Abstract
Background & Aims
Drug-induced liver injury (DILI), especially from antimicrobial agents, is an important cause of serious liver disease. Amoxicillin-clavulanate (AC) is a leading cause of idiosyncratic DILI, but little is understood about genetic susceptibility to this adverse reaction.
Methods
We performed a genome-wide association study using 822,927 single-nucleotide polymorphism (SNP) markers from 201 White European and US cases of AC-DILI and 532 population controls, matched for genetic background.
Results
AC-DILI was associated with many loci in the major histocompatibility complex. The strongest effect was with a human leukocyte antigen (HLA) class II SNP (rs9274407, P=4.8×10−14), which correlated with rs3135388, a tag SNP of HLA-DRB1*1501-DQB1*0602 that was previously associated with AC-DILI. Conditioned on rs3135388, rs9274407 is still significant (P=1.1×10−4). An independent association was observed in the class I region (rs2523822, P=1.8×10−10), related to HLA-A*0201. The most significant class I and II SNPs showed statistical interaction (P=0.0015). High-resolution HLA genotyping (177 cases and 219 controls) confirmed associations of HLA-A*0201 (P=2×10−6) and HLA-DQB1*0602 (P=5×10−10), and their interaction (P=0.005). Additional, population-dependent effects were observed in HLA alleles with nominal significance. In an analysis of auto-immunerelated genes, rs2476601 in the gene PTPN22 was associated (P=1.3×10−4).
Conclusions
Class I and II HLA genotypes affect susceptibility to AC-DILI, indicating the importance of the adaptive immune response in pathogenesis. The HLA genotypes identified will be useful in studies of the pathogenesis of AC-DILI, but have limited utility as predictive or diagnostic biomarkers because of the low positive-predictive values.
Keywords: Hepatotoxicity, GWAS, pharmacogenomics, MHC, Side Effect
Introduction
Idiosyncratic liver toxicity due to a prescribed drug is usually referred to as drug-induced liver injury (DILI). Most DILI involves reactions which appear unrelated to drug dose or concentration. Though comparatively rare, DILI is a serious clinical problem with up to 10% of cases with simultaneous severe elevations in alanine transaminase (ALT) and bilirubin developing liver failure.1 A prospective study estimated the standardized incidence rate of symptomatic hepatic adverse drug reactions at 8.1 per 100,000 people in France.2 In the US, 13% of acute liver failure cases are due to idiosyncratic hepatotoxicity with 75% of those dying or requiring emergency liver transplantation.3
Amoxicillin-clavulanate (AC) is among the most commonly prescribed antimicrobials worldwide. This drug is generally well tolerated and, while liver injury can occur rarely, the overall risk benefit is favourable. DILI following AC administration (AC-DILI), which appears to be primarily due to the clavulanate component,4 is an important cause of idiosyncratic DILI in the US and Europe,5–7 and represents 17% of all DILI-related hospitalizations.6, 8 While most patients with AC-DILI make a full recovery,5, 9 cases of acute liver failure leading to death or liver transplantation have been reported.10 The mechanism of AC-DILI is unknown. Three previous studies from Northwestern Europe11–13 reported an association between AC-DILI and the human leukocyte antigen (HLA) class II allele DRB1*1501, with odds ratios (OR) ranging between 2.6 and 10. A further study of 27 Spanish cases did not observe a significant association with DRB1*15, but did report a significantly higher frequency of DQB1*06.14 The differences in findings between the Spanish and Northwestern European studies may be due to use of low-resolution genotyping, population-specific linkage-disequilibrium patterns, population stratification or to a larger proportion of hepatocellular cases in the Spanish study.15
We conducted a genome-wide association (GWA) study to investigate whether additional common genetic variants affect susceptibility to AC-DILI. A group of well-phenotyped cases (n=201) with high causality scores were assembled from several multicenter collections together with 532 genetically-matched population controls. Our study confirms the DRB1*1501 association and identifies additional HLA class I and II associations.
Materials and Methods
Case recruitment
Cases (n=211) were recruited in four separate studies (DILIGEN,16 Spanish DILI Registry,6 DILIN7 and EUDRAGENE17) which used similar inclusion criteria. All participants provided written informed consent and each study had been approved by the appropriate ethical review boards.
DILIGEN
Between October 2004 and April 2009, 78 cases of AC-DILI of European origin from centers throughout the UK were collected. Inclusion criteria were suspected liver injury due to AC with either: (a) clinically apparent jaundice or bilirubin > 2.4 mg/dL (after exclusion of cases due to hemolysis), or (b) ALT >5× upper limit of normal (ULN) or (c) alkaline phosphatase (ALP) >2× ULN plus bilirubin above ULN. Cases were identified by searching histological databases and discharge records at UK Regional Liver Units for cases of DILI or cholestasis/hepatitis of unknown etiology. Direct contact with gastroenterologists was also made by advertising the study through professional societies.
EUDRAGENE
Adult (≥18 years) AC-DILI cases identified retrospectively and prospectively from European pharmacovigilance centre adverse drug reaction reports were collected from November 2006 to August 2009. Of the 25 cases included in this study, 15 were from Spain, 7 from France and 3 from Italy.
Case definitions included (i) acute liver injury defined as presenting symptoms suggestive of liver disorder (nausea, vomiting, abdominal pain and/or jaundice) referred to a specialist or admitted to hospital, and (ii) ALT >2× ULN or (iii) aspartate aminotransferase (AST), ALP and total bilirubin >2× ULN.
Spanish DILI Registry
Fifty two cases of AC-DILI patients ≥18 years were selected from those submitted to the Spanish DILI Registry, a collaborative network set up in 1994 to prospectively identify cases of DILI in a standardized manner. Inclusion laboratory criteria for AC-DILI cases in this study were (i) clinically apparent jaundice or bilirubin > 2.4 mg/dL (after exclusion of cases due to hemolysis), or (b) ALT ≥5× ULN or (c) ALP ≥2× ULN. Detailed description of the operational structure of the registry, data recording, and case ascertainment has been reported elsewhere.6
DILIN
Details of the DILIN prospective study have been described previously.18 A total of 65 eligible AC-DILI cases were recruited between August 2004 and April 2009 from five DILIN clinical sites in the US. Of these, 56 cases of European ancestry and ≥18 years were included in the current study. Inclusion laboratory criteria were a) serum AST or ALT >5× ULN on two separate occasions, b) serum ALP >2× ULN on two consecutive occasions, or c) serum total bilirubin > 2.4 mg/dL in the absence of a competing cause of hyperbilirubinemia. Patients were excluded if there was known or suspected acetaminophen overdose, if there was a history of bone marrow or liver transplant prior to DILI onset or if there was a prior history of immune-related liver disease such as autoimmune hepatitis.
