Summary
Despite the use of standardized protocols in, multicentre, randomised clinical trials (RCTs), outcome may vary between centres. Such heterogeneity may alter the interpretation and reporting of the treatment effect. Below, we propose a general frailty modelling approach for investigating, inter alia, putative treatment-by-centre interactions in time-to-event data in multi-centre clinical trials. A correlated random effects model is used to model the baseline risk and the treatment effect across centres. It may be based on shared, individual or correlated random-effects. For inference we develop the hierarchical-likelihood (or h-likelihood) approach which facilitates computation of prediction intervals for the random effects with proper precision. We illustrate our methods using disease-free time-to-event data on bladder cancer patients participating in an European Organization for Research and Treatment of Cancer (EORTC) trial, and a simulation study. We also demonstrate model selection using h-likelihood criteria.
Keywords: Correlated random effects, Focussed model selection, Frailty models, Hierarchical likelihood, Prediction interval, Random treatment-by-centre interaction
1. Introduction
In this paper we focus on multi-centre trials with time to event endpoints. We are interested in investigating potential heterogeneity in outcomes between centres. In this context, the use of proportional hazards (PH) frailty models with random effects, rather than PH models with fixed (centre) effects, are useful [1–4].
Our approach is to model: (a) the between-centre variation in the baseline risk and (b) the treatment effect across centres [2, 5], using random effects. Thus, our model incorporates a random centre effect and a random treatment-by-centre interaction. These two random components (or frailty terms) have usually been assumed to be independent [2, 5]. However, independence may be un-necessarily restrictive [6–8]. In particular, Legrand et al. [4] has recommended using correlated random effects. Furthermore, our approach also models individual-specific frailty terms [9–11], because covariates specified in the protocol, or involved in the minimization procedure [12, p.71], may not account for all prior differences between patients. Thus, by deploying classical frailty concepts, we hope to improve conventional strategies for analyzing RCTs trials.
Usually, inference in frailty models requires a marginal-likelihood approach, whereby the random effects are integrated out of the joint density consisting of response variables and random effects. This may involve the evaluation of analytically intractable integrals over the random effect distributions. To avoid these difficulties, several methods (e.g. Monte Carlo EM and Markov chain Monte Carlo) have been suggested [13, 14], but these remain computationally intensive, particularly when the number of random components is large or when their correlation structure is modelled [15–19].
Another important issue is that of estimating the standard errors for the prediction of random effects, which is required in order to construct 100(1 − α)% prediction intervals. Plots based on these intervals are useful, especially when investigating the heterogeneity of random centre and treatment effects. However, estimating the standard errors of random effects using “plug-in” methods, such as empirical Bayes (EB, [13, 20]), may underestimate the true variability of the estimated random effects. Thus, the development of an integral method of inference for frailty models is required.
Accordingly, we propose a unified method of inference within the h-likelihood framework [21–23]. The h-likelihood consists of data, parameters and unobserved random effects, and obviates integration over the random-effect distributions. Thus, the h-likelihood can be used directly for inference on random effects, while the marginal likelihood cannot because it eliminates them by integration. The h-likelihood approach also gives a statistically efficient estimation procedure for various random-effect models [11, 19, 24–26]. We derive, via the h-likelihood approach, improved methods for estimating the standard errors of the predictor of the random effects and the frailty parameters. In particular, we emphasize inference on the random effects rather than on just estimating the frailty parameters. Predictions and their intervals are useful in investigating heterogeneity over centers. We illustrate the methodology by analyzing time to first recurrence in patients with bladder cancer from an EORTC trial [27] and by a simulation study. We also employ the data to illustrate model selection using criterion [11] based on h-likelihood.
The paper is organized as follows. In Section 2 we review a formulation of frailty models, present an extension, and show how to interpret the random-effect terms. The h-likelihood estimation procedure for fitting the model is derived and an improved method for estimating the standard-error of the frailty parameters is proposed. Next, a new prediction method for random effects is proposed in Section 3. The new method is illustrated using the bladder cancer data set in Section 4. A simulation study is conducted to evaluate the performance of the proposed method in Section 5. And, finally, we discuss the approach in Section 6. The technical details are given in Appendices.
2. The model and estimation
2.1. Model formulation and interpretation
In general, suppose that data consist of right censored time-to-event observations collected from q centres. Let Tij (i = 1, …, q, j = 1, …, ni, n = Σi ni) be the survival time for the jth observation in the ith centre (or cluster) and let Cij be the corresponding censoring time. Then observable data become yij = min{Tij, Cij} and δij = I(Tij ≤ Cij), where I(·) is the indicator function.
Denote by vi a s-dimensional vector of unobserved log-frailties (random effects) associated with the ith cluster. Given vi, the conditional hazard function of Tij is of the form
| (1) |
where λ0(·) is a unknown baseline hazard function,
is the linear predictor for the hazards, and xij = (xij1, …, xijp)T and zij = (zij1, …, zijs)T are p × 1 and s × 1 covariate vectors corresponding to fixed effects β = (β1, …, βp)T and log-frailties vi, respectively. Here zij is often a subset of xij. Alternatively, it may be the constant (unity) representing a cluster effect on the baseline hazard [13]. In this paper, we assume the normal distribution for vi:
which is useful for modelling multi-component [11, 28] or correlated frailties [13, 29]. Here the covariance matrix Σi = Σi(θ) depends on θ, a vector of unknown parameters. We note that the formulation of model (1) is actually the same as that of Vaida and Xu [13] but that their covariance matrix for Σi is diagonal [8].
Equation (1) includes some well-known models. Let vi0 be a random baseline intercept (representing the random baseline risk) and let vi1 be a random slope (i.e. random treatment effect or random treatment-by-center interaction). If in model (1) zij = 1 and vi = vi0 for all i, j, it becomes a random intercept or shared model [30, 31] with
| (2) |
where vi0 ~ N (0, Σi) with for all i. Let β1 be the effect of primary covariate xij1 such as the main treatment effect and let βm (m = 2, …, p) be the fixed effects corresponding to the covariates xijm. Our two random components lead to a bivariate model [8, 32] with
| (3) |
which is easily derived by taking zij = (1, xij1)T and vi = (vi0, vi1)T in (1). Here
Model (3) allows a correlation term, ρ = σ01/(σ0σ1), between two random effects (vi0 and vi1) within a centre and potentially extends the independent frailty model in which ρ = 0 [2, 5].
Furthermore, we note the model (1) may be asymmetric (or unbalanced) as it does not contain a generic individual-specific frailty term, vij to match the individual-level fixed effects, xij. Following Ha et al. [11], the one-component model (1) with can be extended easily to a two-component model with
| (4) |
where and are independent, and and are random-covariate vectors corresponding to and , respectively. In fact, the model (4) can be written as in (1) by taking and . Thus, the extension of results from one-component model (1) to two-component model (4) is straightforward [11, 21].
