Abstract
Methylation of cytosines is a pervasive feature of eukaryotic genomes and an important epigenetic layer that is fundamental for cellular differentiation processes and control of transcriptional potential. DNA methylation patterns can be inherited and influenced by the environment, diet and aging, and disrupted in diseases.
Complete DNA methylomes for several organisms are now available, helping clarify the evolutionary story of this epigenetic mark and its distribution in key genomic elements. Nonetheless, a complete understanding of its role, the mechanisms responsible for its establishment and maintenance, and its cross talk with other components of cellular machiney remains elusive.
Keywords: DNA methylation, epigenetics, gene regulation
Introduction
Methylation at the carbon 5 position of cytosines (5meC) constitutes an important epigenetic layer that contributes to the definition of transcriptional and regulatory potential of genomic DNA [1, 2]. DNA methylation is a typical characteristic of most eukaryotes and some of its features are conserved in many species. While cytosine methylation is a stable modification of the genomic DNA that can be inherited, it also dynamically changes during the lifespan of certain cells and tissues of an organism and it is susceptible to diet and other environmental influences. Indeed, it is essential for the correct onset of differentiation processes and for defining tissue specific transcriptional profiles, and can be dysregulated in disease states. Recently, methods have been developed for the genome-wide detection of 5meC, and complete maps for several organisms are available, including humans. Currently available high-throughput data combined with results from classic genetic experiments are beginning to clarify the roles of DNA methylation in a variety of processes. Nonetheless, the mechanisms for the establishment and maintenance of epigenetic patterns and the complex interplay of DNA methylation with other epigenetic and regulatory layers are not well understood [3]. The correct reprogramming of DNA methylation is critical when considering regenerative medicine for the generation of induced Pluripotent Stem Cells (iPSCs) with full differentiation potential. However, base-resolution iPSCs DNA methylomes are not yet available. Several studies have shown that methylation profiles of iPSCs are aberrant with respect to those of Embryonic Stem Cells (ESC) and these differences may decrease or restrict the differentiation potential [4, 5]. Additionally, aberrant methylation in iPSCs was observed to be inherited from the progenitor cell type [6]. In general, additional genome-wide high-resolution data from both healthy and diseased cells will be required to shed light on the dynamic variation of these marks, their role in healthy cells, and their relevance in diseases.
Methods for determining genome-wide profiles of DNA methylation
Recently, a plethora of new methods have been developed for the determination of genome-wide DNA methylation patterns [7]. These developments are beginni to contribute to our comprehension of the role of this epigenetic mark in bot development and disease states. Traditionally, DNA methylation could be determined only for specific loci through Sanger sequencing of bisulfite converted and PCR amplified genomic DNA fragments. While sodium bisulfite has no effect on 5meC, it specifically converts cytosine to uracil, and during PCR amplification of bisulfite treated DNA, uracil is replaced with thymine.
Several methods have been developed which enable capture of genome-wide profiling of DNA methylation. These can be divided into three types: 1) enrichment of methylated genomic DNA fragments, 2) digestion with methylation-sensitive restriction enzymes (RE) and 3) sequencing of bisulfite converted DNA. Each of these methods have been scaled for the analysis of genome-wide profiles with quantification of methylation being based on either microarrays or high-throughput DNA sequencing.
Methylated DNA Immunoprecipitation (MeDIP) is the most common method based on enrichment, where an antibody specific for 5meC is used to capture methylated genomic DNA fragments [8, 9]. This method can provide relatively cheap and reasonably comprehensive genome-wide data, but the resolution is limited and the enrichment is not linearly related to the actual methylation level [10]. Exemplifying methods based on methylation sensitive RE, the HpaII tiny fragment Enrichment by Ligation-mediated PCR assay (HELP) can be used to determine genome-wide patterns based on the combined activity of HpaII and MspI restriction enzymes (RE) [11]. The main disadvantages of this approach are in the resolution of the data and the bias due to the non-uniform distribution of RE cutting sites. The only methods that currently provide genome-wide base-resolution methylation information are based on high-throughput sequencing of bisulfite converted DNA [12, 13]. While these methods (BS-Seq and MethylC-Seq) are still relatively expensive for large genomes (currently ~$10,000 for 30x coverage of the human genome), the cost of sequencing is dramatically decreasing at greater than Moore’s law pace (doubling every 18 months), meaning that soon the cost of enrichment will be significantly greater than the cost of sequencing.