Causality assessment
Diagnosis of DILI was done by expert hepatologists at each of the collaborating centers. In addition, the cases were evaluated by application of the Council for International Organizations of Medical Science (CIOMS) scale, also called the Roussel Uclaf Causality Assessment Method (RUCAM). The pattern of liver injury was classified according to the International Consensus Meeting Criteria.19 Only cases having at least possible causality (score≥3) were included in the study.
DNA preparation from cases
For DILIGEN and Spanish DILI Registry cases, DNA was prepared as described previously.14, 16 EUDRAGENE DNA was extracted at Erasmus Medical Centre genotyping laboratory, Netherlands using standard procedures. DILIN DNA was extracted from lymphocytes and stored at the NIDDK biosample repository at Rutgers University, Piscataway, NJ.
Controls
Genotyped controls (n=532) from the Population Reference Sample (POPRES)20 were matched to the cases using principal component analysis (PCA). Based on the first two principal components that capture much of the genetic substructure among Europeans, 306 controls were selected as Northwestern Europeans (predominantly POPRES UK) and 160 controls were selected as Spanish (predominantly POPRES Spanish and Portuguese).
Genotyping
Genome-wide analysis
Genome-wide genotyping of the European DILI cases and POPRES controls was performed by Expression Analysis, Inc (Durham, NC) and of the US cases by the Center for Human Genome Variation, Duke University. All subjects were genotyped using the Illumina Human1M-Duo BeadChip, containing 1,072,820 markers. A total of 822,927 markers, 201 cases (77 DILIGEN, 19 EUDRAGENE, 49 Spanish DILI Registry and 56 DILIN) and 532 controls passed quality control (QC). (see Supplemental Materials and Methods).
HLA genotyping
High resolution genotyping of HLA-A, B, DRB1, DQA1 and DQB1 was performed on a subset of genotyped subjects, including 182 cases (74 DILIGEN, 49 Spanish DILI Registry and 59 DILIN) and 219 POPRES controls (genetically matched to cases as described above) by Histogenetics (Ossining, New York) as described in the Supplemental Materials and Methods.
Statistical analysis
The statistical association of each marker in the GWA study was determined by logistic regression with gender and first two principal components as covariates under an additive model and Fisher’s exact allelic test within sub-population analyses. GWA analyses were carried out with PLINK.21 We inferred the genomic inflation factor by converting p-values from regression test to chi-square values, then taking the median (0.4687) divided by the expected median value (0.456) of chi-square distribution with 1 degree of freedom.
HLA haplotypes were inferred using Beagle version 2.1.22 Associations of HLA alleles were tested using logistic regression, including the first two principal component scores as covariates to control for population stratification. To test for independent effects within regions having multiple associated variants, we included one or more variants as a covariate and tested the significance of other variants by adding them step-wise. All detailed analyses were performed with R.23 Additional statistical analyses of HLA alleles were conducted by logistic regression using glm function (generalized linear model) and heterogeneity testing with meta.MH function (Mantel-Haenszel meta-analysis) from the rmeta package (http://cran.r-project.org/web/packages/rmeta/index.html) adjusting for gender and principal components when feasible.
Results
Clinical characteristics of the cases
Clinical details of the 201 cases included in this study are summarized in Table 1. As in previous studies of AC-DILI, there were more male than female cases. The average age at onset was 61±14 years. Four cases underwent liver transplantation. Most cases (70%) were characterized as cholestatic or mixed at presentation and 88% had bilirubin levels > 2.4 mg/dL. Most causality scores (93%) suggested DILI was either probably or highly likely due to AC. There were some significant differences in the pattern of disease and other phenotypic characteristics among the four collections (Table S1).
Table 1.
Clinical characteristics of AC-DILI cases
| Cohorts | DILIGEN | EUDRAGENE | Spanish DILI Registry | DILIN | All (%) | |
|---|---|---|---|---|---|---|
| N | 77 | 19 | 49 | 56 | 201 | |
| Sex | M | 39 (51%) | 11 (58%) | 27 (55%) | 36 (64%) | 113 (56%) |
| F | 38 (49%) | 8 (42%) | 22 (45%) | 20 (36%) | 88 (44%) | |
| Age | Mean (±stdev) | 61 (± 14) | 68 (± 14) | 60 (± 18) | 59 (± 14) | 61 (± 14) |
| Total days on drug | Mean (±stdev) | 8 (± 6) | 7 (± 5) | 11 (± 9) | 12 (± 8) | 10 (± 7) |
| Days to DILI onset | Mean (±stdev) | 15 (± 14) | 11 (± 9) | 16 (± 12) | 32 (± 19) | 20 (± 17) |
| Pattern of liver injury | Cholestatic | 36 (47%) | 6 (32%) | 17 (35%) | 20 (36%) | 79(39%) |
| Mixed | 18 (23%) | 5 (26%) | 17 (35%) | 23 (41%) | 63 (31%) | |
| Hepatocellular | 19 (25%) | 0 | 15 (31%) | 10 (18%) | 44 (22%) | |
| Unknown | 4 (5%) | 8 (42%) | 0 | 3 (5%) | 15 (7%) | |
| Causality (CIOMS) | Unlikely | 0 | 0 | 0 | 0 | 0 |
| Possible | 7 (9%) | 1 (5%) | 0 | 6 (11%) | 14 (7%) | |
| Probable | 25 (32%) | 12 (63%) | 23 (47%) | 37 (66%) | 97 (48%) | |
| Highly probable | 45 (58%) | 6 (32%) | 26 (53%) | 13 (23%) | 90 (45%) | |
| Peak Bilirubin (mg/dL) | Mean (±stdev) | 12.5 (±9.8) | 10.8 (±12.2) | 12.3 (±9.5) | 16.5 (±11.8) | 13.5 (±10.6) |
| Peak ALT (U/L) | Mean (±stdev) | 455 (±465) | 342 (±268) | 548 (±668) | 575 (±713) | 500 (±584) |
| Peak ALP (U/L) | Mean (±stdev) | 453 (±253) | 438 (±171) | 417 (±286) | 456 (±293) | 444 (±268) |
Genome-wide analysis
GWA analysis was carried out on 201 DILI cases and 532 controls for 822,927 markers that passed QC. PCA showed that all cases clustered within Northwestern, Western and Southern Europe (Figure S1), consistent with previous studies on Europeans.24 Based on these genetic patterns, the US-derived DILIN samples were predominantly of Northwestern European origin with genotype patterns correlating well with both UK controls and cases. The genomic inflation factor (λ) of our study was 1.03, which indicates no problems with population stratification.