In order to interpret the fixed and random effects, we consider model (3) with a single binary-treatment indicator, xij. Then,
Now, the time-dependent relative risk for treatment becomes
which is free of time t and holds for all patients in centre i. Here exp(β1) is the usual expression for the relative risk in a standard PH model. Thus, ψij(t|x = 1, x = 0), represents a random multiplicative divergence from the standard relative risk in a PH model which is homogeneous with respect to centres. Note that exp(β1 + vi1) is often called the treatment hazard ratio in the ith centre [2, 5]. We also have that
Thus vi1 means the random deviation of the ith centre from the overall treatment effect. Similarly, in order to interpret vi0 we consider the model without the covariate xij
whence,
which is free of time t and holds for all patients in centre i, and vi0 represents the random deviation of the ith centre from the overall underlying baseline risk.
2.2. H-likelihood estimation
We now show how to derive the h-likelihood estimation procedure for fitting a correlated semiparametric model (1) and also propose how to obtain valid standard-error estimates for frailty parameters (i.e. dispersion parameters).
Since the functional form of λ0(t) is unknown, following Breslow [33], we approximate the baseline cumulative hazard function by a step function with jumps at the observed death times [23, 34];
where y(k) is the kth (k = 1, …, D) smallest distinct death time among the yij’s, and λ0k = λ0(y(k)).
Following Ha et al. [23], the hierarchical log likelihood (h-likelihood) for frailty models (1) is defined by
| (5) |
where
ℓ1ij = ℓ1ij(β, λ0; yij, δij|vi) is the logarithm of the conditional density function for yij and δij given vi,
is the logarithm of the density function for vi with parameters θ, Λ0(·) is the baseline cumulative hazard function, and . Here β = (β1, …, βp)T, , λ0 = (λ01, …, λ0D)T, d(k) is the number of deaths at y(k) and R(k) = R(y(k)) = {(i, j) : yij ≥ y(k)} is the risk set at y(k). In (5) log likelihood of the ith cluster is the logarithm of the joint density of (yi, δi) and vi, where yi = (yi1, …, yini)T and δi = (δi1, …, δini)T. As the number of λ0ks can increase with the number of events, the function λ0(t) is potentially of high dimension. Accordingly, for estimation of (β, v) Ha et al. [23] proposed the use of the profiled h-likelihood h* from which λ0 is eliminated:
| (6) |
where
are solutions of the estimating equations, ∂h/∂λ0k = 0, for k = 1, …, D. Note here that does not depend on λ0. Thus Lee and Nelder’s [21–22] h-likelihood procedure for hierarchical generalized linear models (HGLMs) can be extended to the frailty models based on h* [11, 19]. Here, for the estimation of frailty parameters θ we use an adjusted profile h-likelihood [18, 35], pβ,v(h*) defined in (A2); the details for estimation procedure are given in Appendix A.
We have shown that the approximated standard-error estimates for β̂ are obtained from the inverse of −∂2h*/∂(β, v)2, given in (7) [18, 23]. In this paper we propose that the approximated standard-error estimates for θ̂ are directly obtained using the inverse of −∂2pβ,v(h*)/∂θ2; the technical details are given in Appendix B. We also show the performance of these estimates by simulation below. Some conceptual differences between h-likelihood and other estimation methods for frailty models (1) are described in Appendix C.
3. Prediction of random effects
In HGLMs location parameters and dispersion parameter are asymptotically orthogonal [21]. Thus, very recently Lee and Ha [36] showed that a proper standard-error (SE) estimate for the prediction interval of random effects v can be computed from the inverse of the information matrix for (β, v) based on the h-likelihood. Here, the SE becomes the squared root of the approximation of the conditional mean-square error of prediction (CMSEP) of Booth and Horbert [37]. This is a general measure of predictive uncertainty and, following Lee and Ha [36], its extension to frailty models is straightforward as shown below.
In frailty models (1), as in HGLMs, location parameters (β, λ0, v) and frailty parameters θ are asymptotically orthogonal. For a moment, assume that θ is known. Accordingly, we need only focus on (β, v) after eliminating λ0 i.e. by using h*. Following Ha et al. [23] and Ha and Lee [18], the asymptotic covariances for β̂ and v̂ − v are obtained from the inverse of information matrix, J(h*; β, v) = −∂2h*/∂(β, v)2, of β and v based on h*:
| (7) |
where X and Z are the n × p and n × q model matrices for β and v whose ijth row vectors are and , respectively, , and the weight matrix W* = W*(β, v) is given in (B4). Here, BD(·) denotes a block diagonal matrix. This means that the SEs of v̂ − v can be computed from the information matrix, J in (7), of the profile h-likelihood h*.
Let yo = (yT, δT)T and . Here y and δ are the vectors of yij’s and δij’s, respectively. Following Booth and Horbert [37], the CMSEP based on yo and ψ is defined by
where v̂ (ψ̂) ≡ v̂ (ψ)|ψ=ψ̂ and v̂ (ψ) is the solution to ∂h/∂v = 0 for a given ψ. Note here that v̂ (ψ) = E(v|yo) asymptotically. Along the lines of Lee and Ha [36], J(h*; β̂, v̂)−1 ≡ J(h*; β, v)−1|β=β̂, v = v̂ gives the first-order approximation to the CMSEP, leading to a SE for v̂ − v:
| (8) |
| (9) |
where varψ(v|yo} = E[{v̂ (ψ) − v}{v̂ (ψ) − v}T|yo] and D(ψ) = E[{v̂ (ψ̂) − v̂ (ψ)}{v̂(ψ̂) − v̂ (ψ)}T|yo] is a nonnegative correction that accounts for the variability of parameter estimates ψ̂. Here ∂v̂/∂ψ = − (−∂2h/∂v∂vT)−1 (−∂2h/∂v∂ψT)|v= v̂. For the SE of prediction of random effects Vaida and Xu [13] and Othus and Li [20] used the EB method based on conditional posterior distribution of v given yo, leading to
| (10) |
which corresponds to the first term on the right hand side of (8) [21, 37]. The EB method can underestimate the SE of v̂ − v because it ignores the term above, D(ψ), which accounts for the inflation of the CMSEP caused by estimating [36, 37]. Following Lee and Ha [36], for the 95% h-likelihood and EB prediction intervals for v we use
where v̂ are obtained from (A1). Here the estimated h-likelihood and EB standard errors, SE(v̂ − v), are also obtained from the square roots of (9) and (10), respectively.
