Abstract
The FoxM1 transcription factor gene is over-expressed in cancer. Its expression is stimulated by oncogenic signaling pathways and reactive oxygen species. It is also a target of regulation by the tumor suppressor genes. The transcriptional activity of FoxM1 depends upon activation by the Cyclin/Cdks as well as Plk1. FoxM1 stimulates expression of several genes involved in the cell cycle progression. Moreover, it supports proliferation of tumor cells by stimulating expression of the antioxidant genes and reducing oxidative stress. A new study provided evidence that FoxM1, in the absence of its inhibitor, the tumor suppressor Arf, drives metastasis of hepatocellular carcinoma (HCC). It induces an EMT-like phenotype in HCC cells, increases cell-migration and induces pre-metastatic niche at the distal organ of metastasis. FoxM1 directly activates genes involved in multiple steps of metastasis. In this review, we discuss the evidence for a master regulatory role of FoxM1 in tumor metastasis.
FoxM1 belongs to a large family of forkhead box (Fox) transcription factors. Unlike the other Fox-transcription factors, FoxM1 is associated with cell-proliferation, and is expressed only in proliferating cells (1,2). In adult mammals, FoxM1 expression is detected mainly in the progenitor and regenerating tissues, and it is over-expressed in various human malignancies. For example, gene expression profiles in carcinomas, including prostate, breast, lung, ovary, colon, pancreas, stomach, bladder, liver and kidney revealed that FoxM1 is over-expressed in all carcinomas (3). Also, high expression of FoxM1 in glioblastoma correlates with the tumorigenicity of the glioma cells (4). Moreover, in breast cancer, over-expression of FoxM1 strongly correlates with poor prognosis (5). Over-expression of FoxM1 in various tumors indicates a strong dependence of the tumor cells on FoxM1, and that is explained partly by its role in cell proliferation.
FoxM1 plays important roles in cell cycle progression (1,2). FoxM1 stimulates expression of Skp2 and Cks1, which are involved in the proteolysis of p27Kip1 and G1/S progression (1). FoxM1 also stimulates expression of a number of genes that are critical for the G2/M progression. Included are Plk1, Aurora B, Cyclin B1, CDC25B, CENP-A and Survivin (1). Therefore, it is not surprising that FoxM1 expression is restricted to proliferating cells. Interestingly, FoxM1 itself is regulated during the cell cycle. The transcriptional activation function of FoxM1 depends upon phosphorylation by Cyclin/Cdks and by the Plk1 kinase. FoxM1 is phosphorylated in the C-terminal activation domain by Cyclin/Cdks, which serves as priming phosphorylation for further phosphorylations by Plk1 (6,7). Mutations of the Cyclin/Cdk or the Plk1 phosphorylation sites render FoxM1 transcriptionally inactive (6,7). The transcriptionally active, phosphorylated-FoxM1 accumulates as the cells progress through the cycle (6–8). At the end of M-phase, FoxM1 becomes dephosphorylated (6), and in early G1 phase of the next cycle it is polyubiquitinated by APC/C-Cdh1 for degradation by the proteasome (8). The degradation of FoxM1 in the early G1 phase is important for regulated entry into S phase (8). Thus, in proliferating cells FoxM1 is synthesized and degraded in every cycle of cell division. Synthesis of FoxM1 in early G1 phase or during a transition from G0 to G1 phase is stimulated by growth factors(8,9).
FoxM1 expression is induced also by oncogenes (Fig. 1 and ref. 9). For example, activated RAS increases expression of FoxM1 and the increase in FoxM1 expression is critical for RAS-induced transformation. RAS increases expression of FoxM1 by inducing the cellular levels of the reactive oxygen species (ROS) (9). In fact, ROS alone was shown to activate expression of FoxM1 (9). Following induction by ROS, FoxM1 functions in a negative feed back loop to attenuate the levels of ROS by stimulating expression of the antioxidant genes Superoxide Dismutase (MnSOD), Catalase and Peroxiredoxin 3 (PRDX3) (9). This ROS-regulatory function of FoxM1 protects proliferating normal or tumor cells from oxidative stress and promotes survival(Fig. 1). Consistent with that notion, tumor cells expressing ROS-inducing oncogenes (such as RAS or Akt) are addicted to FoxM1 for their survival (9). Moreover, the tumor cells over-expressing FoxM1 are resistant to apoptosis or premature senescence induced by oxidative stress, which has strong implications in resistance to chemotherapy. In that regard, it is noteworthy thatFoxM1 over-expression in breast cancer cells was shown to confer resistance to Cisplatin, Herceptin and Paclitaxel (10,11). Interestingly, those studies indicated additional pathways through which FoxM1 over-expression confers drug-resistance.