Alternative methods to target specific regions of the genome have also been developed. Some of these methods allow choosing of the target regions, like padlock-probe based targeting or enrichment methods coupled with promoter tiling arrays [4, 9]. Another method, Reduced Representation Bisulfite Sequencing (RRBS) relies on a combination of RE fragment size selection, and bisulfite sequencing, in order to fractionate the genome and enrich for fragments with high CpG content regions prior to sequencing [14].
In a recent study the coverage, resolution, cost and concordance of four sequencing-based methylation profiling methodologies were evaluated [15]. The concordance between these technologies was quite high when comparing methods based on sequencing of bisulfite converted DNA (up to 82% and 99% for mC in CG and non-CG sequence context, respectively), and when comparing enrichment methods (99%), while regions assessed by all four methods were 97% concordant. The authors also showed the power of integration of two complementary methods, in that enriching for hyper- and hypo-methylated regions, along with histone methylation, RNA, and SNP could allow for assessment of allele-specific epigenetic states.
The choice of the best-suited method depends inevitably on required data resolution, cost, size of the genome and throughput in the number of samples [7].
Methylation types
In prokaryotes a methyl group can be added to both cytosines and adenine in palindrome target sequences [16] while in eukaryotes DNA methylation is restricted to cytosines. The most common sequence context where this epigenetic mark is found is the CpG (or CG) di-nucleotide (mCpG or mCG). This di-nucleotide is under represented in eukaryotic genomes because of the mutagenic effect of this modification [17]. In fact the few regions that are GC and CpG rich, named CpG Islands (CGI), are usually depleted of 5meC [17]. CGI are offen within upstream gene regulatory regions and their presence, altered methylation status, and the overall CpG content of the promoter regions are highly predictive of the transcriptional potential of the downstream gene [18].
In several eukaryotic organisms 5meC can also be found in other sequence contexts [16]. In general, this type of modification is named non-CG methylation. In particular 5meC in the CHG and CHH sequence context were found (mCHG and mCHH, respectively; H being A, C or T). Recently, pervasive non-CG methylation was found in the human genome, even if in restricted differentiation stages [19, 20] (see following sections).
Certain genomic regions are enriched in 5meC within specific sequence contexts. For example, mCHH are highly enriched in A. thaliana transposons [13]. Indeed, in some organisms different enzymes and pathways are responsible for these alternative types of methylation, while for others this is still open to debate [21].
Recently, the presence of an additional DNA methylation modification, 5-hydroxy-methylcytosine (5hmC), was found in mouse Purkinje neurons and the brain [22]. This base can be detected using thin-layer chromatography, while standard RE and bisulfite conversion based methods appear not to be able to reveal it [23]. Importantly, TET proteins can oxidize 5meC to 5hmC, which is poorly recognized by DNMT1 and can be converted to cytosine, providing a possible pathway for passive de-methylation [24].