4. Practical example
4.1. The data and correlated model
The duration of the Disease Free Interval (DFI) in non muscle invasive bladder cancer patients, treated in various centres in Europe, is analysed. The DFI is defined as the time from randomization to the date of the first recurrence. Patients without recurrence at the end of the follow-up period were censored at their last date of follow-up. Patients were enrolled in 7 studies conducted by the EORTC [27]. For simplicity of analysis, we consider only 410 patients from 21 centres included in EORTC trial 30791 (Table 1). The two covariates of interest are: CHEMO xij1 (0=No, 1=Yes) and TUSTAT xij2 (0=Primary, 1=Recurrent). Notice that xij1 is the main treatment covariate. Patients with missing values for xij2 were excluded. The numbers of patients per centre varied from 3 to 78, with mean 19.5 and median 15. Of the 410 patients, 204 patients (49.8 per cent) without recurrence were censored at the date of last follow up.
Table 1.
Numbers of patients and recurrences per centre in the bladder cancer data
| Centre | np | ne | rc | Centre | np | ne | rc |
|---|---|---|---|---|---|---|---|
| 1 | 3 | 1 | 0.67 | 12 | 15 | 11 | 0.27 |
| 2 | 3 | 2 | 0.33 | 13 | 18 | 8 | 0.56 |
| 3 | 4 | 4 | 0 | 14 | 18 | 9 | 0.50 |
| 4 | 5 | 2 | 0.60 | 15 | 21 | 10 | 0.52 |
| 5 | 5 | 5 | 0 | 16 | 27 | 17 | 0.37 |
| 6 | 6 | 4 | 0.33 | 17 | 28 | 15 | 0.46 |
| 7 | 7 | 3 | 0.57 | 18 | 30 | 12 | 0.60 |
| 8 | 8 | 4 | 0.50 | 19 | 42 | 13 | 0.69 |
| 9 | 11 | 5 | 0.55 | 20 | 52 | 18 | 0.65 |
| 10 | 14 | 8 | 0.43 | 21 | 78 | 46 | 0.41 |
| 11 | 15 | 9 | 0.40 | ||||
np = No. of patients in centre; ne = No. of events; rc = 1 − (ne/np); Centres ordered by increasing np.
For the purpose of analysis, we consider the three submodels of (3):
M1 (Cox): Cox model without frailties (basic hazard),
M2 (Indep): Cox models, with two independent frailty terms (ρ = 0),
M3 (Corr): Cox models, with two correlated frailty terms (ρ ≠ 0).
Models M2 and M3 contain the random baseline risk vi0 and the random treatment-by-centre interaction term, vi1xij1. The models were fitted using SAS/IML. The results are summarized in Table 2. In all three models the two fixed effects (βj, j = 1, 2) are significant. In particular, the use of chemotherapy (CHEMO = 1) significantly prolongs the time to first recurrence as compared to patients who do not receive chemotherapy (CHEMO = 0): see also [4]. The two nested models (M1 and M2) ignoring random components or their correlation show similar results for βj (j = 1, 2). However, the absolute magnitude and SE of the estimate for the main treatment effect β1 in M1 and M2 are smaller than those for the correlated model (M3). In M2 and M3, the variances ( and ) indicate the amount of variation between centres in the baseline risk and in the treatment effect, respectively. Here, the estimate of is relatively larger than that of . This does not seem surprising since differences in outcome according to treatment effect are typically smaller than differences due to patient characteristics which often vary across centres. However, care may be necessary in comparing the two variances because these two values should not be interpreted on the same scale.
Table 2.
Results for fitting the four models to the bladder cancer data
| Model | β̂1 (SE) | β̂2 (SE) | (SE) | (SE) | σ̂01 (SE) | [ρ̂] |
|---|---|---|---|---|---|---|
| M1 (Cox) | −0.667 (0.170) | 0.509 (0.144) | – | – | – | |
| M2 (Indep) | −0.695 (0.175) | 0.544 (0.149) | 0.070 (0.058) | 3 × 10−12 (1 × 10−4) | – | |
| M3 (Corr) | −0.757 (0.191) | 0.532 (0.150) | 0.161 (0.178) | 0.036 (0.170) | −0.068 (0.149) | [−0.893] |
| M4 (B) | −0.695 (0.175) | 0.544 (0.149) | 0.070 (0.058) | – | – |
M1: Cox model without frailties; M2: independent frailty model with ρ = 0;
M3: correlated frailty model with ρ ≠ 0; M4: shared frailty model with random baseline risk (B) only; β1 and β2, effects of treatment and tumor status, respectively; and , the variances of random baseline risk and random treatment effect, respectively; σ01 and ρ, the corresponding covariance and correlation with ρ = σ01/(σ0σ1);
SE, the estimated standard error for parameters.
Moreover, the correlated model M3 explains the degree of dependency between the two random components (i.e. the random centre effect v0 and the random treatment-by-centre interaction v1). The estimate of ρ (ρ̂ = −0.893) gives a large negative value, indicating that the two predicted random components (v̂0 and v̂1) have a strong negative correlation. It is clear from the plot (not shown) of v̂1 against v̂0 that as vi0 increases (i.e. the baseline risk increases), vi1 decreases. Note here that exp(vi1) represents the ratio of treatment hazard rate in the ith centre (i.e. exp(β1 + vi1)) to overall hazard rate (i.e. exp(β1)). In particular, the estimate of β1 in M3 is negative; we see that a decreasing value of vi1 corresponds to an increased treatment effect. Thus, the negative correlation leads to the conclusion that treatment confers more benefit in centres with a higher baseline risk. This is consistent with the findings by Turner et al. [6] and Rondeau et al. [8] in the context of meta-analysis.
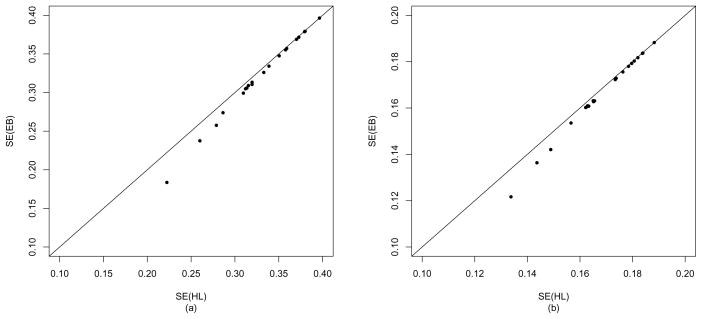
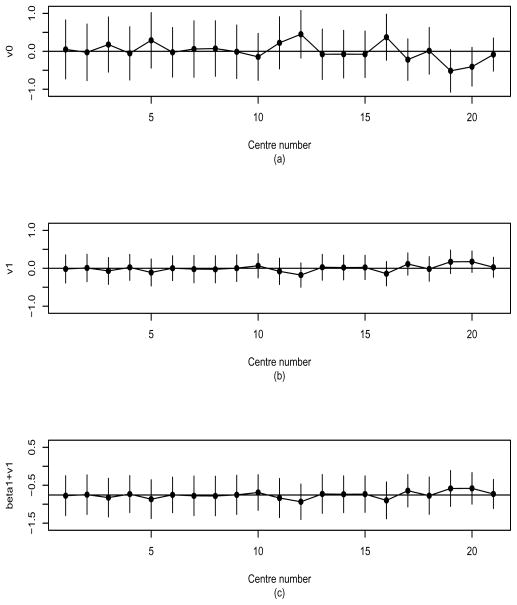
Figure 1 compares SE estimates of h-likelihood (HL) versus EB under M3. As expected, the EB estimates are smaller than HL estimates in both vi0 and vi1, leading to a lower coverage probability of prediction interval than the nominal level. Accordingly, below, we conduct detailed analyses of the random effects using HL. Figure 2 shows the estimates and 95% prediction intervals for the random effects in the 21 centres using M3. It shows the variations of the two random components (vi0, vi1) over centres, ordered by the number of patients entered. In particular, Figure 2(a) shows that centres 12 and 19 provide the highest and the lowest baseline risk, respectively. From Figure 2(b) we see that the corresponding centres give lowest and highest treatment hazards, respectively, which leads in this case to a negative correlation (ρ̂ = −0.893), as shown in Table 2.