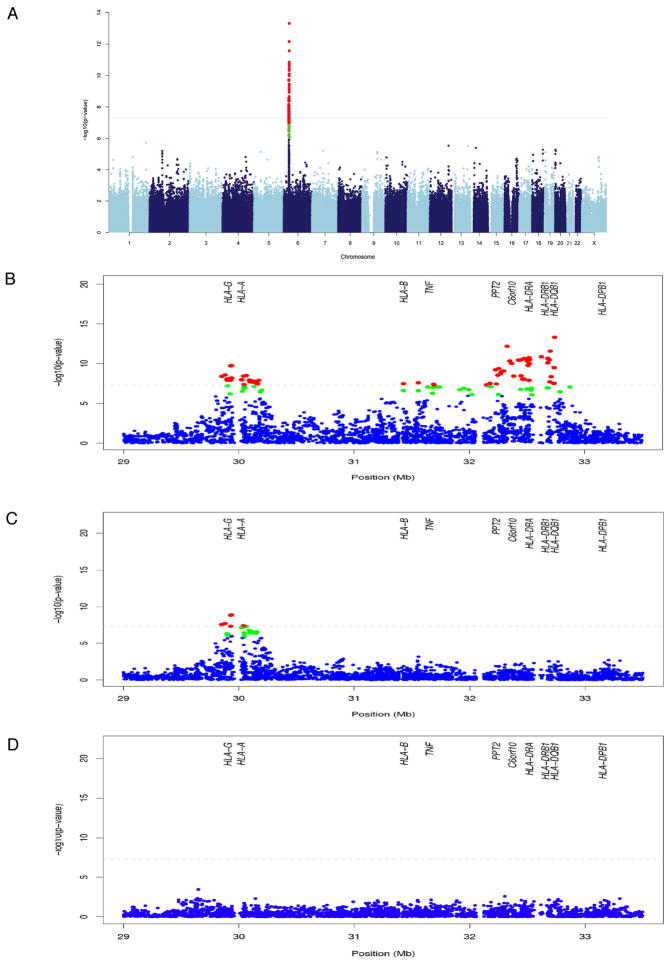
Case-control association analysis revealed one marked peak of association with several dozen genome-wide significant SNPs within the major histocompatibility complex (MHC) on chromosome 6p21.3 (Figure 1A). Quantile-quantile (Q-Q) plots (Figure S2) suggested that no SNPs outside the MHC region showed a genome-wide significant association with DILI. The most significant associations localized within the class II and class I regions (Figure 1B). The top SNP associated with AC-DILI was rs9274407 (p=4.8 × 10−14 with an estimated additive OR of 3.1 (95% confidence interval (CI)=2.3–4.2); Table 2). This SNP is located within the HLA-DQB1 gene and had relatively high linkage disequilibrium (LD)(r2 = 0.76) with rs3135388, one of the other top associated SNPs that is strongly correlated with (i.e. a tag for) DRB1*1501 and the haplotype DRB1*1501-DQB1*0602.25 The top SNP (rs9274407) was significantly associated conditioned on the tag SNP (rs3135388) (p=0.00011), whereas rs3135388 was not significant conditioned on rs9274407 (p=0.93).
Figure 1. AC-DILI genome-wide association results.
Each point represents association analysis results for a single SNP with chromosome position on the x axis and -log10 p-value on the y axis. All 201 DILI cases and 532 population controls were included in the analysis. SNPs with p-values smaller than 10−6 and 10−7 are highlighted in green and red, respectively. Panel A shows the results for the entire genome. Panels B to D show enlarged sections of the MHC region. Panel B is an enlargement of the results presented in A, panel C shows the analysis of each SNP in this chromosome region conditioned on the top class II SNP (rs9274407) and panel D analysis of each SNP conditioned on the top class I and II SNPs (rs9274407 and rs2523822). Positions of a range of MHC genes are shown in B to D.
Table 2.
Summary of top-associated SNPs from the genome-wide analysis together with conditioning analysis
| SNPs | HLA Allele tagged | HLA Class | Chr | Position (Build 36) | P-value | OR (95% CI) | P-value conditioned on rs9274407 | P-value conditioned on rs3135388 |
|---|---|---|---|---|---|---|---|---|
| rs9274407 | II | 6 | 32740809 | 4.8×10−14 | 3.1 (2.3–4.2) | N/A | 0.00011 | |
| rs9267992 | II | 6 | 32328375 | 6.8×10−13 | 3.1 (2.3–4.2) | 0.07 | 0.0034 | |
| rs3135388 | DRB1*1501-DQB1*0602 | II | 6 | 32521029 | 3.5×10−11 | 2.8 (2.1–3.8) | 0.93 | N/A |
| rs2523822 | HLA-A*0201 | I | 6 | 29936639 | 1.8×10−10 | 2.3 (1.8–2.9) | 1.2×10−9 | 2.1×10−10 |
The data shown is for all 201 DILI cases and 532 controls
Several SNPs from the HLA class I region were also genome-wide significant, the most significant being rs2523822 (p=1.8×10−10, OR=2.3, CI=1.8–2.9), previously reported as a tag for A*0201.25 Conditioning on rs9274407, the association peak within class I remained genome-wide significant, whereas none of the other class II region SNPs remained significantly associated (Figure 1C, Figure S2, Table 2). We found a statistically significant interaction (p=0.0015, OR=2.3, CI=1.4–3.8) between rs9274407 and rs2523822, indicating that the increased risk when both minor alleles were present was larger than expected based on their individual effects. Conditioned on both rs9274407 and rs2523822, no additional remarkable associations within the MHC were observed (Figure 2D).