Establishment and maintenance of DNA methylation patterns
Most species where this epigenetic mark is present have two general types of enzymes: de novo and maintenance DNA methyltransferases (DNMTs) [21]. The mechanism by which methylation patterns are established, maintained and inherited is not completely clarified in any species though [25]. The common paradigm is that de novo DNMTs (DNMT3A and DNMT3B in human) establish methylation patterns early in embryonic development [26]. These enzymes have the same activity on both hemi- and un-methylated DNA and are down-regulated but still expressed after cell differentiation [27]. Maintenance enzymes (DNMT1 in human) then copy the newly established DNA methylation patterns through each cellular division [28]. DNMT1 has a preference for hemi-methylated DNA while showing some de novo activity [29, 30]. The picture seems to be more complex though, since genetic experiments with gene knock-outs for these enzymes suggest that the concerted activity of de novo and maintenance enzymes is required for the complete and correct establishment of methylation profiles [25]. Nevertheless, their expression varies greatly during the differentiation processes, suggesting that the requirement for their activity is not constant. Moreover, these enzymes appear not to be freely available in the cells but rather associated with chromatin complexes or at the DNA replication fork [27]. Finally, these enzymes require accessory proteins and show a complex interplay with nucleosome positioning and particular histone marks [31]. These findings help to explain why not all cytosines are methylated. A revised model points to the possibility that the overall methylation level of a region might be copied rather than the exact methylation status of each individual cytosine [27].
In general, the complex cross-talk of these enzymes with other components of the transcriptional machinery has to be considered. In fact, as discussed in the following sections, despite DNA methylation being usually considered a repressive mark, its role can vary greatly in different genomic contexts, in associations with other regulatory mechanisms.
Methylomes and evolution
The role of DNA methylation in different genomic contexts can be remarkably different, and alternative mechanisms are available for establishing 5meC patterns in different genomic regions. The density of possible methylation sites, as well as the distribution of 5meC, is not uniform in the genome. In particular, control of methylation levels in promoters, gene bodies, regulatory features, and transposable and repetitive elements to be critical.
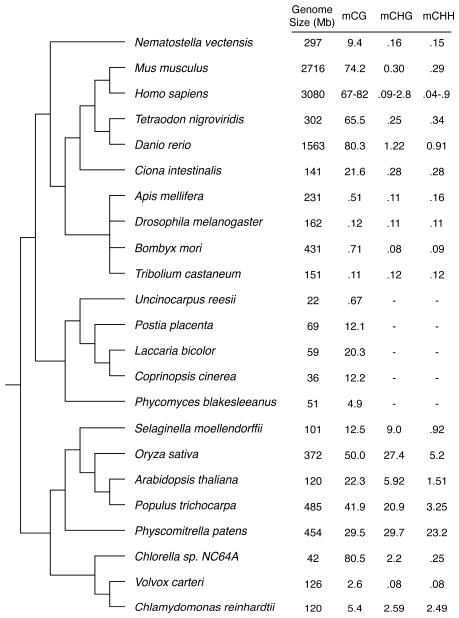
Recently, base-resolution sequencing of several eukaryotic methylomes has provided initial insights into the evolutionary history of DNA methylation and its distribution in these key genomic regions. Currently, the complete methylomes for 23 organisms are available (10 animals, 8 plants and 5 fungi; Fig. 1) [13, 19, 32, 33]. In general the DNA methylation landscape can be either continuous along the genome, or constituted by a series of heavily methylated DNA domains interspersed with domains that are methylation free. When considering the data available for a limited set of loci in an even wider set of organisms, the current perspective is that the evolutionary history of DNA methylation in gene bodies and transposons may be independent. These genomic regions are present in both plants and vertebrates; however, transposon methylation is only conserved in fungi while gene body methylation only occurs in invertebrates [16]. Gene body methylation seems to be a property inherited from ancient genomes, while transposon methylation appears to be related to the degree of sexual outcrossing [27]. In general, for those organisms where the methylation of certain genomic elements was present, their methylation pattern is quite conserved. However, the presence of non-CG methylation is less common, but when present, it is always at a lower level than mCG [13, 19, 32, 33].
Fig. 1.
Methylation levels in 23 eukaryotic organisms. The organisms are organized according to their evolutionary distance. Tree topology is determined from the NCBI Taxonomy (http://www.ncbi.nlm.nih.gov/guide/taxonomy/) and displayed using TreeView X (http://darwin.zoology.gla.ac.uk/~rpage/treeviewx/index.html). The genome size is indicated together with the percentage of methylated sites within three sequence contexts: CpG, CHG and CHH (H being A, C or T).