Figure 1.
Standard error (SE) estimates of empirical Bayes (EB) versus h-likelihood (HL) analyses for random effects in the bladder cancer data; (a) v0 and (b) v1 in correlated model (M3).
Figure 2.
Random effects of 21 centres in the bladder cancer data and their 95% prediction intervals, under correlated model (M3); (a) baseline risk (vi0); (b) treatment-by-centre interaction (vi1); (c) log treatment hazard ratio (bi1 = β1 + vi1). Centres are sorted in increasing order of number of patients.
Figures 2(a) and 2(b) also give the prediction intervals for the random baseline risk (vi0) and the random treatment-by-centre interaction (vi1), respectively. Overall, the lengths of the intervals are seen to decrease as the number of patients per centre increases, particularly for Figure 2(a): see also [13]. Figure 2(a) indicates substantial variation in the baseline risk across centres. However, Figure 2(b) shows overall homogeneity in the effect of treatment across centres, that is, there is little treatment-by-centre interaction in this data set. Thus, in this multicentre trial there is little difference in the treatment effects across centres and the treatment is shown to be effective, while there appears to be substantial variation in the baseline risk of DFI across centres. These results suggest that the treatment effect may be generalized to a broader patient population as in the findings by Yamaguchi and Ohashi [2].
In addition, the prediction intervals for the log treatment hazard rates (i.e. bi1 = β1 + vi1) in the different centres are also useful to check the variations over centres. Similarly, the 95% prediction interval of bi1 is given by
where b̂i1 = β̂i1 + v̂i1 and . Here var(b̂i1 − bi1) = var(β̂1) + var(v̂i1 − vi1) + 2cov(β̂1, v̂i1 − vi1) is obtained from J−1 in (7). Figure 2(c) shows wider interval lengths than in Figure 2(b) due to the additional variance and covariance terms, but again confirms there is little difference in the treatment effects over centres.
4.2. Model selection
A thorough analysis will involve us in enlarging the potential model space beyond M1, M2 or M3. We consider a number of extensions below and show how to select an appropriate model using a Akaike information criterion (AIC) [11] based on the focussed extended restricted likelihood (ERL, [35]);
Notice that is a deviance based on the ERL pβ,v(h*) in (A2) which eliminates (β, v) from h*, the profile h-likelihood from which the nuisance function λ0(t) has already been eliminated. Thus, is a function only of the frailty parameters θ and is used to select the frailty structure best supported by the data. Here pT is the number of frailty parameters (i.e. the parameters governing the frailty distribution), not the number of all fitted parameters or frailties. Notice that the focussed is a sharper model selection tool than the more usual unfocussed AIC [11].
Recall that vi0 and vi1 are the random baseline risk and random treatment effect of the ith centre, respectively. For the purpose of analysis, we consider the following five models including M1–M3, λij(t|v) = λ0(t) exp(ηij) with ηij allowing several frailty structures in models M2–M5: Here (vi0, vi1) ~ BN means that and ρ = Corr(vi0, vi1); (vi0, vi1) ~ IN also means BN with ρ = 0.
where B and T denote random baseline risk and random treatment effect, respectively. Here M3 is our full model and the others are various simplifications of it by assuming null components, i.e. M1 (vi0 = 0, vi1 = 0), M2 (ρ = 0), M4 (vi1 = 0) and M5 (vi0 = 0). For ease of comparison and ranking of candidate models, we have set the smallest AIC to be zero and the other AIC values are shifted accordingly. In Table 3 we report the AIC differences, not the AIC values themselves. The deviance from model M2 is very similar to that obtained in M4 because in M2 the variance of the vi1 is very small, i.e. in Table 2. If the AIC difference is larger than 1 the choice can be made [38, p.84]. Under this empirical criterion, we note that selects M4 as an appropriate model; its estimation results are also presented in Table 2. In particular, it clearly rejects more complex models M2 and M3 than M4, indicating that it reflects model complexity properly [11].
Table 3.
Deviance results: AICs for selecting frailty structures in the bladder cancer data
| Model | pT | ||||
|---|---|---|---|---|---|
| M1 (Cox) | 2196.2 | 0 | 1.2 | ||
| M2 (Indep) | 2193.0 | 2 | 2.0 | ||
| M3 (Corr) | 2192.7 | 3 | 3.7 | ||
| M4 (B) | 2193.0 | 1 | 0 | ||
| M5 (T) | 2194.2 | 1 | 1.2 | ||
| M6 (I) | 2195.6 | 1 | 2.6 | ||
| M7 (B+I) | 2192.3 | 2 | 1.3 | ||
| M8 (T+I) | 2193.5 | 2 | 2.5 | ||
| M9 (Indep+I) | 2192.3 | 3 | 3.3 | ||
| M10 (Corr+I) | 2192.1 | 4 | 5.1 | ||
AIC, differences where the smallest AIC is adjusted to be zero; T, random treatment effect (vi1); I, individual random effect (vij); Indep, B & T are independent; Corr, B & T are correlated; , the number of frailty parameters.