Fig. 1.
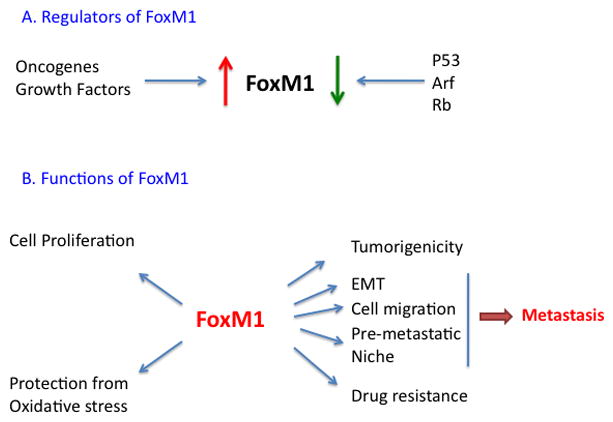
A, schematic diagram indicating FoxM1 expression is stimulated by oncogenes and growth factors and inhibited by p53. Rb and p19Arf inhibit activity of FoxM1. B, schematic diagram indicating FoxM1 stimulates expression of genes involved in cell division, attenuation of oxidative stress, tumorigenicity and drug resistance. A new study (20) demonstrated that FoxM1 could stimulate expression of genes involved in various steps of tumor metastasis, including epithelial to mesenchymal-like transition, cell-migration and pre-metastatic niche formation.
The functions of FoxM1 in expression of the cell-division genes, and in attenuation of oxidative stress are significant for cancer development and progression. Consistent with that, expression and the transcriptional activity of FoxM1 are regulated by the tumor suppressor genes (Fig. 1). For example, expression of FoxM1 is regulated by p53 (12,13). It was suggested that the G2/M checkpoint function of p53 relies on inhibition of FoxM1 expression (12). FoxM1 is regulated by p19Arf. P19Arf binds to FoxM1 and re-localizes FoxM1 to the nucleolus, thereby, inhibiting expression of the FoxM1 activated genes (14). The regulation of FoxM1 by p19Arf is significant as there is new genetic evidence, discussed below, that p19Arf inhibits tumor metastasis induced by FoxM1.
Metastasis of tumor involves a series of interrelated events (See ref. 15 for a comprehensive review). Briefly, the initial steps involve vascularization of the primary tumor for aggressive growth through secretion of angiogenic factors, increased motility and invasion of the tissue stroma through secretion the matrix metalloproteinases and other changes in the tumor cells, such as epithelial to mesenchymal like transition. The invasive tumor cells penetrate the blood vessels (intravasation) to enter the circulation or migrate through the lymphatic channels.
The tumor cells also associate with bone marrow derived cells, endothelial cells, stromal cells and others, which provide a supportive microenvironment for the tumor cells. The circulating tumor cells extravasate into the parenchyma of a distal organ where they undergo metastatic growth. Interestingly, several in vitro studies on FoxM1 implicated its involvement in the early steps of metastasis. For example, FoxM1 was shown to stimulate invasion and angiogenesis of pancreatic cancer cells through induction of matrix metalloproteinase genes MMP-2 and MMP-9, as well as vascular endothelial growth factor (VEGF) (16). Similar functions of FoxM1 in stimulating expression of the MMP genes were described also in glioma (17). Moreover, over-expression of FoxM1 coincides with metastasis of prostate cancer (18). However, a direct in vivo evidence for a role of FoxM1 in tumor metastasis was lacking. That evidence came from in vivo studies of FoxM1 in hepatocellular carcinomas.
