Abstract
miRNAs are a new class of endogenous small RNAs that negatively regulate gene expression at the posttranscriptional level. Accumulating experimental evidence shows that miRNAs regulate cellular apoptosis, proliferation, differentiation, and migration. Dysregulation of miRNA expression leads to various human diseases including cancer and cardiovascular disease. miRNA maturation is regulated at multiple steps by different mechanisms, including miRNA editing, hairpin loop binding, self-regulation, and cross-talk with other signaling pathways. Vascular cell movement plays a pivotal role in the development of various cancers and cardiovascular diseases. miRNAs have been found to regulate vascular cell movement. Presently the chemically synthesized antagomir, miRNA mimics have been widely used in investigating the biological functions of miRNA genes. The viral vectors, including adenoviral, lentiviral, and adeno-associated viral vectors, have been used to efficiently overexpress or knockdown miRNAs in vitro and in vivo. Therefore, targeting vascular cell movement using miRNA-based drug or gene therapy would provide a novel therapeutic approach in the treatment of cancers and vascular diseases.
Keywords: miRNA, vascular, cell movement, therapy
miRNA maturation and regulation
miRNAs are a new class of endogenous small RNAs that are associated with a variety of human diseases, including cancer, heart failure, vascular disease, and diabetes et al. In the last several years, miRNA has been extensively investigated, it is now recognized that miRNA plays a key role in regulating fundamental cellular processes, including cell proliferation, apoptosis, differentiation, and migration. miRNA genes are located in the intron, non-coding exon, and intergenic regions of genomes, and are initially transcribed by polymerase II into primary miRNAs (pri-miRNAs). The pri-miRNAs are subsequently cleaved in the nucleus by a microprocessor complex composed of Drosha and DGCR8(Pasha) into a stem-loop precursor-miRNA (pre-miRNA) which has a length of approximate 70 nucleotides (nt). Pre-miRNAs are in turn transported by exportin-5 into the cytoplasm, where they are cleaved by a RNase III family member Dicer, into 22-nt mature miRNAs. The mature miRNAs are able to be incorporated into the RNA-induced silencing complex (RISC) that contains PACT, Dicer, Ago2, and TRBP. RISC funtions to degrade mRNA or block protein translation by either binding mRNA through a perfect complementary match or through an imperfect match in the 3′ untranslated region (UTR) [1,2,3]. Regulation of miRNA maturation at multiple steps in miRNA biogenesis includes the following mechanisms(Fig. 1).
Fig. 1. miRNA maturation and regulation.
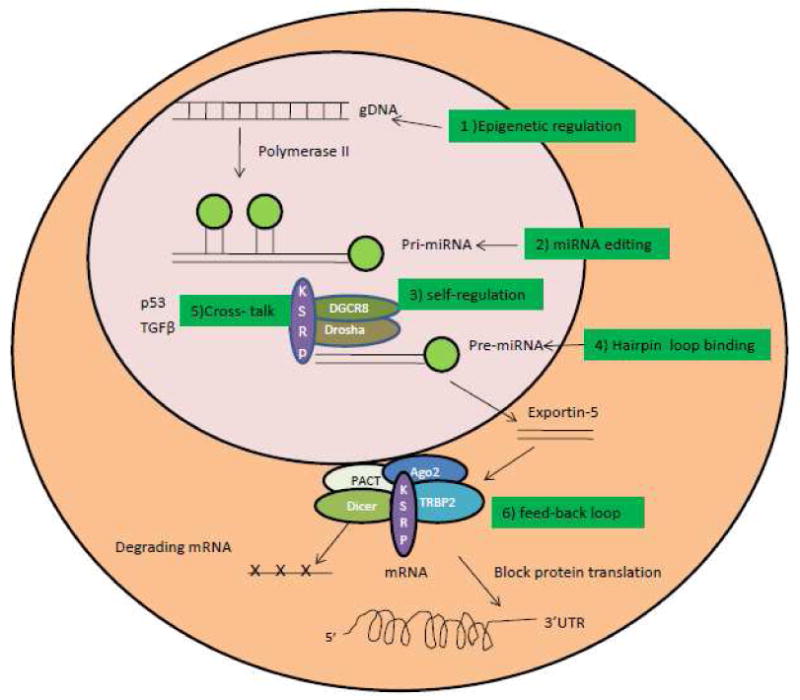
miRNA is transcribed by a polymerase II promoter from genomic DNA (gDNA) and forms primary miRNA, which is processed by a microprocessor complex containing Drosha, DGCR8 (Pasha), and KSRP into precursor miRNA (pre-miRNA). pre-miRNA is transported into the cytoplasm by exportin-5. In the cytoplasm, the RNase III enzyme Dicer cleaves the pre-miRNA into 22-nt mature miRNA, then degrades the targeted mRNA and blocks protein translation by incorporating the miRNA into RISC containing the Dicer, Ago2, PACT, and TRBP2 protein complex. miRNA maturation is regulated through different mechanisms, including epigenetic regulation, miRNA editing, microprocessor self regulation, hairpin loop binding, cross-talk, and feedback loops.
