bounds (20% and 62%) on the proportion of informative criteria whose semantics ERGO Annotation can fully capture. Fortunately, as we will describe in the Discussion, the utility of ERGO Annotations does not depend on the annotations expressing the full meaning of the eligibility criteria. Using ERGO Annotations that capture partial meaning of the criteria will affect the recall and precision of the query results, but for many purposes (e.g., screening a large database to find subjects who are potentially eligible for a study or identifying studies focusing on particular diseases), the queries may be sufficiently generalized so that ERGO Annotations could be functionally adequate.
ERGO Annotations could potentially fully specify the meaning of all non-temporal informative eligibility criteria (61%, including the 25% that are elementary criteria). Therefore, ERGO can fully specify 25–61% of the informative criteria and could partially represent the meaning of the rest.
VII.B. Performance of automated encoding process
For our initial feasibility study, we evaluated our NLP tools by comparing their noun phrase outputs against the manually derived ERGO Annotation for each test criterion. The manual standard is the most specific noun phrase as discussed in Section V.A. An exact match occurs when the noun phrase output is a valid ERGO annotation exactly matching or semantically equivalent to the manual standard. A match is when the output is a valid ERGO Annotation but not semantically equivalent to the manual standard (e.g., the output is “Asthma” when the manual standard is “Severe Asthma”). In this case, for simple statements, a match is a generalization of the manual standard, and is at least as specific as the root noun in the statement. Similarly, a match for a complex statement is always a generalization of the manual standard. A non-match is when the output is not a valid ERGO Annotation of the manual standard. For a comparison criterion, an exact match requires that the acquired annotation include not only the maximally specific noun phrase but also the comparison operator and the quantity components.
We constructed our test set of criteria by searching ClinicalTrials.gov with “heart disease” on January 12, 2009 and selecting the second, sixth, eighth, and tenth open interventional studies. (Trial #2 and #4 had several criteria in common, so we excluded #4.) The four trials yielded 60 distinct criteria, of which we removed six because they were too vague or non-discriminating. After rewriting, the criteria were decomposed into unique 100 simple and 13 comparison statements, yielding 113 atomic statements.
We manually annotated the atomic statements (Table 4) to generate the reference set of ERGO Annotations. We then applied our automated method for generate ERGO Annotations to the atomic statements and manually compared the output to the reference standard. Using the first procedure described in Section VI.A, we obtained 54 (47.8%) exact matches, 22 (19.5%) matches, and 37 (32.7%) non-matches. Using the second procedure described in Section VI.A, we obtained 68 (60%) exact matches, 6 (5%) matches, and 39 (35%) non-matches. Most of the new exact matches were the result of special code written to improve the recognition of comparison criteria. MMTx and the combination of OBA/OpenNLP gave us roughly similar results.
Table 4.
Manually annotating the criteria with the maximally specific ERGO Annotations.
| Atom | ERGO Annotations |
|---|---|
| adult | adult [NP] |
| age >= 18 years | age [NP], >=, 18 years |
| age <= 75 years | age [NP], <=, 75 years |
| coronary heart disease | coronary [modifier] heart disease [NP] |
| coronary heart disease risk factors | coronary [modifier] heart disease [modifier] risk factors [NP] |
| previous exposure to any cholesteryl-ester transfer protein inhibitor | previous [modifier] exposure to cholesteryl-ester transfer protein inhibitor [NP] |
| previous exposure to any cholesteryl-ester transfer protein vaccine | previous [modifier] exposure to cholesteryl-ester transfer protein vaccine [NP] |
| poorly controlled diabetes | poorly controlled [modifier] diabetes [NP] |
VIII. Discussion
ERGO Annotation changes the problem of representing eligibility criteria from formal encoding in some expression language to classifying and decomposing criteria and identifying noun phrases in simple and comparison criteria. Our prior study of 1000 randomly selected eligibility criteria and the ERGO expression language inform the categorization and decomposition of criteria into simple and comparison criteria connected by Boolean and other semantic connectors. ERGO Annotation aims to capture the basic semantics of criteria by identifying linguistic noun phrases and formalizing them as terminological expressions, in place of detailed modeling based on some information model.