Role of FoxM1 in hepatocellular cancer development was studied using a mouse strain (FoxM1 fl/fl) in which the FoxM1 alleles were floxed. The FoxM1 alleles were specifically deleted in the adult liver by mating the mice with a transgenic strain that expresses Cre recombinase under the control of albumin promoter (14). Interestingly, deletion of FoxM1 had very little effect on the survival of the mice, indicating the FoxM1 function is not critical for the normal hepatocytes (14). However, when the mice were subjected to diethylnitrosamine (DEN)/phenobarbital (PB) liver carcinogenesis protocol, a well established carcinogenesis protocol for liver cancer in which mice develop liver cancer (hepatocellular carcinoma, HCC) by 9 to12 months with high penetrance, the mice harboring deletion of the FoxM1 alleles in the liver did not develop HCC. The observations demonstrated an essential role of FoxM1 in HCC development (14). Moreover, when FoxM1 was deleted after development of HCC, there were significant decreases in the sizes of HCC, suggesting that FoxM1 is a potential molecular target for HCC therapy. Gusarova et al. (19) extended the observations further by using a cell-penetrating form of a peptide derived from p19Arf that was previously shown to inhibit FoxM1 (14). Residues between 26 and 44 of p19Arf when injected in mice bearing HCC induced apoptosis of the HCC cells without having a significant effect on the neighboring normal hepatocytes.
While FoxM1 is essential for HCC development, over-expression of FoxM1 alone did not have significant effect on HCC development (20 and references therein). Therefore, Park et al. (20) decided to study the effect of FoxM1 over-expression in the absence of p19Arf, a potent inhibitor of FoxM1. These authors generated a bi-transgenic strain (FoxM1bTg;Arf−/−) in which FoxM1 was expressed from the Rosa26 promoter in Arf −/− background. When that strain was subjected to DEN/PB liver carcinogenesis protocol, the mice developed very aggressive HCC. More interestingly, the HCC in the FoxM1bTg;Arf−/− background, unlike the single transgenics, were highly metastatic. Over 70% of the HCC in FoxM1bTg;Arf−/−mice exhibited metastasis to the lung. The extent of metastasis was reduced significantly when one copy of Arf was present (FoxM1bTg;Arf+/−mice), indicating that p19Arf inhibits FoxM1-induced metastasis. The authors also demonstrated that ectopic expression of FoxM1 in Arf−/− HCC cells, which were non-metastatic, induced metastatic ability in experimental metastasis assays (20).
The mechanistic studies by Park et al. suggested that FoxM1 could function as a master activator of metastasis, as it induced various steps of metastasis (Fig. 1 and ref. 20). For example, FoxM1-induced metastasis of HCC involved epithelial to mesenchymal-like transition (EMT-like) of the HCC cells. Also, ectopic expression of FoxM1 in cells expressing lower levels of Arf could induce EMT-like changes (20). There was a loss of E-cadherin expression and that was accompanied by an increase in the level Snail, a repressor of E-cadherin expression. The EMT-like changes could be related to increased activation of the Akt-signaling pathway in HCC because Akt-pathway has been shown to stabilize Snail (20 and references therein). A recent study using E-cadherin promoter-luciferase construct indicated that expression of FoxM1 could activate transcription driven by the E-cadherin promoter (21). To explain the apparent discrepancy with the observation by Park et al. (20), the authors of that study suggested that the Snail-mediated repression of E-cadherin and other mechanisms might be dominant in tumor cells, as they studied the E-cadherin promoter activity in normal kidney cells. Also, the level of Arf could be a factor. Clearly, further analyses on the endogenous promoter will be required to resolve the basis of the discrepancy. Nevertheless, in addition to EMT-like changes, expression of FoxM1 increased cell-migration. Interestingly, FoxM1 was shown to transcriptionally activate Stathmin, which increases cell motility by destabilizing microtubules. In the HCC cells, FoxM1 increased expression of VEGF, an activator of angiogenesis(20). In addition to activating the mechanisms that allow HCC cells to escape the primary tumor sites, FoxM1 stimulated pathways that are involved in pre-metastatic niche formation (20 and references therein). It was shown that FoxM1 could bind to the promoters of lysyl oxidase (LOX) and lysyl oxidase-like 2 (LOXL2) and could stimulate their expression. LOX and LOXL2 were shown to be involved in generating pre-metastatic niche at the distal organ of metastasis (22). Park et al. demonstrated the presence of pre-metastatic niche in the lung sections of their FoxM1bTg;Arf−/− mice harboring HCC. The non-tumorous lung sections contained Cd11b+ and c-kit+ cells and exhibited evidence for collagen deposition. Moreover, using mouse xenograft models Park et al. showed that the Arf−/− HCC cells upon over-expression of FoxM1 became highly tumorigenic, as they developed tumors when injected subcutaneously in mice. Those tumors secreted LOX and LOXL2 to induce pre-metastatic niche in the lung (20). Interestingly, inhibition of LOX or LOXL2 inhibited pre-metastatic niche and metastasis without affecting the increase in tumorigenicity by FoxM1.