1. Epigenetic regulation
miRNA genes located near CpG islands in the genome were found to be affected by hypermethylation of the neighboring chromatin. Moreover, some miRNA genes themselves were found to be methylated in various cancers [4,5,6,7]. Silencing of the miRNA genes by methylation leads to upregulation of the targeted genes by those miRNAs, thereby causing the cancer or cardiovascular disease [8,9,10].
2. miRNA editing
Conversion of pri-miRNAs into pre-miRNA was also found to be subjected to RNA editing that converts adenosine to inosine (A --> I RNA editing), which leads to the accumulation of edited pre-miRNAs, thus blocking the processing by RISC and miRNA maturation. miR-151 and miR-142 are two miRNAs that were shown to be regulated by RNA editing [11,12].
3. Self-regulation in miRNA biogenesis pathway
A self-regulating miRNA biogenesis pathway was also reported in which Drosha and DGCR8 regulate each other. The Drosha-DGCR8 complex degrades the mRNA of DGCR8, whereas DGCR8 stabilizes the Drosha via a feedback loop mechanism [13].
4. Hairpin loop binding
The hairpin loop of pri-miRNA or pre-miRNA has been shown to play a key role in regulating miRNA maturation. The mutation in the hairpin loop region of pri-miRNA or pre-miRNA leads to reduced efficiency in miRNA maturation [14,15]. The KH-type splicing regulatory protein KSRP (also known as KHSRP), a RNA binding protein, was found to interact with the microprocessor in the nucleus and RISC in the cytoplasm to promote miRNA maturation as a shuttle protein. Interestingly, KSRP was shown to regulate a subset of miRNAs by binding the hairpin loop of pre-miRNA [15].
5. Cross-talk between the miRNA biogenesis pathway and other cellular signaling pathways
p53 was found to interact with the Drosha processing complex through association with DEAD-box RNA helicase p68 and to promote the cleavage of pri-miRNAs to pre-miRNAs [16]. TGF-β was shown to promote the cleavage of pri-miR-21 into pre-miR-21 through the DROSHA/DGCR8/p68 complex, thus facilitating miR-21 maturation [17].
6. A regulatory feedback loop between miRNA and target gene
miRNAs are regulated by transcription factors that bind to the promoter regions of miRNA genes, thus activating miRNA expression. In some cases, the miRNA can bind to the 3′ UTR of the transcription factor mRNA and thereby inhibit its expression [18,19,20,21,22]. This feedback regulatory mechanism is able to fine-tune the miRNA and transcription factor gene expression levels, thereby maintaining the cellular homeostasis.
Some miRNA maturation does not truly follow the biogenesis pathway. Recently, miR-451 maturation was found to require Ago2 but was independent of Dicer cleavage [23,24]. However, in our recent work, we found that miR-451 was still downregulated in vascular smooth muscle cells (VSMCs) from VSMC-specific Dicer knockout mice. More interestingly, some miRNAs were even upregulated the VSMCs isolated from VSMC-specific Dicer knockout mice (Y. Pan, unpublished data). Taken together, these studies suggest that maturation of miRNA may be regulated differentially in a cell context-dependent manner.