Human methylomes
Base-resolution global maps of DNA methylation in humans remained elusive for a long time because of the difficulties of performing comprehensive analysis on a multiple-gigabase genome. Recent improvements in the throughput of sequencing technologies, simultaneous reduction of the cost, and coupling of bisulfite conversion with cutting edge sequencing methods, has now enabled acquisition of the complete human methylomes for several cell types: human embryonic stem cells (hESC), fetal and neonatal fibroblasts, and fibroblastic differentiated derivative of hESC [19, 20]. The choice of these particular cell types is related to the relevance of DNA methylation in the onset and regulation of cellular differentiation processes. These maps demonstrated for the first time the feasibility of applying such genome-wide methods on genomes of this size, and now provide reference maps against which other maps of healthy and diseased cells can be compared.
Non-CG methylation
A striking finding from the first human methylomes is the relative abundance of non-CG methylation in hESC [19, 20]. Indeed, one quarter of the 5meC was present in the CHG or CHH sequence context, with some preference for CA di-nucleotides in both sequence contexts. Moreover, non-CG methylation was lost during the differentiation process but can be re-established at the same loci upon generation of iPSCs [19]. While the re-appearance of non-CG methylation in iPSCs was shown for several loci, more comprehensive experiments will be necessary to fully evaluate how the DNA methylome is restored in these cells compared to ESCs. Non-CG methylation was distributed non-randomly in the genome and was particularly enriched in the gene bodies, with increasing levels corresponding to more highly transcribed genes [19]. Non-CG methylation was also particularly enriched in genes related with processing of mRNA and in genes having higher pre-mRNA levels [19]. These findings point to the potential involvement of DNA methylation with the splicing machinery [19, 20]. This hypothesis is also supported by the patterns of CG methylation at the exon-intron boundaries [20]. Enzymes responsible for the deposition of mCG were shown to have a preference for targeting CG di-nucleotides with a relative spacing of 8 bp [34]. Interestingly, the same result was observed for 5meC in CHG and CHH sequence contexts [19]. These findings suggest that the same enzymes may be responsible for non-CG methylation, even if patterns for 5meC at relative distances multiple of 8 bp are not found as clearly as for mCG. Interestingly, methylation at non-CG was much lower level than mCG as only 25% of the sequencing reads for a given non-CG residue were methylated, compared to 80-90% for mCG [19]. Finally, mCHH was slightly more enriched on the antisense strand in gene bodies, and the potentially symmetric mCHG was 98% hemi-methylated [19]. For all of these reasons, it is necessary to acquire deep sequence coverage for those samples where non-CG methylation is expected. Finally, further research will be necessary to clarify the role of non-CG methylation and its relevance for pluripotency.
Methylation of regulatory elements
Biophysical studies of DNA methylation reveal that this base modification plays a important role in repressing accessibility of the transcriptional machinery to the DNA [35]. Indeed, some transcription factors (TF) such as Sp1 are known to be methylation sensitive, even if this seems to be dependent on the considered condition or tissue [36, 37]. Moreover, not all TFs have methylation sites in their binding sites (TFBS) or methylation does not affect their binding.
Recently, binding of TFs and other proteins important for general control of transcription (TAF1 and P300) or for the stem cell biology (SOX2, 0CT4, NANOG and KLF4) was profiled by ChIP-seq in both hESC and fetal fibroblasts [19]. The regions immediately surrounding these TFBS showed depletion of non-CG 5mC in human ESCs, while mCG were much less depleted for many factors [19].
Enhancer regions, as defined by H3K4me1 and H3K27ac ChIP-seq sites, were also profiled and these elements also showed depletion of non-CG 5mC in human ESCs. Interestingly, the enhancers present in fibroblasts showed mCpG depletion, while enhancers shared by both cell types showed depletion of 5meC in the CpG and non-CG context in differentiated and un-differentiated cells, respectively. These findings would suggest that cells in different states of differentiation use different DNA methylation mechanisms to mark these important regulatory features [19].