Our attempts to use OWL and a relational database to solve the three use cases described above clarified the roles that each technology can play in each use case. We formulated the first two use cases—searching for an eligibility criterion in a classification hierarchy, and searching for studies whose eligibility criteria satisfy a conjunction of criteria—as classification problems that are best solved using description logic technology, as we illustrated using OWL queries. We showed an easily implementable user interface that a clinician with some training in the use of terminologies can potentially utilize to formulate queries to search for studies that satisfy fairly complex criteria.
The third use case—screening a database of patient information to find potentially eligible subjects—is best done with a conventional technology like relational databases. We illustrated how ERGO Annotations can be turned into SQL queries, after making certain assumptions about the patient information model and terminology. As in guideline-based decision support systems (DSS), there is no way to avoid this step if your task is to match abstract criteria to concrete patient data. This type of matching has been done before in DSS that derive their data from EHRs. Tools are available to support the mapping process [18] [19]. For example, KDOM [18] can map terms in criteria to terms in an EHR by going through an intermediate global-as-view schema such as the HL7 RIM virtual medical record [20] or the simple data model that we used here to generate SQL queries, like those illustrated in Section VI.B.
For a given criterion classified as “simple” in our categorization, the ERGO Annotation is always more generic than the criterion's intended meaning, simply because there are aspects of criteria, such as temporal ordering and assessment and measurement procedures, that are not captured in ERGO Annotations. For example, a criterion such as “Asthma within last 6 months” will be annotated with “Asthma” (UMLS CUI C0004096). If the criterion is an inclusion criterion, the use of formalized ERGO Annotation (as an OWL or SQL expression) to resolve queries for studies or to screen eligible patients introduces the possibility of false positives and thus decreases the precision of ERGO Annotations. On the other hand, when a concept is negated or when a criterion is used as an exclusion criterion, ERGO Annotations become too restrictive and have the effect of introducing false negatives. In this case the use of ERGO Annotation may decrease the recall of queries. We attempt to minimize such problems by defining the notion of a preferred ERGO Annotation for a criterion, one that minimizes both false positives (by taking the most specific annotation for a positive concept or inclusion criterion) and false negatives (by taking the least specific annotation for a negated or exclusion criterion).
If we ignore the modifiers and semantic connectors, ERGO Annotations collapse into Boolean combinations of terminology codes that are more generic than ERGO Annotations with modifiers. For the use case of screening subjects for studies, using such terminology codes for inclusion criteria has the effect of introducing additional false positives. For negated concepts and exclusion criteria, using only terminology codes introduces additional false negatives. For the use case of querying for studies, the terminological expressions that index studies and allow formulation of queries are Boolean combinations of terminology codes. A simpler reasoner than a full-strength description-logic reasoner can resolve such queries. If we take the automated matches and exact matches from our feasibility study as the ERGO Annotations to use in indexing eligibility criteria and studies, we obtain a system that is intermediate between the full ERGO Annotation system and the simplified system described here.
The advantages of ERGO Annotation over formal expression languages like Arden Syntax [6] or GELLO [7] are twofold: the scalability of its annotation process, and the possibility of reasoning about eligibility criteria. Annotating eligibility criteria is much more scalable because no knowledge of arcane syntax is required, and annotators can be assisted by automated tools to detect noun phrases and recognize terms from standard vocabularies. To explain, any formalization of natural language clinical text into a computable representation necessarily involves decomposition of complex linguistic phrases into stylized expressions. This decomposition requires some clinical and linguistic knowledge as well as an understanding of formal languages and terminologies. The type of preprocessing described in Section V.B is needed regardless of the chosen target representation, be it GELLO, Arden Syntax, or ERGO Annotation. The training required for using ERGO Annotation is less than for using other expression languages because an ERGO annotator does not have to know the details of patient data representation and the syntax of the expression language. Instead, she only needs to focus on the principal noun phrases in the eligibility criteria, to find appropriate terminological codes for the concepts and relationships, and to construct annotations for complex statements using the rules described in this paper. For the criterion “Presence of azotemia within the last 3 months,” instead of writing expressions shown in Figure 1, she only has to annotate the criterion with the terminology code for “azotemia.”