Dysregulation of miRNA maturation and vascular cell movement
Dysregulation of the miRNA maturation pathway may lead to a pathological process and disease state as a result of the alteration of the miRNA expression profile. Knockout of either Dicer or Ago2 in mice results in embryonic mortality [25,26]. Recently, we created a conditional knockout mouse in which exon 23 of Dicer was deleted in VSMCs. These mice displayed a severe phenotype including developmental delay and embryonic lethality as a result of extensive hemorrhage in multiple organs such as the brain, skin, and liver (Y. Pan et al., unpublished data). However, another group reported a less severe phenotype than we observed when they specifically deleted exons 20 and 21 of Dicer in VSMCs of mice. In Dicerexon20/21 knockout mutants, there was no difference in body size whereas we found body size of mutants was smaller during embryonic developmental stages relative to wild type mice. Moreover, hemorrhage was observed only in the liver compared unlike the widespread hemmorhage we observed [27]. Exons 20 and 21 of Dicer encode the first RNA binding domain, whereas exon 23 encodes the second RNA binding domain. Both domains are known to coordinately bind to double-stranded RNA to enable the RNA cleavage. Knockout of Dicer in vascular endothelial cells resulted in reduced postnatal angiogenic response to a variety of stimuli, including exogenous vascular endothelial growth factor (VEGF), limb ischemia, and wound healing [28]. Knockdown of Dicer and Drosha in endothelial cells significantly reduced capillary sprouting and tube formation. It was also found that knockdown of Dicer inhibited migration of VSMCs, whereas knockdown of Drosha had no effect [29]. Furthermore, it was found in adherent human umbilical vein endothelial cells that silencing of Dicer markedly impaired migration and sensitized the apoptosis caused by serum withdrawal-induced apoptosis, whereas overexpression of Dicer reduced the cellular apoptosis[30].
We are just beginning to understand the regulation of miRNA in vascular biology. In particular, the miRNA biogenesis and regulatory pathways in the vascular system have not yet been well characterized. The roles of DGCR8 (Pasha), Ago2, KSRP, PACT, and TRBP in cancers and vascular diseases are still elusive. The combination of RNA interference and gene knockout should reveal the roles of miRNA by allowing investigation of key regulators in miRNA maturation.
miRNA, gene targets, and vascular cell movement
miRNA functions by targeting multiple genes and forming a complex regulatory network that includes positive and negative feedback loop mechanisms between miRNA and the target genes. Interaction between miRNA and the target genes regulates cell proliferation, apoptosis, differentiation, and migration, and is necessary to maintain basic cellular homeostasis. Dysregulation of the miRNA-mediated regulatory network causes aberrant phenotypic alterations. Recent studies show that vascular cells, including vascular endothelial cells and VSMCs, were also tightly regulated by miRNA genes. Vascular cell migration plays a pivotal role in cancer and cardiovascular diseases, and is necessary to both tumor metastasis and atherosclerosis by promoting angiogenesis and vascular wall remodeling. Several miRNA clusters and individual miRNAs have been shown to be involved in vascular cell migration, thus implicating their involvement in cancer and vascular diseases.
The extensively investigated miR-17/92 cluster contains six individual miRNAs: miR-17, miR-18a, miR-19a, miR-20a, 19b-1, and miR-92a-1. Loss of this cluster in mice leads to postnatal death due to septal defect of the heart [31,32]. This cluster was found to be highly expressed in endothelial cells and played an antiangiogenic role by blocking endothelial cell movement and targeting for DICER-mediated degradation the mRNAs of several proangiogenic proteins, including Janus kinase 1, integrin subunit alpha5 and p21 [33,34]. Interestingly, miR-17/20 was not found to affect tumor angiogenesis in this study, which is in contrast to another report that the miR-17/92 cluster promotes neovascularization in a Myc-induced tumor model [35]. The different findings suggest that the role of the miR-17/92 cluster target genes in angiogenesis may be dependent on the cellular context, which has different target genes. For example, the miR-17/92 cluster targets antiangiogenic thrombospondin-1 (Tsp1) and connective tissue growth factor (CTGF) in Myc-induced tumors [35]. The role of miR-17/92 cluster in the VSMCs has not been defined yet. It is not clear whether miR-17/92 cluster plays a role in phenotypic modulation by regulating VSMC differentiation marker gene expression or by regulating VSMC proliferation and migration. However, we found that deletion of Dicer in mouse VSMCs led to downregulation of miR-18a, 19a, 19b-1, and 92a-1, whereas miR-17 and miR-20a were upregulated, suggesting a role of the miR-17/92 cluster in vascular wall remodeling by using different regulatory mechanism in terms of individual miRNA expression.