However, the Tij may also depend on the individual-specific random effects as in Ha et al. [11]. If this is the case, some of the observed variation between centres is attributable to the heterogeneity between patients. We account for this properly, by introducing an appropriate patient-specific frailty component. Let vij be the random effects of the jth patient in the ith centre, satisfying vij ~ N (0, σ2). The extra random term vij, which is matched with individual-level event time Tij and fixed effect xij, can be viewed as modelling heterogeneity between patients at the individual patient level [9]. Accordingly, we consider the following additional models:
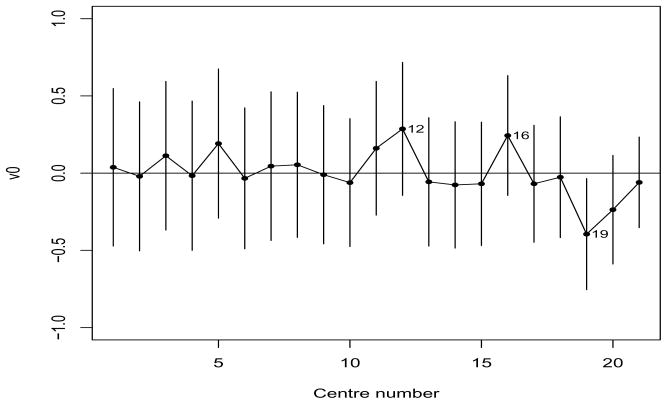
where I denotes individual random effect. Now, M10 is the full model which combines models M3 and M6 and the others are various simplifications of it as before, i.e. M9 (ρ = 0), M8 (vi0 = 0), M7 (vi1 = 0) and M6 (vi0 = 0, vi1 = 0). Note that M6 has independence between the survival times within centres. However, comparing model M6 with M4, M6 is rejected. We also see that additional random effects vij for B, T, Indep and Corr do not lead to any improvement in deviances. Here, the again rejects the additional complexity implied by models M7–M10. Thus, for the bladder-cancer data set the focussed AIC chooses M4 as the best model among those considered. Under M4 the predicted random effects (i.e. random baseline risks) and 95% prediction intervals for each centre are plotted in Figure 3. It shows substantial variations in the baseline risk over centre as evident in Figure 2(a). In particular, the three centres (12, 16) and 19 stand out as having the highest and lowest baseline risks, respectively. Note that although we report the SEs of the σ2s, one should not use them for testing σ2 = 0 [13]. Now we are also interested in testing the hypothesis , no centre effect (i.e. no variation in random-baseline risk). Such a null hypothesis is on the boundary of the parameter space, so that the critical value of an asymptotic distribution is 2.71 at 5% significant level [25, 39, 40]. The difference in deviance (−2pτ (h*) in Table 3) between M1 and M4 is 3.2(> 2.71), indicating that the centre effect is significant, i.e. .
Figure 3.
Random baseline risks (vi0) of 21 centres in the bladder cancer data and their 95% prediction intervals, under the final model (M4). Centres are sorted in increasing order of number of patients.
5. Simulation study
Numerical studies, using 200 replications of simulated data, were conducted to evaluate the performance of the proposed method. Here we consider the two interesting models (2) and (3), which correspond to M4 and M3, respectively. The structure of bladder-cancer data in Table 1 is assumed in order to generate the data from each model. That is, the simulated data structures consist of the total patients n = 410 coming from 21 centres, with the number (ni) of different patients.
Firstly, data are generated from the model (2) with λ0(t) = 1 and the two different binary covariates, the main treatment xij1 and xij2;
Here xij1 and xij2 are generated from a Bernoulli distribution with success probability 0.5, respectively. The corresponding true parameters are β1 = −0.5 and β2 = 0.5. The random effects vi0 are also generated from with and 1.0. The corresponding censoring times were, respectively, generated from exponential distribution with parameter values empirically determined to achieve approximately the right censoring rate in each centre of Table 1.
For the 200 replications we computed the mean, standard deviation (SD), the mean of the estimated SE for β̂j (j = 1, 2) and , respectively. The corresponding SEs are, respectively, obtained from J−1 in (7) and {−∂2pτ (h*)/∂θ2}−1 in (B2). The results of fitting the model (2) are summarized in Table 4. Here, to save space we report only the results about which give similar results to . Overall, the h-likelihood estimates of βj and perform well even though the simulated data consist of somewhat high censoring. In Table 4 SD is the estimates of the true {var(ξ̂)}1/2, and SEM is the average of SE estimates for ξ̂, where . Our SE estimates work well as judged by the very good agreement between SEM and SD.
Table 4.
Simulation results for the estimation of fixed parameters over 200 replications under random baseline (M4) and correlated frailty models (M3) when the true distribution of log-frailty is correctly specified or misspecified (i.e. a mixture of two bivariate normals).
| Fitted model | Setting | Parameter | True | Mean | SD | SEM | |
|---|---|---|---|---|---|---|---|
| M4 | Correct |
|
|||||
| β̂1 | −0.5 | −0.505 | 0.150 | 0.150 | |||
| β̂2 | 0.5 | 0.504 | 0.156 | 0.149 | |||
|
|
1 | 1.005 | 0.426 | 0.418 | |||
|
| |||||||
| M3 | Correct | , σ01 = −0.1 | |||||
| β̂1 | −0.5 | −0.506 | 0.191 | 0.186 | |||
| β̂2 | 0.5 | 0.494 | 0.153 | 0.148 | |||
|
|
0.2 | 0.211 | 0.138 | 0.138 | |||
|
|
0.2 | 0.212 | 0.221 | 0.209 | |||
| σ̂01 | −0.1 | −0.104 | 0.135 | 0.133 | |||
| ρ̂ | −0.5 | −0.493 | – | – | |||
| Correct | , σ01 = −0.5 | ||||||
| β̂1 | −0.5 | −0.480 | 0.261 | 0.283 | |||
| β̂2 | 0.5 | 0.502 | 0.155 | 0.153 | |||
|
|
1 | 1.021 | 0.472 | 0.457 | |||
|
|
1 | 1.029 | 0.569 | 0.559 | |||
| σ̂01 | −0.5 | −0.519 | 0.398 | 0.397 | |||
| ρ̂ | −0.5 | −0.506 | – | – | |||
| Misspecified | , σ01 = −0.1 | ||||||
| β̂1 | −0.5 | 0.502 | 0.198 | 0.185 | |||
| β̂2 | 0.5 | 0.488 | 0.155 | 0.148 | |||
|
|
0.2 | 0.209 | 0.140 | 0.144 | |||
|
|
0.2 | 0.208 | 0.211 | 0.205 | |||
| σ̂01 | −0.1 | −0.102 | 0.139 | 0.138 | |||
| ρ̂ | −0.5 | −0.490 | – | – | |||
| Misspecified | , σ01 = −0.5 | ||||||
| β̂1 | −0.5 | −0.476 | 0.309 | 0.293 | |||
| β̂2 | 0.5 | 0.512 | 0.156 | 0.153 | |||
|
|
1 | 1.048 | 0.483 | 0.461 | |||
|
|
1 | 1.089 | 0.600 | 0.588 | |||
| σ̂01 | −0.5 | −0.525 | 0.415 | 0.404 | |||
| ρ̂ | −0.5 | −0.491 | – | – | |||
SD, standard deviation of estimates over 200 simulations, is defined by {Σi (κ̂(i) − κ̄)2/199}1/2, where κ̂(i) is the estimate of κ in the ith replication and κ̄ = Σi κ̂(i)/200 is the mean of κ̂(i)’s, and , or σ01.
SEM, the mean of estimated standard errors over 200 simulations.