ERGO Annotation does trade expressiveness for the scalability of the annotation process. For example, at current time, ERGO Annotations do not capture temporal requirements that an eligibility criterion may impose on a medical condition or therapy (e.g., “Presence of azotemia within the last 3 months”). On the other hand, criteria encoded in Arden Syntax or GELLO are of no use for our first two use cases. Searching for eligibility criteria or for studies that target a patient cohort defined by a set of criteria requires (1) a way to index and classify the eligibility criteria, and (2) a reasoner that can resolve queries by checking subsumption relationships between the query expression and the eligibility expressions that index the studies. Expression languages like Arden Syntax and GELLO are not designed for these tasks.
ERGO Annotations expressed as OWL expressions can complement ASPIRE's sets of standardized eligibility codes by giving them an ontological foundation. Instead of needing to enumerate standard codes such as “Breast Cancer Estrogen-Receptor Status (Positive/ Negative/Unknown)”, we can associate criteria codes with their corresponding ERGO Annotations organized in a classification hierarchy, making semantic relationships among the codes explicit.
At a basic level, our work may contribute to reducing the variability of eligibility criteria texts. Recall that a full 36% of the criteria in our sample of 1000 from ClinicalTrials.gov were incomprehensible or “underspecified.” Variant criteria such as “treated appropriately for dyslipidemia“ and “Current treatment with statin therapy unless the study doctor determines statins are not appropriate for the subject“ in the context of heart failure trials may in fact target similar subjects. The rewrite rules developed for this project can help study authors write eligibility criteria more clearly and uniformly. A standard library of eligibility criteria can reduce unnecessary variability in the target populations of studies, thus making study results more comparable.
IX. Limitations
One limitation of our method is that we have not modeled the temporal aspects of eligibility criteria. A large proportion of the eligibility criteria surveyed in our study have some kind of temporal constraint or comparison (Table 1). Our current work will extend the ERGO Annotation formalism to include temporal comparisons of the form (Noun_phrase1 temporal comparison operator Noun_phrase2). We will interpret Noun_phrase1 and Noun_phrase2 as representing sets of events that have associated time stamps. A temporal comparison like (radiation therapy before chemotherapy) means the presence of some radiation therapy whose associated time stamp is before that of some chemotherapy a patient has received. We are currently exploring the logical implications of this extension. It does not capture all types of temporal constraints and comparisons we see in eligibility criteria, but is a first step in a rich research direction.
Compared to the state of art in identifying maximal noun phrases in radiology reports, where the recall rate can be 82% or higher [11], our best recall rate (counting both exact matches and matches) of 67% shows relatively low recall at generating correct ERGO Annotations. This is not surprising, given the preliminary NLP techniques used in this early work whose objective was to demonstrate the feasibility of automating this part of the annotation process. Using Me-LEE [11] without any initial training, Borlawsky and Payne obtained similar results (14% of the criteria completely and correctly parsed and 62% partial parses) [21]. Much can be done to improve the recognition rate of the tools. For example, the statistical NLP parsers can be trained on eligibility text. Furthermore, we will evaluate the use of advanced biomedical NLP tools such as ChartIndex [12].
The evaluation described in Section VII.B is limited by the lack of a true gold standard for ERGO Annotations of eligibility criteria. Ideally, the noun phrases, modifiers, and semantic connectors that constitute ERGO Annotations should come from controlled terminologies that have been harmonized to provide consistent semantics. Individuals not involved in the development of ERGO Annotations should establish the reference annotations, and criteria for matches should be established prior to the development of automated methods. The reference ERGO Annotations manually created for the feasibility study do not require terms from controlled vocabularies to be found for all primitive noun phrases. The semantic connectors (e.g., “evidenced by” and “caused by”) that link clinical statements and terminological modifiers that refine noun phrases represent relationships that should be standardized if we are to have a rich compositional language for expressing clinical concepts and statements. Several groups have adopted their own sets of standard relationships. HL7 uses a collection of Act Relationships (e.g., “has component” and “has reason”) that plays exactly the same role as our semantic connectors [22]. SNOMED CT has a standard set of qualifiers, such as severity and finding site, for post-coordinating terms [23]. The Open Biomedical Ontology Foundry has proposed a Relations Ontology for biomedicine [24]. None of these efforts, however, are coordinated or mature enough for the purpose of encoding eligibility criteria in a standard way.