The miR-143/145 cluster is highly expressed in normal VSMCs, whereas the expressions of both miR-143 and miR-145 are decreased in acute and chronic vascular stress and human aortic aneurysms [36]. miR-143 and miR-145 cooperatively target multiple genes, including Kruppel-like factor 4 (Klf4), Elk-1 (a member of the ETS oncogene family) [37], angiotensin-converting enzyme (ACE) [38], and serum response factor (SRF) as well as its coactivator, myocardin, to regulate VSMC phenotype [39]. Loss of the miR-143/145 cluster promotes neointima formation [36,37,38]. However, it was shown in a separate study that loss of the miR-143/145 cluster led to reduced neointima formation as a response to vascular injury [39]. miR-145 was also found to target the hematopoietic transcription factor Fli1 and to block migration in response to growth factor gradients in microvasculature [40]. miR-143 was found to inhibit VSMC migration by targeting versican protein, which is also involved in platelet-derived growth factor induced smooth muscle cell migration [41]. The miR-143/145 cluster was identified as a tumor suppressor and was downregulated in various cancers, including prostate, colon, gastric, bladder, chronic lymphocytic leukemias, and B-cell lymphomas. It functions as a tumor suppressor by targeting extracellular signal-regulated kinase 5 (ERK5) [42], c-Myc, mucin-1 [43,44], and fascin homologue 1 (FSCN1) [45]. Extensive studies on the miR-143/145 cluster indicated that this cluster plays a very important role in the development of vasculature and tumor growth by inhibiting vascular cell movement and tumor angiogenesis.
The miR-221/222 cluster is highly expressed and has been found to inhibit cell migration and angiogenesis by targeting the stem cell factor receptor c-kit, transcriptional factor Stat5a, and ZEB2, a modulator of the epithelial-mesenchymal transition in vascular endothelial cells [29,46,47,48]. miR-221 was identified as a positive regulator in platelet-derived growth factor (PDGF)-mediated signaling in VSMCs. Knockdown of miR-221 expression in VSMCs significantly inhibited PDGF-induced migration and proliferation as well as differentiation by targeting p27 and c-kit [49]. The miR-221/222 cluster has also been shown to promote tumorigenesis by targeting p27, p57, and PTEN in a variety of malignancies including gastric cancer, prostate cancer, chronic lymphocytic leukemia, and glioma [50,51,52,53,54,55,56]. Thus, the miR-221/222 cluster inhibits endothelial cell movement by targeting c-kit, whereas it promotes VSMC proliferation by targeting p27.