Differential methylation
While non-CG methylation was absent in differentiated cells, widespread differences were found in the distribution and levels of CG methylation. Hundreds of differentially methylated regions (DMRs) hyper-methylated in fibroblasts have been identified, many associated with genes important for stem cells functions [19]. On the other hand, large hypo-methylated regions have also been identified when comparing differentiated cells to human ESCs. These have been termed Partially Methylated Domains (PMDs). PMDs might also be expected on the chromosome X, given that the cells examined were derived from a female and that the dosage compensation occurs on sex chromosome. In fact PMDs cover 80% of chromosome X. Nonetheless, surprisingly almost 40% of autosomes were found in the PMD state. Genes in these domains were down-regulated compared to human ESCs, and PMDs were also enriched in histone repressive marks [19]. Interestingly, large blocks of H3K9me2 were present in differentiated cells but not in embryonic cells in the mouse. These same regions were found to overlap human PMDs and shown too be lost in certain cancer cells [38, 39].
CG DNA methylation is usually considered a repressive mark [16]. Indeed, methylation in the context of promoters is inversely related with the transcriptional level of the downstream gene. However, CG methylation in gene bodies is positively correlated with the gene transcriptional level, indicating that the meaning of this epigenetic mark is rather complex and context dependent [40]. Genome-wide studies showed that gene-body non-CG methylation is also clearly positively correlated with gene expression [19]. Rather, gene bodies in human ESCs are always highly methylated, even in poorly expressed genes [19]. All of these findings suggest that the positive correlation between gene-body CG methylation and transcriptional expression could be re-interpreted as loss of DNA methylation upon differentiation and the formation of repressed chromatin blocks [19], rather than expecting a positive effect of gene-body methylation on gene expression. In agreement, there is evidence in A. thaliana for association between gene body methylation and transcriptional elongation, suggesting a scenario where transcription contributes to maintaining or enhancing DNA methylation levels [41].
Data analysis
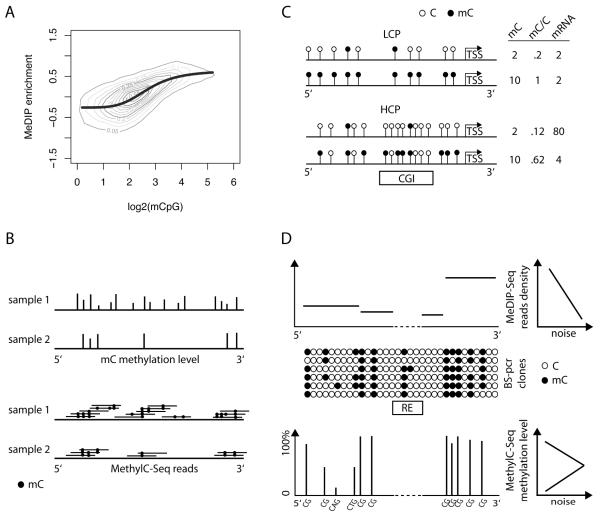
There are specific pitfalls related to the analysis of DNA methylation data. Some of these issues are specific to the methodology chosen for detecting 5meC. For example, a problem for enrichment methods is that the enrichment signal is not linearly related to the actual methylation level (Fig. 2A]. Fortunately, several methods were developed to correct for this bias [10, 42, 43]. When analyzing base-resolution data the challenges are in dealing with non uniform coverage within a sample and comparing samples with different overall sequencing depth (Fig. 2B]. This is particularly critical for the detection of non-CG methylation. In fact, as discussed in a previous section, the level of methylation for 5mC in this sequence context is rather low, and only 25% of the sequencing reads for a given non-CG residue are methylated in average. Rather, alignment of bisulfite-converted reads and bisulfite conversion errors do not represent a serious issue, since a high proportion of reads can be mapped, covering most of the genome.
Fig. 2.