In our preliminary evaluation, we mapped recognized entities into UMLS CUIs because of UMLS's comprehensive coverage of biomedical terms and because existing NLP tools use UMLS as the default source of terminology. Our use cases, however, require us to evaluate subsumption relationships among different terms and possibly compositional terms. UMLS CUIs themselves are not organized along subsumption relationships. The “broader” and “narrower” relations only give broader and narrower concepts in specific terminologies. To properly manage subsumption relationships among ERGO Annotations, ideally the terms and relationships in eligibility criteria should be mapped to reference terminologies, like SNOMED CT, that guarantee appropriate subsumption classification of terms and that provide the mechanism to post-coordinate new terms from primitive terms and their modifiers.
Ultimately, ERGO Annotations will be judged by their utility in satisfying use cases that clinicians and biomedical investigators find important. A formal evaluation of ERGO Annotation requires (1) that we annotate a large number of studies with ERGO Annotations, (2) that we demonstrate the ability of a clinician or investigator to query for studies with better recall and precision than using existing methods, and (3) that we show the use of ERGO Annotation facilitates screening potentially eligible study subjects. The scope of our project does not permit us to perform such an evaluation. Nevertheless, if the eligibility criteria of a large collection of studies can be annotated cheaply and consistently using manual or automated methods, the computational tractability of ERGO Annotations for the three use cases discussed in the paper (Section V.C) will prove this approach worthy.
X. Conclusion
We defined a formal representation called ERGO Annotation for annotating eligibility criteria and demonstrated the capture of eligibility semantics that supports queries of sufficient richness to enable three important use cases in clinical research: classifying an eligibility criterion in a library of criteria; finding studies that use particular criteria; and identifying patients who are potentially eligible for a study. The ERGO Annotation representation requires no knowledge of complex expression languages. We have tested the feasibility of using a semi-automated approach for transforming text-based eligibility criteria into the formal representation. ERGO Annotation and our semi-automated approach provide an expressive ontological and methodological foundation for computable representations of eligibility criteria.
Supplementary Material
Acknowledgements
This work has been supported in part by NLM grant R01-LM-06780.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Search performed on June 27, 2010.
All eligibility criteria of trial NCT00655473 and tables showing how they were pre-processed and rewritten area available as part of the online appendix.
OpenNLP: http://opennlp.sourceforge.net/
Stanford Parser: http://nlp.stanford.edu/software/lex-parser.shtml
Apple Pie Parser: http://nlp.cs.nyu.edu/app/
References
- [1].Campbell MK, Snowdon C, Francis D, Elbourne D, McDonald AM, Knight R, et al. Recruitment to randomised trials: strategies for trial enrollment and participation study. The STEPS study. Health Technol Assess. 2007 Nov;11(48):iii, ix–105. doi: 10.3310/hta11480. [DOI] [PubMed] [Google Scholar]
- [2].Boissel JP, Amsallem E, Cucherat M, Nony P, Haugh MC. Bridging the gap between therapeutic research results and physician prescribing decisions: knowledge transfer, a prerequisite to knowledge translation. Eur J Clin Pharmacol. 2004 Nov;60(9):609–16. doi: 10.1007/s00228-004-0816-2. [DOI] [PubMed] [Google Scholar]
- [3].Weng C, Tu SW, Sim I, Richesson R. Formal Representations of Eligibility Criteria: A Literature Review. JBI. 2009 doi: 10.1016/j.jbi.2009.12.004. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].OWL Working Group OWL 2 Web Ontology Language: Structural Specification and Functional-Style Syntax. 2009 [Google Scholar]
- [5].Niland J. ASPIRE: Agreement on Standardized Protocol Inclusion Requirements for Eligibility. 2007 [cited 2008 August 20]; Available from: http://hsspcohort.wikispaces.com/space/showimage/ASPIRE+CDISC+Intrachange+July+10+2007+Final.ppt.