The miR-15b/16-2 cluster, located in chromosome 3 in humans, 2 in rats, and 3 in mice, is highly conserved in vertebrates. The miR-15a/16-1 cluster, is located in chromosome 13 in humans and 14 in mice, and we have recently identified the miR-15a/16-1 cluster in rats, which is also highly conserved and located on chromosome 15 [57]. Human miR-15a and miR-16-1 are clustered within 0.5 kb at 13q14 [58]. Both miR-15a and miR-16-1 are deleted or downregulated in more than two-thirds of chronic lymphocytic leukemia (CLL) cases [58,59]. The miR-15b/16-2 cluster is located in the intron of the SMC4 gene. Although the miR-15b/16-2 cluster functions as a tumor suppressor in gastric cancer, melanoma, and glioma [60,61,62], it has been less investigated than the miR-15a/16-1 cluster. We found that miR-15b and miR-15a have a very similar expression pattern in various tissues. Moreover, the expression of miR-15a,miR-15b, and miR-16 is higher than that of miR-15a*, miR-15b*, and miR-16* in vivo [57]. Knockout of the miR-15a/16-1 cluster in mice led to CLL at advanced ages. The miR-15b/16-2 cluster knockout mouse has not been generated yet. Thus, it is still unknown whether this cluster plays a synergistic role on the phenotype of miR-15a/16-1 cluster knockout mice, and it will be interesting to determine whether double knockout mice (both clusters deleted) will show a more severe phenotype than mice with only one cluster deleted. miR-16 was identified as an important regulator of the proangiogenic growth factor VEGF in Hela cells [63]. miR-15b and miR-16 were also found to regulate angiogenesis by targeting several angiogenic factors such as uPAR, Cox2, VEGF, and Met in nasopharyngeal carcinoma cells [63,64]. miR-15a was recently found to be regulated by peroxisome proliferator-activated receptor delta (PPARΔ) at the transcriptional level by targeting Bcl-2 in cerebral vascular endothelial cells, which suggests that miR-15a has a significant role in stroke-related vascular dysfunction [65].
miR-126 is an endothelial cell-specific miRNA and has been found to regulate angiogenesis. miR-126 knockout mice display leaky vessels, hemorrhaging, and partial embryonic mortality, due to a loss of vascular integrity and to defects in endothelial cell proliferation, migration, and angiogenesis. miR-126 promotes the vascular signaling of VEGF and fibroblast growth factor by targeting Spred-1 and phosphoinositol-3 kinase regulatory subunit 2 (PIK3R2/p85-β) [59,66]. However, silencing miR-126 expression also promotes VEGF-induced angiogenesis in the absence of blood flow and transcriptional factor Klf2a [67]. It was also found that knockdown of miR-126 using antagomir impairs ischemia-induced angiogenesis [68]. In lung cancer cells, miR-126 was found to target VEGF-A, thus functioning as a tumor suppressor gene [69]. In prostate cancer, miR-126 is naturally deficient as a result of the deletion of its host gene's epidermal growth factor-like domain 7 (Egfl7). Enforced expression of miR-126 in prostate cancer LNCaP cells inhibits migration and invasion by targeting prostein[70]. Therefore, miR-126 is a proangiogenic gene and promotes vascular endothelial cell movement, but is still able to function as a tumor suppressor gene in various cancers.
miR-21, an oncogenic miRNA that is upregulated in various cancers, was found to be induced by hypoxia in pulmonary artery smooth muscle cells (PASMCs). miR-21 promotes PASMC proliferation and migration by targeting programmed cell death protein 4 (PDCD4), Sprouty homologue 2 (Drosophila; SPRY2), and PPARα [71]. miR-21 was also found to be upregulated in serum of atherosclerosis patients compared to serum from healthy people[72].
miR-26a is a recently identified VSMC-specific miRNA that promotes VSMC proliferation and migration while it inhibits apoptosis and differentiation by targeting SMAD-1 and SMAD-4, thereby regulating the TGF-β signaling pathway[73]. miR-26a has also been identified as a tumor suppressor gene that is specifically downregulated in various malignancies such as prostate cancer, colon cancer, lymphoma, and glioma [74,75,76,77].
miR-132 was identified as an angiogenic switch that targets p120RasGAP in the endothelium and thereby leads to Ras activation that supports neovascularization. miR-132 was highly expressed in the endothelium of human tumors and hemangiomas but was undetectable in normal endothelium. Vessel-targeted nanoparticle delivery of anti–miR-132 suppressed angiogenesis and tumorigenesis[78].
In summary, miR-17/92, miR-143/145,miR-221/222,miR-15a/16-1 or miR-15b/16-2, miR-126, miR-21, miR-26 and miR-132 involved in the regulation of vascular cell movement by targeting multiple genes, thus dysregulated the multiple cellular pathways and led to the cancer or cardiovascular disease(Fig.2).
Fig. 2. miRNAs involved in vascular cell movement.