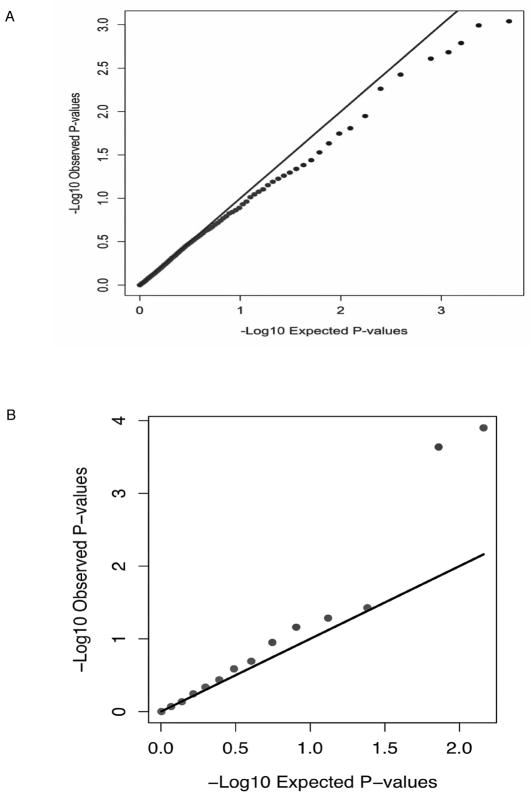
Figure 2. Additional Q-Q plots for ADME and autoimmune genes.
A shows a Q-Q plot for ADME genes and B a Q-Q plot for GWAS hits for autoimmune disease genes. The autoimmune disease SNPs studied related to type 1 diabetes, Crohn’s disease, celiac disease, systemic lupus erythematosus, ankylosing spondylitis, primary biliary cirrhosis, systemic sclerosis, juvenile arthritis and Grave’s disease. All show an association with at least one of these diseases with p ≤ 5×10−8.27
A total of 14 of the 201 cases in the study had shown “possible “ rather than “probable” or “highly probable” causality when scored by the CIOMS system.19 To assess whether exclusion of these cases from the analysis would affect the overall findings, we recalculated p values for the two SNPs with the lowest p values (Table 2). No significant alteration in these p values was seen (data not shown).
To consider the potential for country-specific effects, genome-wide analyses were carried out on 3 separate subgroups within the original 201 cases, namely 74 UK, 51 US cases with Northwestern European genetic backgrounds and 46 Spanish DILI Registry cases with Spanish genetic background (Figure S2). Appropriately matched POPRES controls (306 Northwestern European for the UK and US and 160 Spanish and Portuguese) were used. As with the combined samples, no SNPs outside the MHC were genome-wide significant and most significant SNPs in the combined sample showed similar estimates of effects within each of the three groups (Table 3). Conditioning analysis on top SNPs, similar to that performed for the entire group shown in Table 2, was also performed on the different groups (Table 3). The association of rs9274407 conditioned on rs3135388 was observed only in the UK group with statistical significance (p=7.3×10−5). Rs3135388 was not significantly associated conditioned on rs9274407 consistently in all three groups but rs2523822 was significantly associated conditioned on either rs9274407 or rs3135388 in all three groups. The interaction between the class I rs252382 and class II rs9274407 which was significant for the entire group was also significant in the UK cases (p=0.013) but not in the US and Spanish cases (p=0.14 and 0.31 respectively).
Table 3.
Top-associated SNPs from the genome-wide analysis in UK, US and Spanish cases and controls together with conditioning analysis on these groups
| Top SNPs |
UK (N = 74/306*) | US (N = 51/306) | Spanish (N = 46/160) | Test of heterogeneity (p-value) |
MAF (case vs control) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p- value |
OR (95% CI) |
p-value conditioned on rs9274407 |
p-value conditioned on rs3135388 |
p- value |
OR (95% CI) |
p-value conditioned on rs9274407 |
p-value conditioned on rs3135388 |
p- value |
OR (95% CI) |
p-value conditioned on rs9274407 |
p-value conditioned on rs3135388 |
nw-EU** | Spanish | ||
| rs9274407 | 1.7×10−8 | 3.3 (2.2 – 5.0) | NA | 7.3×10−5 | 7.9×10−6 | 3.0 (1.9 – 4.8) | NA | 0.12 | 0.0019 | 2.9 (1.5 – 5.6) | NA | 0.99 | 0.93 | 0.36 vs. 0.15 | 0.21 vs. 0.081 |
| rs9267992 | 7.6×10−6 | 2.7 (1.8 – 4.1) | 0.4 | 0.23 | 9.6×10−6 | 3.0 (1.9 – 4.9) | 0.38 | 0.075 | 6.0×10−6 | 4.7 (2.4 – 9.1) | 0.0019 | 0.0072 | 0.37 | 0.31 vs. 0.14 | 0.24 vs. 0.063 |
| rs3135388 | 1.4×10−5 | 2.6 (1.7 – 4.0) | 0.097 | NA | 3.9×10−5 | 2.8 (1.8 – 4.6) | 0.89 | NA | 0.00011 | 4.3 (2.1 – 8.9) | 0.11 | NA | 0.51 | 0.30 vs. 0.14 | 0.18 vs. 0.05 |
| rs2523822 | 8.1×10−6 | 2.3 (1.6 – 3.4) | 1.7×10−5 | 7.3×10−6 | 8.3×10−6 | 2.7 (1.8 – 4.1) | 4.5×10−5 | 2.6×10−5 | 0.0051 | 2.0 (1.2 – 3.2) | 0.032 | 0.043 | 0.65 | 0.49 vs. 0.28 | 0.43 vs. 0.28 |
MAF=minor allele frequency
Though the Q-Q plots (Figure S3) suggested that no SNPs outside the MHC region were significantly associated with AC-DILI genome-wide, we also assessed whether there was any indication of a contribution from either genes concerned with drug absorption, distribution, metabolism and excretion (ADME)26 or, in view of the strong HLA associations, non-HLA genes known to be involved in autoimmune diseases generally.27 We extracted all SNPs within 10 kb of a list of 130 ADME genes.26 A total of 4961 such SNPs were included in our analysis. Figure 2A shows the Q-Q-plot of p-values for these SNPs and indicates that no significant associations occur. For the autoimmune disease genes, we extracted SNPs associated with these diseases with reported p-value smaller than 5×10−8 (see Figure 2B).27 Among these SNPs (213 in total), 158 had been genotyped and analyzed in our study. Figure 2B shows the Q-Q-plot of p-values of these SNPs, excluding SNPs from the MHC region. We found two SNPs, rs2476601 and rs6679677 in strong LD with each other to be associated with AC-DILI (for rs2476601, p=1.3×10−4, OR=2.1, 95% CI=1.5–3.2)(Table S2). Rs2476601 is a non-synonymous SNP in PTPN22, the gene encoding the lymphoid-specific protein tyrosine phosphatase, nonreceptor type 22 involved in T-cell-receptor signaling, and has been reported to be associated with multiple autoimmune diseases.28 Although the association was not genome-wide significant, it had a p-value of 0.023 after Bonferroni correction for all the published GWA associations with autoimmune diseases that were genotyped in our study.27 The association of the PTPN22 SNPs with DILI appeared stronger in the UK and US cases than in those from Spain (Table S2).