Next, data were generated from the model (3) with λ0(t) = 1:
| (11) |
The random effects vi0 and vi1 are generated from the bivariate normal distribution with four combinations of frailty parameters; and (1.0, 1.0, 0.5), leading to σ01 = −0.1, −0.5, 0.1, 0.5, respectively. The remaining simulation schemes including (xij1, xij2) and (β1, β2) are the same as before. The results of fitting the model (3) with ρ = −0.5 are also given in Table 4. Though not reported here, we found the similar results for ρ = 0.5. Overall, our approach again works well. However, the estimates of the frailty parameters ( , σ01) are slightly biased when the variances are large as in .
In addition, we investigated the performance of our h-likelihood procedure when the normal assumption of log-frailties vi0 and vi1 in (11) is violated. For linear mixed models Ha et al. [41] and Verbeke and Lesaffre [42] have shown that misspecifying the normal random-effect distribution has little effect on the fixed effects estimates. Following Verbeke and Lesaffre [42], for simplicity we consider a mixture (Johnson and Kotz, [43, p.73]) of two bivariate normal distributions. That is, vi = (vi0, vi1)T are generated from one of the following two cases:
Two non-normal distributions with Cases 1 and 2 have been chosen such that E(vi) = 0 and such that var(vi) equals the random-effect variance-covariance parameters in the first and second settings given in M3 of Table 4, respectively. Note that Cases 1 and 2 produce unimodal and bimodal distributions, respectively (not shown). The results in the third and fourth settings given in M3 of Table 4 again confirm that the h-likelihood method gives robust results for the estimation of parameters, particularly for β, when the distribution of frailty is misspecified.
The SAS/IML program for a correlated model (11) with a simulated data set is available from the website: http://stat.snu.ac.kr//hglmlab.
6. Discussion
We have shown that the proposed method provides a unified framework for the inference. The data-directed simulation results have demonstrated that our procedure performs well for the estimation of parameters, including the estimated SEs. Using h-likelihood, we have also shown how to investigate potential sources of the heterogeneity related to treatment effect over centres in multi-centre clinical trial. The proposed method can be also employed when studying such heterogeneity in a meta-analysis [8] which combines survival data from different clinical trials.
The heterogeneity of treatment effect could also arise in other situations besides treatment-by-centre interaction. For example, it could arise in the case that the treatment effect affects the variances of the frailty terms [44]; a simple dispersion model is a model (2) with allowing a regression model for , given by where xij1 is a main treatment covariate. Pan and MacKenzie [45, 46] have developed appropriate structural dispersion methods for testing this hypothesis in the repeated measures setting with Gaussian response variables and with and without random effects. Thus, we are currently working on an extension of our method to models with structural dispersion.
In the data set in Section 4 we coded the main treatment as xij1 = 0, 1 to indicate control or treatment group. However, the coding of may give a flexible covariance structure for the random effects [6, 8]. Though not reported here, both codings give similar estimation results for all random-effect models (M2–M10) considered and select M4 as the best model. Furthermore, we investigated how the small size (e.g. ni = 3) of some centres in Table 1 influences the inference results. Here, the centres, centre numbers 1 and 2, with less than 4 patients were combined into one new centre. We have also observed (not shown) that the results obtained from fitting a correlated model (M3) under the combined data set are very similar to those of M3 in Table 2.
The focussed in Section 4.2 is a criterion for the frailty parameters only and it cannot be used for model selection involving the β parameter because the restricted likelihood eliminates the β. However, if β is the subject of the model selection process we may use the AIC based upon an adjusted profile h-likelihood pv(hp) in (C1) [11]. Thus, there is clearly scope for further research on the development of a criterion for selecting the best model globally. We have ignored missing covariates in the data set analysed because their frequency is too small (i.e. 4/414=1%), but the original data set with 7 studies includes more missing covariates. The development of h-likelihood methods for frailty models allowing for missing covariates would be an interesting topic for future work.
Acknowledgments
The authors thank the European Organization for Research and Treatment of Cancer Genito-Urinary Tract Cancer Group for permission to use the data from EORTC trial 30791 for this research. This work was supported by the Korea Research Foundation Grant funded by the Korean Government (KRF-2008-521-C00057). Professor MacKenzie was supported by the BIO-SI project (SFI 07/MI/012) and was funded wholly by ENSAI, Rennes, France when this paper was completed. This publication was also supported by grants number 5U10 CA011488-38 through 5U10 CA011488-39 from the National Cancer Institute (Bethesda, Maryland, USA) and by the EORTC Charitable Trust. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute. Professor Legrand was supported by IAP research network grant nr. P6/03 of the Belgian government (Belgian Science Policy).
Appendix A
H-likelihood estimation procedure
With h* in (6) we estimate the fixed parameters (β, θ) and random effects v as follows. Ha et al. [23] further showed that given θ the estimation of τ = (βT, vT)T is obtained by solving
| (A1) |
Here the first partial derivatives, ∂h/∂τ, are given by the simple forms:
where μij = Λ0(yij) exp(ηij). Next, for the estimation of the frailty parameters θ, we use Lee and Nelder’s [21] adjusted profile h-likelihood [18] which eliminates (β, v) from h*, defined by
| (A2) |
where τ̂ = τ̂(θ) = (β̂T(θ), v̂T(θ))T and J(h*; τ) = −∂2h*/∂τ2 is an information matrix for τ with a detailed form in (7). The restricted maximum likelihood (REML) estimator for θ are obtained by solving iteratively
| (A3) |
Note here that
where Σ = BD(Σ1, …, Σq) is the q × q block diagonal matrix and Ĵ = Ĵ(θ) = J(h*; τ)|τ=τ̂(θ). Note also that in implementing (A3) we allow the ∂v̂/∂θ term [18, 24, 26]; the computations of the ∂Ĵ/∂θ term including the ∂v̂/∂θ term are given in Appendix B.
In summary, the estimates of τ and θ are obtained by alternating between the two estimating equations (A1) and (A3) until convergence is achieved [18, 23]. The two equations are, respectively, solved using the Newton-Raphson method with the corresponding Hessian matrices, −∂2h*/∂τ2 and −∂2pτ (h*)/∂θ2. After convergence, we directly compute the estimates of var(τ̂ − τ) and var(θ̂) using the inverses of −∂2h*/∂τ2 and −∂2pτ (h*)/∂θ2, respectively.