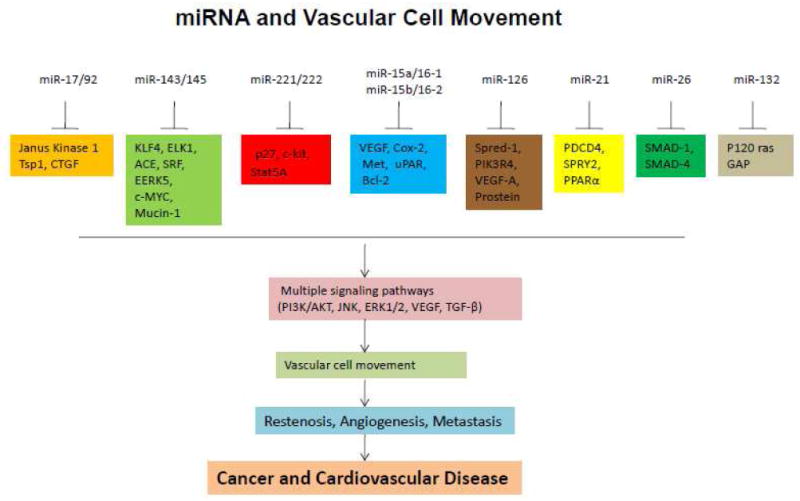
miRNAs, including miR-17/92, miR-143/145, miR-221/222, miR-15a/16-1 or miR-15b/16-2, miR-126, miR-21, miR-26, and miR-132, are involved in vascular cell movement. These miRNAs target different genes in a cellular context-dependent manner, thus regulating different signaling pathways that impact cancers and cardiovascular diseases.
miRNA-based drug and therapeutic strategy in cancer and vascular diseases
The extensive investigation of various miRNA genes in the past several years has revealed that miRNA expression is dysregulated in various cancers and cardiovascular diseases. Some miRNAs are upregulated, whereas others are downregulated in disease states. The dysregulation of miRNA expression alters fundamental cellular processes, including proliferation, apoptosis, differentiation, and migration. Vascular cell movement plays a major role in angiogenesis, tumor invasion, metastasis, and vascular diseases such as restenosis after angioplasty and atherosclerosis. As described above, several miRNAs and miRNA clusters are involved in vascular cell movement, which suggests a novel strategy of targeting those vascular miRNAs, thus treating the cancers and vascular diseases by inhibiting vascular cell movement.
Antagomir- and miRNA mimic-based drug development and vascular cell delivery
miRNAs are often up- or downregulated in cancer or cardiovascular disease. The miRNA based drug is required to reduce or enhance the miRNA expression for the particular therapeutic purpose. So far, antagomir (also called anti-miR) or blockmir has been used to silence miRNA expression, whereas mimics of miRNA are used to simulate upregulation in vitro and in vivo. To develop miRNA-based drugs to modulate miRNA expression, the current strategy is to synthesize chemically modified, cholesterol-conjugated single-stranded RNA analogues complementary to or the same as the mature miRNAs for antagomir or mimics, respectively. For example, an antagomiR-16-based drug was designed as 5′-csgsccaauauuuacgugcugscsusas-Chol-3′. The lower case letters represent 2′-OMe-modified nucleotides; the subscript ‘s’ represents a phosphorothioate linkage; and Chol represents cholesterol linked through a hydroxyprolinol linkage. This modified antagomir or mimic-like drug can be directly delivered in vivo and has been shown to be very efficient, stable, and specific in silencing miRNA expression [68,79,80,81]. To target vascular cell movement using miRNA-based drugs, antagomirs or mimics must be specifically delivered into the vascular cells. Recently, an integrin alpha-nu-beta3 (αvβ3)-targeted nanoparticle delivery of anti-miR-132 showed very promising results in inhibiting angiogenesis and tumor metastasis [78,82]. The integrin αvβ3 is preferentially expressed in diseased vascular endothelial cells and thus specifically targets tumor vasculature.
miRNA-based gene therapy
To harness miRNA's therapeutic potential, miRNA genes can be overexpressed or silenced using vector delivery, including plasmid and viral vectors. In particular, adenoviral, lentiviral, or retroviral vectors and adeno-associated viral vectors are highly efficient in delivering genes. Currently more than 2,000 gene therapy-based clinical trials are underway; these include targeting important genes or shRNA in cancers and cardiovascular diseases. However, miRNA-based gene therapy is just in its infancy. The only gene therapy in clinical trial based on miRNA therapy is miR-122, which is a liver-specific miRNA. The miR-122-based gene therapy is currently in phase 2 clinical trials[83]. Several other studies targeting miRNA let-7 or miR-16 in cancer by gene therapy are in progress, but have not yet entered clinical trials.