HLA analysis
To further investigate the observed associations within the MHC, high resolution genotyping of HLA-A, B, DRB1, DQA1 and DQB1 was performed on a subset of 177 cases and 219 genetically-matched controls. The relationship between HLA genotypes and the top associated SNPs within Northwestern European and Spanish cases and controls was investigated (see Table S3). There were substantial differences in LD between Northwestern Europeans and Spanish for rs9274407 and DQB1*0602 and for rs2523822 and A*0201 that were reflected in subsequent associations. In particular, rs2523822/C showed poor correlation with A*0201 in Spanish, especially among cases (r2=0.64) compared with Northwestern Europeans (r2=0.96 for cases) (Table S3). DQB1*0602 was the most significantly associated HLA allele with DILI overall (p=1.4×10−6; OR=3.3, CI=2.0–5.7) and within each group. DRB1*1501 was in near-perfect LD with DQB1*0602 in both Northwestern Europeans and Spanish, and the association of either allele with AC-DILI was statistically similar. A summary of key findings conditioned on DQB1*0602 is provided in Table 4. DQA1*0102, an allele included in though not exclusive to the DRB1*1501-DQB1*0602 haplotype, was slightly more significant than DQB1*0602 in Northwestern Europeans but not in Spanish. Conditioned on DQB1*0602, DQB1*0402 was associated in Northwestern Europeans with nominal significance, but not in Spanish. This is consistent with the SNP results, where rs9274407 conditioned on rs3135388 was an indicator of a DQB1*0402 association in Northwestern Europeans (Table S3). Unlike its tag SNP (rs3135388), DQB1*0602 was associated in the Spanish group conditioned on rs9274407 with nominal significance (p=0.01).
Table 4.
Analysis of combined SNP and HLA data by conditioning on DQB1*0602
| Conditioned on PC1 + PC2 + sex, and | Allele | All (177 cases, 219 controls) | Northwestern European (130 cases, 107 controls) | Spanish (47 cases, 121 controls) | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | ||
| DQB1*0602 | 4.2 (2.7 – 6.6) | 4.6 × 10−10 | 3.3 (2.0 – 5.7) | 1.4 × 10−6 | 7.4 (3.1 – 17.8) | 7.4 × 10−6 | |
| + DQB1*0602 | DRB1*1501 | 0.8 (0.1–5) | 0.72 | 1.4 (0.2–10) | 0.77 | N/A | 0.26 |
| + DQB1*0602 | DQA1*0102 | 2.0 (1.2 – 4.2) | 0.02 | 2.9 (1.1 – 7.2) | 0.02 | 1.8 (0.7–5) | 0.37 |
| + DQB1*0602 | DQB1*0402 | 2.3 (0.9–6) | 0.1 | 7.5 (1.5–38) | 4.9 × 10−3 | 0.9 (0.1 – 6) | 0.4 |
| + DQB1*0602 | A*0201 | 2.2 (1.6 – 3.2) | 1.9 × 10−6 | 2.9 (1.8 – 4.4) | 7.9 × 10−7 | 1.6 (0.88–2.8) | 0.13 |
| + DQB1*0602 | B*1801 | 2.0 (1.0 – 3.9) | 0.05 | 0.98 (0.36 – 2.6) | 0.95 | 4.0 (1.5–11) | 0.004 |
| + DQB1*0602 and A*0201 | rs2523822/C | 3.9 (1.5 – 11) | 0.01 | 0.86 (0.15 – 4.9) | 0.86 | 7.9 (2.1–30) | 0.004 |
| + DQB1*0602 and A*0201 | rs9274407/Minor | 2.4 (0.94 – 5.9) | 0.07 | 9.7 (1.9–49) | 1 × 10−3 | 0.67 (0.11–4.1) | 0.65 |
| +rs9274407/Minor | DQB1*0602 | 1.8 (0.68–5.0) | 0.23 | 0.37 (0.071–1.9) | 0.2 | 11 (1.6–85) | 0.01 |
The class I allele A*0201 was associated (p=2×10−6, OR=2.2, CI=1.6–3.2) with DILI in the combined cases. Consistent with the SNP results, there was a significant statistical interaction between A*0201 and DQB1*0602 in the combined cases (p=0.0048, OR=3.3, CI=1.4–7.7) together with the UK and US cases individually (p=0.012, OR=3.9, CI 1.4–11 and p=0.03, OR=5.4, CI 1.2–25). However, A*0201 was not as strongly associated in Spanish as in Northwestern Europeans. A test of heterogeneity on the effect of A*0201 gave a p-value of 0.071, suggesting a population-dependent effect (Figure S4). This inconsistency with the result of rs2523822 could be explained by the much lower LD between rs2523822 and A*0201 in Spanish cases (Table S3). Additionally, B*1801 was associated in Spanish cases with nominal significance independently of A*0201 and DQB1*0602, but not in Northwestern Europeans.