Pitfalls in the analysis of DNA methylomes. (A) MeDIP-chip enrichment is not linearly related to the DNA methylation level (graph created using MEDME [10]). (B) The mC calls determined for two samples with different sequencing depth; the height of the bars corresponds to the methylation level (the proportion of methylated over total reads for a given cytosine); a differentially methylated region can be improperly identified if the depth is not considered; in addition higher methylation levels could be computed in the sample with lower coverage. (C) two promoters with the same number of mC but different density of available methylation sites (LCP and HCP : low and high CpG content promoter, respectively); the increase of mC from 2 to 10 in the two promoters can have remarkably different effect on the transcription of the downstream gene (TSS: transcriptional start site; CGI: CpG island). (D) signal and signal-to-noise for enrichment methods (MeDIP-seq) and for bisulfite sequencing methods (MethylC-Seq); RE is a repetitive region that cannot be covered.
Once the final set of 5meC calls, or an estimate derived from the enrichment level, is available, one has to consider both the absolute and relative methylation level. The absolute level represents the density of 5meC. On the other hand, this information has to be considered with respect to the total number of available methylation sites, and in general the CG content of the loci under consideration. As an example, a promoter containing a hypo-methylated CGI can have the same 5meC density as a fully methylated low CpG content promoter, in absolute terms, but their functional consequences are remarkably different (Fig. 2C]. Indeed hypo-methylation at CGI is likely to be associated with transcriptional expression of the downstream gene. In general, it was shown that transcription of genes downstream of low CG content promoters does not correlate with their upstream-region DNA methylation level. However, the methylation status of high and especially intermediate CpG content promoters is critical for transcriptional repression [18].
A substantial difference between enrichment and bisulfite-sequencing based methods is that for the former the count of reads for given loci determines the methylation level, while for the latter the read depth is related to the library reparation, sequencing and mapping process. Also, the ability to call a 5meC is directly related to the sequencing depth in bisulfite sequencing-based experiments (Fig. 2B]. Related with this, the methylation level, the proportion of reads with 5meC over the set of reads covering a specific base, is a measure whose variability decreases with increasing depth. Finally, since the methylation level is a finite scale ß-distributed measure, the variance of measurements with a mean near the mid-range can be much larger than the variance of measurements with a mean close to the limits (0 and 1; Fig. 2D). These are important pitfalls to keep in mind when determining differential methylation levels.
In general, few tools are currently available for the analysis of epigenomics data, in particular high-throughput DNA methylation data [7]. Pipelines for both low- and high-level analyses must be developed. Low-level analysis can be defined as read mapping algorithms and 5meC calling, taking into account bisulfite conversion level, sequencing errors and multiple testing issues. Higher-level analysis involves the determination of absolute and relative methylation levels across genomic regions based on genome annotation, clustering, visualization and integration with other heterogeneous data types. Particularly important is the determination of differentially methylated regions, since DNA methylomes vary between different cell types, as a function of differentiation stage, age, and as a result of disease states. Methods for the identification of differentially methylated regions must also take into account that genomic regions of very different scale might exist, spanning from megabase size PMDs to small DMRs that constitute only few differentially methylated cytosine bases.
Future perspectives
Technologies for high-throughput detection of the sites of DNA methylation in genomes are being constantly improved and with equally significant reductions in cost. Three important limitations might be overcome in the near future. Single cell sequencing analysis would eliminate many problems of interpreting DNA methylomes, which currently are the produce of a mixed population of cells/chromosomes, each with heterogeneous methylation status [44, 45]. Similarly, of “amplification-free” methods would greatly reduce the problem of the uneven distribution of bisulfite converted reads [44, 45]. Finally, direct detection of 5meC, without use of bisulfite conversion, would eliminate the issue of the degradation and loss of material following the chemical conversion [44-47] as well as increase the ability to map the sequencing reads onto the genome.