Appendix B
The computation of −∂2pτ (h*)/∂θ2
The adjusted profile h-likelihood in (A2) can be expressed as
where τ = (βT, vT)T, ĥ = h*|τ= τ̂ (θ) = h*(τ̂(θ), θ) and Ĵ = J(h*; τ)|τ= τ̂(θ) = J(τ̂ (θ), θ). Since
| (B1) |
we have
| (B2) |
We now show how to compute equation (B2). Following Lee and Nelder [22] and Ha and Lee [18], we allow for ∂v̂/∂θr in computing the two equations, (B1) and (B2), but not for ∂β̂/∂θr. Then we have
since (∂h*/∂v)|v = v̂ = 0: see also Appendix 2 of Ha et al. [23]. Along the lines of Appendix C of Lee and Nelder [21], we can show that
where Ŵ is given in (B3), U = Σ−1 and . From these results the first term on the right hand side (RHS) of (B2) becomes
where . From (7) we have
| (B3) |
where Ŵ = W*|τ= τ̂(θ) = W *(τ̂(θ), θ). Note here that following Appendix B of Ha and Lee [18], W* = W*(β, v) is given by
| (B4) |
where W1 = diag{Λ̂0ij exp(ηij)} is the n × n diagonal matrix with Λ̂0ij = Λ̂0(yij) and W2 = (W3M)C−1(W3M)T is the n × n symmetric matrix. Here W3 = diag{exp(ηij)}, is the D × D diagonal matrix, and M = (M1, …, MD)T is the n × D indicator matrix whose (ij, k)th element is 1 if yij ≥ y(k) and 0 otherwise. Notice that Λ̂0ij and λ̂0k also depend on (β, v) only and that the corresponding matrix forms are available in Ha and Lee [18]. Thus, the two derivatives in the second term on the RHS of (B2) are computed as follows:
Here and are calculated by the following procedures.
since ∂W*/∂θr = 0, and
where
and ∂W*/∂v and ∂2W*/∂v2 can be calculated by repeatedly differentiating (B4) with respect to v.
Appendix C
Comparison of different estimation methods
Ha et al. [19, 23] have showed that the profile h-likelihood h* in (6) is proportional to the penalized partial likelihood hp [PPL, 17], which uses the partial likelihood [47–48] for ℓ1ij in h; h* = hp + constant. The h-likelihood and PPL procedures are the same for the estimation of β and v, given frailty parameters θ, but are different for that of θ. For the estimation of θ, the h-likelihood method uses the restricted likelihood pβ,v(h*), whereas the PPL method uses an adjusted profile h-likelihood
| (C1) |
where J(hp; v) = −∂2hp/∂v2, which is a Laplace approximation to the marginal likelihood [19]; notice that pv(hp) − pv(h*) = constant. However, the PPL ignores the ∂v̂/∂θ term in solving the score equations ∂pv(hp)/∂θ = 0; this leads to an underestimation of the parameters and/or SEs, particularly when the cluster size ni is small [14, 18, 19].
Recently, the penalized maximum likelihood approach [8], which penalizes the baseline hazard λ0(t) in marginal likelihood, has been proposed for the inference of parameters, but it can not directly use for inference of frailties because it eliminates them by integration as in standard marginal-likelihood approach [13, 34]. Furthermore, Bayesian approaches [4, 7] have been also suggested. Legrand et al. [4] proposed a Bayesian approach using a Laplace integration technique to approximate the marginal posterior density, π(θ|y, δ); it can be shown that under uniform priors (i.e. flat priors) for β and θ, log{π(θ|y, δ)} ≃ pβ,v(h*). Thus, we see that the h-likelihood method is equivalent to Legrand et al.’s method under uniform priors - a choice, however, which is unlikely to be adopted in practice by Bayesians. Komarek et al. [7] also proposed to use a Markov chain Monte Carlo algorithm but in an accelerated failure time model with Gaussian random effects.
Contributor Information
Il Do Ha, Email: idha@dhu.ac.kr, Department of Asset Management, Daegu Haany University, Gyeongsan, 712-715, South Korea.
Richard Sylvester, Email: richard.sylvester@eortc.be, European Organisation for Research and Treatment of Cancer, Brussels, Belgium.
Catherine Legrand, Email: catherine.legrand@uclouvain.be, Institut de statistique, biostatistique et sciences actuarielles (ISBA), Université catholique de Louvain, Louvain-la-neuve, Belgium.
Gilbert MacKenzie, Email: gibert.mackenzie@ul.ie, CREST, ENSAI, Rennes, France and Centre for Biostatistics, University of Limerick, Ireland.
References
- 1.Andersen PK, Klein JP, Zhang M-J. Testing for centre effects in multi-centre survival studies: a monte carlo comparison of fixed and random effects tests. Statistics in Medicine. 1999;18:1489–1500. doi: 10.1002/(sici)1097-0258(19990630)18:12<1489::aid-sim140>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi T, Ohashi Y. Investigating centre effects in a multi-centre clinical trial of superficial bladder cancer. Statistics in Medicine. 1999;18:1961–1971. doi: 10.1002/(sici)1097-0258(19990815)18:15<1961::aid-sim170>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Glidden DV, Vittinghoff E. Modelling clustered survival data from multicentre clinical trials. Statistics in Medicine. 2004;23:369–388. doi: 10.1002/sim.1599. [DOI] [PubMed] [Google Scholar]
- 4.Legrand C, Ducrocq V, Janssen P, Sylvester R, Duchateau L. A Bayesian approach to jointly estimate centre and treatment by centre heterogeneity in a proportional hazards model. Statistics in Medicine. 2005;24:3789–3804. doi: 10.1002/sim.2475. [DOI] [PubMed] [Google Scholar]
- 5.Gray RJ. A Bayesian analysis of institutional effects in multicenter cancer clinical trial. Biometrics. 1994;50:244–253. [PubMed] [Google Scholar]
- 6.Turner RM, Omar RZ, Yang M, Goldstein H, Thompson SG. A multilevel model framework for meta-analysis of clinical trials with binary outcomes. Statistics in Medicine. 2000;19:3417–3432. doi: 10.1002/1097-0258(20001230)19:24<3417::aid-sim614>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Komarek A, Lesaffre E, Legrand C. Baseline and treatment effect heterogeneity for survival times between centers using a random effects accelerated failure time model with flexible error distribution. Statistics in Medicine. 2007;26:5457–5472. doi: 10.1002/sim.3083. [DOI] [PubMed] [Google Scholar]
- 8.Rondeau V, Michiels S, Liquet B, Pignon JP. Investigating trial and treatment heterogeneity in an individual patient data meta-analysis of survival data by means of the penalized maximum likelihood approach. Statistics in Medicine. 2008;27:1894–1910. doi: 10.1002/sim.3161. [DOI] [PubMed] [Google Scholar]
- 9.Yau KKW, Kuk AYC. Robust estimation in generalized linear mixed models. Journal of the Royal Statistical Society, Series B. 2002;64:101–117. [Google Scholar]
- 10.Song X-Y, Lee S-Y. Model comparison of generalized linear mixed models. Statistics in Medicine. 2006;25:1685–1698. doi: 10.1002/sim.2318. [DOI] [PubMed] [Google Scholar]
- 11.Ha ID, Lee Y, MacKenzie G. Model selection for multi-component frailty models. Statistics in Medicine. 2007;26:4790–4807. doi: 10.1002/sim.2879. [DOI] [PubMed] [Google Scholar]
- 12.Friedman LM, Furberg CD, DeMets DL. Fundamentals of clinical trials. Springer; New York: 1998. [Google Scholar]
- 13.Vaida F, Xu R. Proportional hazards model with random effects. Statistics in Medicine. 2000;19:3309–3324. doi: 10.1002/1097-0258(20001230)19:24<3309::aid-sim825>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Gamst A, Donohue M, Xu R. Asymptotic properties and empirical evaluation of the NPMLE in the proportional hazards mixed-effects model. Statistica Sinica. 2009;19:997–1011. [Google Scholar]
- 15.Abrahantes JC, Legrand C, Burzykowski T, Janssen P, Ducrocq V, Duchateau L. Comparison of different estimation procedures for proportional hazards model with random effects. Computational Statistics and Data Analysis. 2007;51:3913–3930. [Google Scholar]
- 16.Duchateau L, Janssen P. The Frailty Models. Springer; New York: 2008. [Google Scholar]
- 17.Ripatti S, Palmgren J. Estimation of multivariate frailty models using penalized partial likelihood. Biometrics. 2000;56:1016–1022. doi: 10.1111/j.0006-341x.2000.01016.x. [DOI] [PubMed] [Google Scholar]
- 18.Ha ID, Lee Y. Estimating frailty models via Poisson hierarchical generalized linear models. Journal of Computational and Graphical Statistics. 2003;12:663–681. [Google Scholar]
- 19.Ha ID, Noh M, Lee Y. Bias reduction of likelihood estimators in semi-parametric frailty models. Scandinavian Journal of Statistics. 2010;37:307–320. [Google Scholar]
- 20.Othus M, Li Y. Marginalized frailty models for multivariate survival data. Harvard University Biostatistics Working Paper Series, paper. 104 http://www.bepress.com/harvardbiostat/paper104.