The frequencies of the five-gene HLA haplotypes in cases and controls are shown in Table S4. The most significant association was observed for A*0201-B*0702-DRB1*1501-DQB1*0602 in Northwestern Europeans (p=0.00074, OR=13) and Spanish (p=0.013, OR=20). However, both DQB1*0602 and A*0201 were significantly associated conditioned on that haplotype. A*0201-B*1801 was significantly associated (p=0.015, OR=6.4) in Spanish only.
Relationship of genotype with clinical features
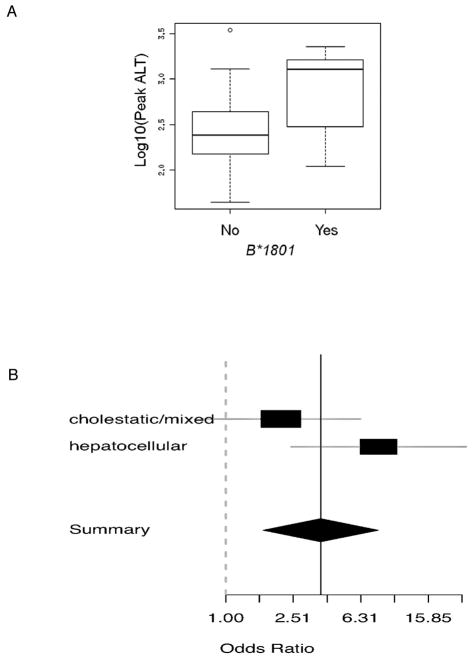
We investigated the relationship between the top-associated HLA alleles and clinical features of AC-DILI, including age-at-onset, pattern of liver damage and disease severity as assessed by magnitude of transaminase or bilirubin elevation (Table S5). B*1801 carriage was significantly correlated with peak ALT values in Spanish (p=0.0056, Figure 3A), but not in Northwestern Europeans (p=0.90). We further investigated whether the effect of B*1801 varied in Spanish cases according to pattern of liver damage (Figure 3B). The estimated effect in hepatocellular cases (OR=8) was much larger than cholestatic/mixed cases (OR=2) but the apparent heterogeneity was not statistically significant (p=0.10). No other notable correlations were observed in either Spanish or Northwestern Europeans.
Figure 3. Relationship between HLA genotype and clinical parameters.
A shows the relationship between HLA-B*1801 and peak ALT in Spanish cases and B a comparison of odds ratios for different liver damage patterns for HLA-B*1801 in Spanish cases and controls.
Clinical predictive values
Assuming the prevalence of AC-DILI is 0.014%,2 the best positive predictive value (PPV) is 0.1% for Northwestern Europeans based on the presence of both A*0201 and DQB1*0602 (frequency of 41% in cases), and 0.13% for Spanish based on the presence of both B*1801 and DQB1*0602 (frequency of 8.5% in cases). In each case these alleles are present at approx. ten-fold higher rates than in the population overall. The best negative predictive value (NPV), expressed for clarity as 1-NPV, is 0.006% for Northwestern Europeans based on the absence of A*0201 (carriage frequency 74% in cases), and 0.007% for Spanish based on the absence of rs2523822/C (carriage frequency 74% in cases), approximately half the assumed rate of AC-DILI in the population overall (Table S6).
Discussion
This study was accomplished through an international cooperation to assemble a considerably larger and more diverse AC-DILI patient collection than previous published studies on the subject.11–13 This resulted in the most powerful and comprehensive investigation into genetic risk factors for AC-DILI and the largest GWA for any rare serious adverse event conducted to date. We confirmed the previous associations of HLA-DRB1*1501 and DQB1*0602 with DILI susceptibility and our larger sample size and GWA approach allowed the identification of additional HLA risk factors together with an apparent statistical interaction between two HLA alleles.
In particular, a novel contribution to disease susceptibility from a common variant (rs2523822) in the region of the HLA-A locus was found. This variant is a tag for the HLA-A*0201 allele in individuals of European ancestry,25 and the effect of rs2523822 and A*0201 were indistinguishable in the Northwestern European subset of this study. When considered as an individual risk factor, the effect of A*0201 was seen only in cases of Northwestern European, and not Spanish, origin. This is in spite of the most strongly associated SNP in this region (rs2523822) showing an effect of similar magnitude in both populations. Nevertheless, the OR for A*0201 in the Spanish subset was also in the same direction as for the other subjects. Comparison of the LD patterns in Northwestern versus Southern European subsets showed that rs2523822 is indeed strongly correlated with A*0201 in Northwestern Europeans (r2=0.96), but not in Spanish (r2=0.64). Given this observation, and the high degree of functional variation in the MHC region, one plausible explanation is that A*0201 is not the actual causal variant underlying the association with rs2523822, but merely happens to be an adequate “tag” for the causal site(s) in Northwestern Europeans, but not in the Spanish. Similarly, since the most significant class II association in Northwestern Europeans rs9274407 was still significantly associated conditioned on HLA-DRB1*1501-DQB1*0602 (or its tag SNP rs3135388) but the opposite was not true, there is a possibility that the causal variant(s) may lie in another class II locus. Further investigation of genetic variation in the MHC region, and functional assessment of these variants in the context of AC-DILI will be necessary to identify the causal site(s) securely. The differences between populations in terms of risk associated with class I HLA allele are somewhat similar to the situation with serious skin rash (SSR) induced by antiepileptic drugs in East Asians where HLA-B*1502 is a risk factor in Han Chinese but not in other groups such as Japanese.29 The complexity of the MHC region in terms of the number of variable sites, and local differences in LD, make definitive claims of causality particularly difficult.
A relatively rare HLA haplotype positive for DRB1*1501 together with DQB1*0602, A*0201 and B*0702 was significantly associated with AC-DILI in both Northwestern Europeans and Spanish. However, conditioning analysis showed that the association was driven by the A*0201 and the DRB1*1501-DQB1*0602 alleles regardless of the cis or trans configuration. We found compelling evidence of a statistical interaction between rs9274407 and rs2523822 (or DQB1*0602 and A*0201). An interaction of this nature appears biologically plausible in view of the complementary roles for the class I and class II HLA gene products in the T-cell response. If validated, this demonstrates the kind of gene-by-gene interaction that has often been speculated on but rarely observed in studies of complex traits in humans.30 On the other hand, without knowing the true causal variants, especially for the class I allele, we cannot exclude the possibility that the interaction between class I and II is due to imperfect tagging of the causal variants by SNPs or classical HLA alleles.