HCC is one of the deadliest malignancies mainly because the current therapeutic approaches are ineffective. For eligible patients, a curative surgery is the preferred method of therapy. However, a unique feature of HCC is intrahepatic metastasis, which makes surgical intervention largely ineffective, and five-year survival following surgery remains very low (23). Therefore, understanding the mechanisms of metastasis of HCC will be important. Intrahepatic metastasis of HCC is associated with loss in the expression of E-cadherin (24). Moreover, increased expression of snail, a regulator of E-cadherin has been correlated with poor prognosis of HCC (25). In addition, over-expression of Stathmin correlates with the aggressiveness of HCC (26). Interestingly, as described above, these changes in E-cadherin, Snail and Stathmin expression were observed also in the HCC of the FoxM1bTg;Arf−/− mice. Moreover, the development of HCC in the bi-transgenic mouse model of Park et al. was associated with fibrosis of the liver, which also is observed during development of HCC in humans. Thus, the bi-transgenic mouse model developed by Park et al. many features of human HCC, and therefore, offers an excellent model to investigate the basis of poor prognosis of HCC and the mechanisms involved in the intrahepatic metastasis of HCC.
Metastasis of HCC was not observed in Arf+/+ background (20). Moreover, in Arf+/− background, the FoxM1-driven metastasis was significantly lower compared to that in Arf−/− background. Clearly, Arf is a potent inhibitor of FoxM1-induced metastasis. It is noteworthy that FoxM1 over-expression and silencing of Arf are common events in cancer. Although FoxM1 is over-expressed in HCC, the extent to which Arf is mutated or silenced in HCC is not clear. One study with 117 HCC samples provided evidence for loss of Arf expression by hypermethylation in about 42% of the samples and loss of heterozygosity in 27% of the samples (27). Also, it is possible that high-level expression of FoxM1 is able to overcome the Arf-regulation and induce development of aggressive HCC. Arf is known to regulate numerous pathways, including activation of p53 (See ref. 28 for a review). Therefore, at this point, it is unclear exactly how Arf inhibits metastasis. Interestingly, a cell-penetrating form of a peptide corresponding to residues between 26 and 44 was shown to inhibit expression of LOX, LOXL2 and Stathmin, which are activated by FoxM1. Moreover, the peptide was able to efficiently inhibit FoxM1-driven metastasis in an experimental metastasis assay (20). Therefore, it is likely that FoxM1 is the major target of Arf-regulation that leads to inhibition of metastasis. However, further studies on the FoxM1/Arfinteraction will be important in determining how Arf inhibits FoxM1-induced metastasis. It is possible that detailed studies on the FoxM1/Arf interaction will lead to development of new therapeutic approaches against aggressive HCCs.
The observations on FoxM1’s role in metastasis of HCC have strong implications on metastasis of other tumors, as over-expression of FoxM1 is a common event in cancer. For example, the correlation between FoxM1 over-expression and metastasis of prostate cancer (18) should be investigated further to establish a causal link. Also, FoxM1 over-expression in breast cancer is considered to be a biomarker for poor prognosis (5). Based on the observations by Park et al. (20), it is tempting to speculate a causal link between FoxM1 over-expression and metastasis of breast cancers. The study by Park et al. has linked FoxM1 to only a limited number of genes that have been implicated in metastasis. It is unclear how FoxM1 overcomes the inhibitory effects of the metastasis suppressor genes. Studies on metastasis suppression have identified a large number of genes (See ref. 29 for a review) that affect metastasis without having effects on the growth of the primary tumors. Also, several micro-RNA genes called metastamir have been characterized that have pro-or anti-metastatic activity (reviewed in 30). Future studies on the connections between FoxM1 over-expression and inactivation of the metastasis suppressor genes as well as those with metastamir will provide valuable insights into the mechanisms of FoxM1 that appears to be a master regulator of metastasis.