- [6].Hripcsak G, Clayton PD, Pryor TA, Haug P, Wigertz OB, Van der lei J. Proc Annu Symp Comput Appl Med Care. Washington, DC: 1990. The Arden Syntax for Medical Logic Modules; pp. 200–4. [Google Scholar]
- [7].Sordo M, Boxwala A, Ogunyemi O, Greenes R. Description and Status Update on GELLO: a Proposed Standardized Object-oriented Expression Language for Clinical Decision Support. Medinfo. 2004:164–8. [PubMed] [Google Scholar]
- [8].Pryor T, Hripcsak G. Sharing MLM's: An Experiment between Columbia-Presbyterian and LDS Hospital. Proc Annu Symp Comput Appl Med Care. 1993:399–403. [PMC free article] [PubMed] [Google Scholar]
- [9].Tu SW, Peleg M, Carini S, Rubin D, Sim I. ERGO: A Template-Based Expression Language for Encoding Eligibility Criteria. 2008 [Google Scholar]
- [10].Tu SW, Musen MA. MedInfo. London, UK: 2001. Modeling Data and Knowledge in the EON Guideline Architecture; pp. 280–4. [PubMed] [Google Scholar]
- [11].Tu SW, Campbell JR, Glasgow J, Nyman MA, McClure R, McClay J, et al. The SAGE Guideline Model: achievements and overview. J Am Med Inform Assoc. 2007 Sep–Oct;14(5):589–98. doi: 10.1197/jamia.M2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ross J, Carini S, Tu SW, Sim I. Medinfo. Capetown, South Africa: 2010. Analysis of Eligibility Criteria Complexity in Randomized Clinical Trials. submitted. [PMC free article] [PubMed] [Google Scholar]
- [13].Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004 Sep–Oct;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon. J Am Med Inform Assoc. 2005 May–Jun;12(3):275–85. doi: 10.1197/jamia.M1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peleg M, Tu SW, Bury J, Ciccarese P, Fox J, Greenes RA, et al. Comparing Computer-Interpretable Guideline Models: A Case-Study Approach. J Am Med Inform Assoc. 2003;10(1):52–68. doi: 10.1197/jamia.M1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the Meta-Map program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- [17].Spackman K. SNOMED RT and SNOMEDCT. Promise of an international clinical terminology. MD Comput. 2000 Nov–Dec;17(6):29. [PubMed] [Google Scholar]
- [18].Peleg M, Keren S, Denekamp Y. Mapping computerized clinical guidelines to electronic medical records: knowledge-data ontological mapper (KDOM) J Biomed Inform. 2008 Feb;41(1):180–201. doi: 10.1016/j.jbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- [19].German E, Leibowitz A, Shahar Y. An architecture for linking medical decision-support applications to clinical databases and its evaluation. J Biomed Inform. 2009 Apr;42(2):203–18. doi: 10.1016/j.jbi.2008.10.007. [DOI] [PubMed] [Google Scholar]
- [20].Johnson PD, Tu SW, Musen MA, Purves I. Proc AMIA Symp. Washington, DC: 2001. A Virtual Medical Record for Guideline-Based Decision Support; pp. 294–8. [PMC free article] [PubMed] [Google Scholar]
- [21].Borlawsky T, Payne PR. Evaluating an NLP-based approach to modeling computable clinical trial eligibility criteria. AMIA Annu Symp Proc. 2007:878. [PubMed] [Google Scholar]
- [22].Health Level Seven HL 7 Reference Information Model. 2009 [cited 2008]; Available from: http://www.hl7.org/library/data-model/RIM/modelpage_mem.htm.
- [23].Cornet R. Definitions and qualifiers in SNOMED CT. Methods Inf Med. 2009;48(2):178–83. [PubMed] [Google Scholar]
- [24].Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, et al. Relations in biomedi-cal ontologies. Genome Biol. 2005;6(5):R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.