We found evidence for an increased frequency of DQB1*0402 in the Northwestern European cases. This allele is associated with increased susceptibility to the autoimmune liver disease primary biliary cirrhosis (PBC).31 PBC occurs predominantly in females, whereas the majority of the DILI patients positive for DQB1*0402 in this study were males and therefore unlikely to be misdiagnosed PBC cases. Xenobiotic exposure is believed to have a role in triggering PBC32 and it is possible that there are common susceptibilities to PBC and AC-DILI.
From previous reports, it appears that the HLA class II DRB1*1501-DQB1*0602 haplotype is also a risk factor for other forms of DILI.14, 33 A second HLA class II haplotype, DRB1*0701-DQA1*0201, is a risk factor for hepatotoxicity relating to ximelagatran34 and lapatinib.35 However, apart from our reported association between flucloxacillin DILI and B*570116 and an association between A*3303 and ticlopidine DILI in Japanese,36 the role of HLA class I genotypes in susceptibility to DILI generally is still poorly understood and merits further investigation. Interestingly, in addition to being possibly associated with DILI in the Spanish, B*1801 was further associated with peak ALT values in these cases, suggesting a possible role for this allele in phenotypic expression. It is worth noting that the only two cases of fulminant liver failure in the Spanish cohort were both B*1801 carriers.
Additionally, the possibility that the genetic determinants of DILI risk detected here using common SNP markers may potentially represent contributions from a larger number of rare genetic variants should be considered.37 This is particularly relevant to rare adverse events such as DILI, for which rare genetic determinants are in principle one of the most parsimonious explanations. Detection of rare variants will require whole genome sequencing which is increasingly feasible.38
Although our GWA study failed to provide any genome-wide significant evidence of a role for non-HLA genes, including ADME genes, in AC-DILI susceptibility, it remains possible that other genes contribute with smaller effects. In particular, the connection between AC-DILI and autoimmune disease is further extended by the association of the PTPN22 SNP rs2476601. This association could also apply to autoimmune-related DILI due to other drugs since the gene product has a general role in regulation of T-cell responses.
This unique study has demonstrated the ability to determine novel genetic risk factors of AC-DILI in diverse populations. We have shown there are improvements in the NPV for the combined HLA risk genotypes compared to our assumed incidence of AC-DILI of 0.014% with carriers of both class I and II risk alleles showing a nearly 10-fold increase in risk. A PPV of just 0.1% means that HLA genotyping will not be an effective means of prospectively identifying those at risk of AC-DILI but our findings have clinical utility in that HLA genotyping may be of value in strengthening AC-DILI diagnoses in view of the high NPVs seen.
Supplementary Material
Acknowledgments
Contributors to sample collection via Spanish DILI network, EUDRAGENE, DILIN and DILIGEN are listed in the Appendix. Drs Lucena, Molokhia, Shen and Daly contributed equally to this manuscript.
Funding: The genome-wide association study and HLA genotyping was funded by the International Serious Adverse Events Consortium with support from Abbott, Daiichi-Sankyo, GlaxoSmithKline, Johnson & Johnson, Novartis, Pfizer, Roche, Sanofi-Aventis, Takeda, Wellcome Trust and Wyeth. The DILIGEN sample collection was part funded by the UK Department of Health (ref PHGX10A) and by UK NIHR funding to the Nottingham Digestive Diseases Centre. The DILIN network is structured as a U01 cooperative agreement with funds provided by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) under grants: 2U01-DK065176-06 (Duke), 2U01-DK065201-06 (UNC), 2U01-DK065184-06 (Michigan), 2U01-DK065211-06 (Indiana), 5U01DK065193-04 (UConn), 5U01-DK065238-08 (UCSF/CPMC). Additional funding is provided by CTSA grants: UL1 RR025761 (Indiana), UL1 RR025747(UNC), and UL1 UL1 RR024986 (UMich). The EUDRAGENE collaboration has received support from the EC 5th Framework program. (QLRI-CT-2002-02757) and the SAEC. MM is funded by a UK NIHR postdoctoral award. The Spanish DILI Registry is part funded by the Spanish Medicine Agency and has received support from SAEC and Boehringer-Ingelheim, Barcelona, Spain. CIBERehd and RETIC are funded by Instituto de Salud Carlos III. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- DILI
drug-induced liver injury
- CI
confidence interval
- OR
odds ratio
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
Footnotes
Conflicts of interest: The authors disclose the following: Dr Fontana acted as a consultant to GlaxoSmithKline. Dr Nelson is an employee of GlaxoSmith Kline. The remaining authors disclose no conflicts.