To construct miRNA-based gene therapy vectors, the primary miRNA genes can be cloned into different viral vectors to enhance miRNA expression(Fig.3a). To silence miRNA expression for gene therapy, the antisense sequence to mature miRNA has been used to construct an antagomir (blockmir), or decoy or sponge viral vector(Fig.3b). It has been shown that silencing of miRNA expression by these vectors were very efficient [78,84,85,86]. We constructed a lentiviral antagomir-21 vector in which the antisense miRNA sequence was driven by human U6 promoter(Fig.3c), and were able to achieve greater than 80% silencing of the endogenous expression of miR-21 in prostate cancer cells [87]. Similarly adenoviral or adeno-associated viral vectors can also be used to construct miRNA gene therapy vectors [88,89,90].
Fig. 3. Lentiviral vector-mediated miRNA gene delivery.

A. Lentiviral vector expressing pri-miRNA gene. miRNA genes is driven by polymerase II promoter and co-expressed with EGFP reporter gene. B. Lentiviral decoy or sponge miRNA vector. Antisense miRNA sequence is embedded by a hairpin loop and forms a pre-miRNA structure and driven by polymerase II promoter, then co-expressed with EGFP reporter gene C. Lentiviral antagomiRNA vector. Antisense miRNA gene against mature sequence is driven by polymerase III promoter human U6.
In conclusion, miRNA regulates gene expression by targeting multiple genes at the posttranscriptional level. miRNA maturation is regulated at primary, precursor, and mature levels by different mechanisms including miRNA editing, RNA protein binding, self-regulation. and cross-talk with other multiple signaling pathways. Vascular cell movements are regulated by miRNA genes in a cell context-dependent manner, such as the specific regulation of vascular endothelial cells by miR-126 or vascular smooth muscle cells by miR-143/145. Although further work is required to ascertain the many function of miRNAs in normal physiology and disease states, it is clear that vascular cell-specific miRNAs represent an important potential target for the treatment of cancers or cardiovascular diseases through drugs or gene therapy.
Acknowledgments
This work was supported by awards HL095957 and HD061420 to J. Yue from National Heart, Lung, and Blood Institute and the Eunice Kennedy Shriver National Institute of Child Health & Human Development, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We appreciate the assistance of Jin Emerson-Cobb and Valentine William of the Physiology Department in editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem. 2010 doi: 10.1093/jb/mvq113. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickstein J, Senyuk V, Premanand K, Laricchia-Robbio L, Xu P, et al. Methylation and silencing of miRNA-124 by EVI1 and self-renewal exhaustion of hematopoietic stem cells in murine myelodysplastic syndrome. Proc Natl Acad Sci U S A. 2010;107:9783–9788. doi: 10.1073/pnas.1004297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, et al. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, et al. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 8.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto Y, Akiyama Y, Otsubo T, Shimada S, Yuasa Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 2010;31:777–784. doi: 10.1093/carcin/bgq013. [DOI] [PubMed] [Google Scholar]
- 10.Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394:1047–1052. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- 11.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 17.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 22.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 26.Morita S, Horii T, Kimura M, Goto Y, Ochiya T, et al. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, et al. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 30.Asada S, Takahashi T, Isodono K, Adachi A, Imoto H, et al. Downregulation of Dicer expression by serum withdrawal sensitizes human endothelial cells to apoptosis. Am J Physiol Heart Circ Physiol. 2008;295:H2512–2521. doi: 10.1152/ajpheart.00233.2008. [DOI] [PubMed] [Google Scholar]
- 31.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 34.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, et al. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–4950. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 35.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsson E, Fredlund Fuchs P, Heldin J, Barkefors I, Bondjers C, et al. Discovery of microvascular miRNAs using public gene expression data: miR-145 is expressed in pericytes and is a regulator of Fli1. Genome Med. 2009;1:108. doi: 10.1186/gm108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Hu G, Zhou J. Repression of versican expression by microRNA-143. J Biol Chem. 2010;285:23241–23250. doi: 10.1074/jbc.M109.084673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachdeva M, Mo YY. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res. 2010;2:170–180. [PMC free article] [PubMed] [Google Scholar]