- 21.Lee Y, Nelder JA. Hierarchical generalized linear models (with discussion) Journal of the Royal Statistical Society, Series B. 1996;58:619–678. [Google Scholar]
- 22.Lee Y, Nelder JA. Hierarchical generalised linear models: a synthesis of generalised linear models, random-effect models and structured dispersions. Biometrika. 2001;88:987–1006. [Google Scholar]
- 23.Ha ID, Lee Y, Song J-K. Hierarchical likelihood approach for frailty models. Biometrika. 2001;88:233–243. [Google Scholar]
- 24.Ha ID, Lee Y. Comparison of hierarchical likelihood versus orthodox best linear unbiased predictor approaches for frailty models. Biometrika. 2005;92:717–723. [Google Scholar]
- 25.Ha ID, Lee Y. Multilevel mixed linear models for survival data. Lifetime Data Analysis. 2005;11:131–142. doi: 10.1007/s10985-004-5644-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y, Nelder JA, Pawitan Y. Generalised Linear Models with Random Effects: unified analysis via h-likelihood. Chapman and Hall; London: 2006. [Google Scholar]
- 27.Sylvester R, van der Meijden APM, Oosterlinck W, Witjes J, Bouffioux C, Denis L, Newling DWW, Kurth K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. European Urology. 2006;49:466–477. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Yau KKW. Multilevel models for survival analysis with random effects. Biometrics. 2001;57:96–102. doi: 10.1111/j.0006-341x.2001.00096.x. [DOI] [PubMed] [Google Scholar]
- 29.Yau KKW, McGilchrist CA. ML and REML estimation in survival analysis with time dependent correlated frailty. Statistics in Medicine. 1998;17:1201–1213. doi: 10.1002/(sici)1097-0258(19980615)17:11<1201::aid-sim845>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 30.McGilchrist CA, Aisbett CW. Regression with frailty in survival analysis. Biometrics. 1991;47:461–466. [PubMed] [Google Scholar]
- 31.Hougaard P. Analysis of multivariate survival data. Springer; New York: 2000. [Google Scholar]
- 32.Longford NT. Random coefficient models. Oxford University Press; New York: 1993. [Google Scholar]
- 33.Breslow NE. Discussion of Professor Cox’s paper. Journal of the Royal Statistical Society, Series B. 1972;34:216–217. [Google Scholar]
- 34.Nielsen GG, Gill RD, Andersen PK, Sørensen TIA. A counting process approach to maximum likelihood estimation in frailty models. Scandinavian Journal of Statistics. 1992;19:25–44. [Google Scholar]
- 35.Lee Y, Nelder JA. Extended-REML estimators. Journal of Applied Statistics. 2003;30:845–856. [Google Scholar]
- 36.Lee Y, Ha ID. Orthodox BLUP versus h-likelihood methods for inferences about random effects in Tweedie mixed models. Statistics and Computing. 2010;20:295–303. [Google Scholar]
- 37.Booth JG, Hobert JP. Standard errors of prediction in generalized linear mixed models. Journal of the American Statistical Association. 1998;93:262–272. [Google Scholar]
- 38.Sakamoto Y, Ishiguro M, Kitagawa G. Akaike information criterion statistics. KTK Scientific Publisher; Tokyo, Japan: 1986. [Google Scholar]
- 39.Self SG, Liang KY. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. Journal of the American Statistical Association. 1987;82:605–610. [Google Scholar]
- 40.Stram DO, Lee JW. Variance components testing in the longitudinal mixed effects model. Biometrics. 1994;50:1171–1177. [PubMed] [Google Scholar]
- 41.Ha ID, Lee Y, Song J-K. Hierarchical-likelihood approach for mixed linear models with censored data. Lifetime Data Analysis. 2002;8:163–176. doi: 10.1023/a:1014839723865. [DOI] [PubMed] [Google Scholar]
- 42.Verbeke G, Lesaffre E. The effect of misspecifying the random-effects distribution in linear mixed models for longitudinal data. Computational Statistics and Data Analysis. 1997;23:541–556. [Google Scholar]
- 43.Johnson NL, Kotz S. Continuous multivariate distributions. John Wiley & Sons; New York: 1972. [Google Scholar]
- 44.Noh M, Ha ID, Lee Y. Dispersion frailty models and HGLMs. Statistics in Medicine. 2006;25:341–1354. doi: 10.1002/sim.2284. [DOI] [PubMed] [Google Scholar]
- 45.Pan JX, MacKenzie G. Regression models for covariance structures in longitudinal studies. Statistical Modelling. 2006;6:43–57. [Google Scholar]
- 46.Pan JX, MacKenzie G. Modelling conditional covariance in the linear mixed model. Statistical Modelling. 2007;7:49–71. [Google Scholar]
- 47.Cox DR. Regression models and life tables (with Discussion) Journal of the Royal Statistical Society, Series B. 1972;74:187–220. [Google Scholar]
- 48.Breslow NE. Covariance analysis of censored survival data. Biometrics. 1974;30:89–99. [PubMed] [Google Scholar]