Role of authors: MIL, MM, YS, TJU, MRN and AKD coordinated the writing of manuscript; MIL, MM, PBW, RJF and AKD coordinated sample collection; YS, TJU, AF, IP, MJD, DBG and MRW performed and coordinated data analysis; GPA, RJA, CPD, MP, PJF and PBW advised on clinical aspects; GPA, RJA, CPD, FR-C, MR-G, JMN, MP, RJF, MM, MG, JFD, EB-G, AC, BHCS, AC, LI, Q-YY, ME, PBW contributed to sample collection and to the final version of the manuscript; PTD and CS advised on immunogenetic aspects and contributed to the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4:489–99. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 2.Sgro C, Clinard F, Ouazir K, et al. Incidence of drug-induced hepatic injuries: A French population-based study. Hepatology. 2002;36:451–455. doi: 10.1053/jhep.2002.34857. [DOI] [PubMed] [Google Scholar]
- 3.Ostapowicz G, Fontana RJ, Schiodt FV, et al. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137:947–54. doi: 10.7326/0003-4819-137-12-200212170-00007. [DOI] [PubMed] [Google Scholar]
- 4.Stricker BH, Van den Broek JW, Keuning J, et al. Cholestatic hepatitis due to antibacterial combination of amoxicillin and clavulanic acid (augmentin) Dig Dis Sci. 1989;34:1576–80. doi: 10.1007/BF01537113. [DOI] [PubMed] [Google Scholar]
- 5.de Abajo FJ, Montero D, Madurga M, et al. Acute and clinically relevant drug-induced liver injury: a population based case-control study. Br J Clin Pharmacol. 2004;58:71–80. doi: 10.1111/j.1365-2125.2004.02133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrade RJ, Lucena MI, Fernandez MC, et al. Drug-induced liver injury: an analysis of 461 incidences submitted to the Spanish registry over a 10-year period. Gastroenterology. 2005;129:512–21. doi: 10.1016/j.gastro.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Chalasani N, Fontana RJ, Bonkovsky HL, et al. Causes, clinical features, and outcomes from a prospective study of drug-induced liver injury in the United States. Gastroenterology. 2008;135:1924–34. 1934, e1–4. doi: 10.1053/j.gastro.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrade RJ, Lucena MI, Kaplowitz N, et al. Outcome of acute idiosyncratic drug-induced liver injury: Long-term follow-up in a hepatotoxicity registry. Hepatology. 2006;44:1581–8. doi: 10.1002/hep.21424. [DOI] [PubMed] [Google Scholar]
- 9.Garcia Rodriguez LA, Stricker BH, Zimmerman HJ. Risk of acute liver injury associated with the combination of amoxicillin and clavulanic acid. Arch Intern Med. 1996;156:1327–32. doi: 10.1001/archinte.1996.00440110099013. [DOI] [PubMed] [Google Scholar]
- 10.Fontana RJ, Shakil AO, Greenson JK, et al. Acute liver failure due to amoxicillin and amoxicillin/clavulanate. Dig Dis Sci. 2005;50:1785–90. doi: 10.1007/s10620-005-2938-5. [DOI] [PubMed] [Google Scholar]
- 11.Hautekeete ML, Horsmans Y, van Waeyenberge C, et al. HLA association of amoxicillin-clavulanate-induced hepatitis. Gastroenterology. 1999;117:1181–1186. doi: 10.1016/s0016-5085(99)70404-x. [DOI] [PubMed] [Google Scholar]
- 12.O’Donohue J, Oien KA, Donaldson P, et al. Co-amoxiclav jaundice: clinical and histological features and HLA class II association. Gut. 2000;47:717–720. doi: 10.1136/gut.47.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson PT, Daly AK, Henderson J, et al. Human leucocyte antigen class II genotype in susceptibility and resistance to co-amoxiclav-induced liver injury. J Hepatol. 2010;53:1049–53. doi: 10.1016/j.jhep.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 14.Andrade RJ, Lucena MI, Alonso A, et al. HLA class II genotype influences the type of liver injury in drug-induced idiosyncratic liver disease. Hepatology. 2004;39:1603–12. doi: 10.1002/hep.20215. [DOI] [PubMed] [Google Scholar]
- 15.Lucena MI, Andrade RJ, Fernandez MC, et al. Determinants of the clinical expression of amoxicillin-clavulanate hepatotoxicity: a prospective series from Spain. Hepatology. 2006;44:850–6. doi: 10.1002/hep.21324. [DOI] [PubMed] [Google Scholar]
- 16.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nature Genetics. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 17.Molokhia M, McKeigue P. EUDRAGENE: European collaboration to establish a case-control DNA collection for studying the genetic basis of adverse drug reactions. Pharmacogenomics. 2006;7:633–8. doi: 10.2217/14622416.7.4.633. [DOI] [PubMed] [Google Scholar]
- 18.Fontana RJ, Watkins PB, Bonkovsky HL, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009;32:55–68. doi: 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danan G, Benichou C. Causality assessment of adverse reactions to drugs--I. A novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol. 1993;46:1323–30. doi: 10.1016/0895-4356(93)90101-6. [DOI] [PubMed] [Google Scholar]
- 20.Nelson MR, Bryc K, King KS, et al. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83:347–58. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. [Accessed March 9, 2011];The R Project for Statistical Computing. http://www.r-project.org.
- 24.Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bakker PI, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmadi KR, Weale ME, Xue ZY, et al. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nat Genet. 2005;37:84–9. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- 27.Hindorff LA, Junkins HA, Mehta JP, et al. [Accessed March 9, 2011];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies/
- 28.Chung SA, Criswell LA. PTPN22: its role in SLE and autoimmunity. Autoimmunity. 2007;40:582–90. doi: 10.1080/08916930701510848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaniwa N, Saito Y, Aihara M, et al. HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics. 2008;9:1617–22. doi: 10.2217/14622416.9.11.1617. [DOI] [PubMed] [Google Scholar]
- 30.Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am J Hum Genet. 2009;85:309–20. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirschfield GM, Liu X, Xu C, et al. Primary Biliary Cirrhosis Associated with HLA, IL12A, and IL12RB2 Variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selmi C, Gershwin ME. The role of environmental factors in primary biliary cirrhosis. Trends Immunol. 2009;30:415–20. doi: 10.1016/j.it.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Singer JB, Lewitzky S, Leroy E, et al. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat Genet. 2010;42:711–4. doi: 10.1038/ng.632. [DOI] [PubMed] [Google Scholar]
- 34.Kindmark A, Jawaid A, Harbron CG, et al. Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J. 2008;8:186–95. doi: 10.1038/sj.tpj.6500458. [DOI] [PubMed] [Google Scholar]
- 35.Spraggs CF, Budde LR, Briley LP, et al. HLA-DQA1*02:01 Is a Major Risk Factor for Lapatinib-Induced Hepatotoxicity in Women With Advanced Breast Cancer. J Clin Oncol. 2011;29:667–73. doi: 10.1200/JCO.2010.31.3197. [DOI] [PubMed] [Google Scholar]
- 36.Hirata K, Takagi H, Yamamoto M, et al. Ticlopidine-induced hepatotoxicity is associated with specific human leukocyte antigen genomic subtypes in Japanese patients: a preliminary case-control study. Pharmacogenomics J. 2008;8:29–33. doi: 10.1038/sj.tpj.6500442. [DOI] [PubMed] [Google Scholar]
- 37.Dickson SP, Wang K, Krantz I, et al. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.