Acknowledgments
Authors apologize to colleagues whose work could not be cited because of the space constraints. The review is dedicated to the memory of Dr. RH Costa. PR is supported by US Public Health Service (PHS) Grants CA 124488 and CA 77637 and by a Merit Review Grant (IO1BX000131) from the Veteran’s Administration.
References
- 1.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 3.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–50. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 5.Bektas N, Haaf A, Veeck J, Wild PJ, Luscher-Firzlaff J, Hartmann A, et al. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer. 2008;8:42–50. doi: 10.1186/1471-2407-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YJ, Dominguez-Brauer C, Wang Z, Asara JM, Costa RH, Tyner AL, et al. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J Biol Chem. 2009;284:30695–707. doi: 10.1074/jbc.M109.007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–82. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. APC/C-Cdh1 mediated proteolysis of the forhead box transcription factor is critical for regulated entry into S phase. Mol Cell Biol. 2008;28:5162–71. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park HJ, Carr JR, Wang Z, Nogueira V, Hay N, Tyner AL, et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 2009;28:2908–18. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, et al. FoxM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–63. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandit B, Halasi M, Gartel AL. p53 negatively regulates expression of FoxM1. Cell Cycle. 2009;8:3425–7. doi: 10.4161/cc.8.20.9628. [DOI] [PubMed] [Google Scholar]
- 14.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talmadge JE, Fidler IJ. AACR Centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 2010;28:297–321. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 17.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–9. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 18.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64–85. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park HJ, Gusarova G, Wang Z, Carr JR, Li J, Kim KH, et al. Deregulation of FoxM1b leads to tumor metastasis. EMBO Molecular Medicine. 2011;3:21–34. doi: 10.1002/emmm.201000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wierstra I. The transcription factor FoxM1c binds to and transactivates the promoter of the tumor suppressor gene E-cadherin. Cell Cycle. 2011;10:760–6. doi: 10.4161/cc.10.5.14827. [DOI] [PubMed] [Google Scholar]
- 22.Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda T, Beppu T, Ishiko T, Horino K, Baba Y, Mizumoto T, et al. Intrahepatic dissemination of hepatocellular carcinoma after local ablation therapy. J Hepatobilliary Pancreat Surg. 2008;15:589–95. doi: 10.1007/s00534-007-1288-4. [DOI] [PubMed] [Google Scholar]
- 24.Inayoshi J, Ichida T, Sugitani S, Tsuboi Y, Genda T, Honma N, et al. Gross appearance of hepatocellular carcinoma reflects E-cadherin expression and early recurrence after surgical treatment. J Gastroenterol Hepatol. 2003;18:673–7. doi: 10.1046/j.1440-1746.2003.03021.x. [DOI] [PubMed] [Google Scholar]
- 25.Myoshi A, Kitajima Y, Kido S, Shimonishi T, Matsuyama S, Kitahara K, et al. Snail accelerates cancer invasion by upregulating MMP expression and is associated with poor prognosis of hepatocellular carcinoma. Br J Cancer. 2005;92:252–8. doi: 10.1038/sj.bjc.6602266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh SY, Huang SF, Yu MC, The TS, Chen TC, Lin YJ, et al. Stathmin overexpression associated with polyploidy, tumor cell invasion, early recurrence and poor prognosis in human hepatoma. Mol Carcinog. 2010;49:476–87. doi: 10.1002/mc.20627. [DOI] [PubMed] [Google Scholar]
- 27.Anzola M, Cuevas N, Lopez-Martinez M, Salz A, Burgos JJ, Martinez de Pancorboa M. P14ARF gene alterations in human hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2004;16:19–26. doi: 10.1097/00042737-200401000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 29.Hurst DR, Welch DR. Metastasis suppressor genes at the interface between the environment and tumor cell growth. Int Rev Cell Mol Biol. 2011;286:107–80. doi: 10.1016/B978-0-12-385859-7.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;19:7495–8. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]