- 44.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70:378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, et al. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883–891. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Song YH, Li F, Yang T, Lu YW, et al. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem Biophys Res Commun. 2009;381:81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, et al. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30:1562–1568. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 49.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 51.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One. 2008;3:e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Zhang C, Kang C, You Y, Pu P, Yang W, et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol. 2009;34:1653–1660. doi: 10.3892/ijo_00000296. [DOI] [PubMed] [Google Scholar]
- 55.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfo L, et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–3959. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 57.Yue J, Tigyi G. Conservation of miR-15a/16-1 and miR-15b/16-2 clusters. Mamm Genome. 2010;21:88–94. doi: 10.1007/s00335-009-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia L, Zhang D, Du R, Pan Y, Zhao L, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 61.Xia H, Qi Y, Ng SS, Chen X, Chen S, et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380:205–210. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- 62.Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2010;126:2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 63.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, et al. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249–254. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hua Z, Lv Q, Ye W, Wong CK, Cai G, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Senturia R, Faller M, Yin S, Loo JA, Cascio D, et al. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci. 2010;19:1354–1365. doi: 10.1002/pro.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, et al. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J Cell Mol Med. 2009;13:1577–1585. doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, et al. MicroRNA-21 plays a Role in Hypoxia-mediated Pulmonary Artery Smooth Muscle Cell Proliferation and Migration. Am J Physiol Lung Cell Mol Physiol. 2010 doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li T, Cao H, Zhuang J, Wan J, Guan M, et al. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2010 doi: 10.1016/j.cca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- 73.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2010 doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Yao Y, Smith LP, Nair V. MicroRNA-26a-mediated regulation of interleukin-2 expression in transformed avian lymphocyte lines. Cancer Cell Int. 2010;10:15. doi: 10.1186/1475-2867-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Theodore SC, Rhim JS, Turner T, Yates C. MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn Dis. 2010;20:S1-96–100. [PMC free article] [PubMed] [Google Scholar]
- 77.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, et al. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- 78.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 80.Li J, Bai H, Zhu Y, Wang XY, Wang F, et al. Antagomir dependent microRNA-205 reduction enhances adhesion ability of human corneal epithelial keratinocytes. Chin Med Sci J. 2010;25:65–70. doi: 10.1016/s1001-9294(10)60024-7. [DOI] [PubMed] [Google Scholar]
- 81.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murphy EA, Majeti BK, Barnes LA, Makale M, Weis SM, et al. Nanoparticle-mediated drug delivery to tumor vasculature suppresses metastasis. Proc Natl Acad Sci U S A. 2008;105:9343–9348. doi: 10.1073/pnas.0803728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 84.Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, et al. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010 doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 87.Yang CH, Yue J, Fan M, Pfeffer LM. IFN Induces miR-21 through a Signal Transducer and Activator of Transcription 3-Dependent Pathway as a Suppressive Negative Feedback on IFN-Induced Apoptosis. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kato Y, Sawata SY, Inoue A. A lentiviral vector encoding two fluorescent proteins enables imaging of adenoviral infection via adenovirus-encoded miRNAs in single living cells. J Biochem. 2010;147:63–71. doi: 10.1093/jb/mvp144. [DOI] [PubMed] [Google Scholar]
- 89.Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, et al. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suzuki T, Sakurai F, Nakamura S, Kouyama E, Kawabata K, et al. miR-122a-regulated expression of a suicide gene prevents hepatotoxicity without altering antitumor effects in suicide gene therapy. Mol Ther. 2008;16:1719–1726. doi: 10.1038/mt.2008.159. [DOI] [PubMed] [Google Scholar]