Several aspects regarding the prevalence and role(s) of DNA methylation must be clarified. A comprehensive list of DNA-binding proteins affected or insensitive to cytosine methylation should be developed. This is particularly critical for TFs, given that they bind upstream gene regulatory regions, where DNA methylation has always been considered a critical factor. Also, the relationship between the methylation of specific regions and the transcriptional potential of nearby genes is still unclear. DNA methylation is only one epigenetic control point. Future studies must attempt to integrate this key mark with all other epigenetic and regulatory mechanisms. For example, there is strong evidence on the relevance of promoter DNA methylation and transcriptional repression of the downstream gene. Induced de-methylation can restore the expression of silenced genes, as well as re-establishment of high methylation levels can suppress them [48]. However, in other cases, variation in the level of DNA methylation may simply be a consequence of closed chromatin structure, as it has been hypothesized for PMDs, where embryonic methylation levels might be lost after differentiation, resulting in accumulation of repressive chromatin marks [19].
Few examples of allelic methylation are available, but this important phenomenon requires more comprehensive analysis [49]. This will require the availability of matched genomes/methylomes, and possibly the use of methods that are not dependent on bisulfite conversion, as this complicates the evaluation of the differential methylation for C-T SNPs.
Finally, a critical area is the application of base-resolution methylome analysis for clinical studies where DNA methylomes of diseased cells can be generated and compared to healthy cells, possibly using matched samples. These studies will be also necessary to better understand the effect of drugs that target the epigenome such as the demethylating agent Decitabine. These drugs are used to de-methylated tumor suppressor genes that have been silenced by DNA methylation [48] but their genome-wide effect is poorly understood.
Acknowledgements
MP is supported by a Catharina Foundation postdoctoral fellowship; work in our laboratory is supported by grants from NIH (U01 ES017166), DOE (FG02-04ER15517), NSF (MCB-0929402) and by the Mary K. Chapman Foundation (JRE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting typesetting and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content and all legal disclaimers that apply to the journal pertain.
List of abbreviations: 5meC, MeDIP, RE, HELP, mCpG, CGI, mCHG, mCHH, 5hmC, DNMT, ESC, hESC, IPS, TF, TFBS, ChIP, DMR, PMD1
- 5meC
- 5-methyl-cytosine
- MeDIP
- methylated DNA immunoDreciDitation
- RE
- restriction enzyme
- HELP
- HpaII tiny fragment enrichment by ligation-mediated PCR
- mCpG
- methyl-CpG
- CGI
- CpG island
- mCHG
- methyl-CHG
- mCHH
- methyl-CHH
- 5hmC
- 5-hydroxy-methylcytosine
- DNMT
- DNA methyl transferase
- ESC
- embryonic stem cells
- hESC
- human embryonic stem cells
- IPS
- induced pluripotent stem cells
- TF
- transaction factor
- TFBS
- transaction factor binding site
- ChIP
- chromatin immunoprecipitation
- DMR
- differentially methylated regions
- PMD
- partially methylated domain.
References
- [1].Holliday R, Pugh JE. DNA modification mechanis during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- [2].Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- [3].Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- [4].Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Doi A, Park I, Wen B, Murakami P, Aryee M, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley G, Feinberg A. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010 doi: 10.1038/nature09342. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Laird P. Principles and challenges of genome-wide DNA methylation analysis. Nature Reviews Genetics. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- [8].Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam W, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- [9].Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- [10].Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Research. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, Hatchwell E, Selzer RR, Richmond TA, Green RD, Melnick A, Greally JM. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Research. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cokus S, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen S. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Meissner A, Mikkelsen T, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein B, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander E. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Harris RA, Wang T, Coarfa C, Zhou X, Xi Y, Nagarajan RP, Hong C, Downey S, Johnson BE, Delaney A, Zhao Y, Olshen A, Ballinger T, Schillebeeckx M, Echipare L, OGeen H, Lister R, Pelizzola M, Chung W, Gu H, Bock C, Gnirke A, Zhang M, Haussler D, Ecker J, Li W, Farnham PJ, Waterland RobertA, Meissner A, Marra M, Hirst M, Milosavljevic A, Costello JF. Sequence-based profiling of DNA methylation: comparisons of methods and catalogue of allelic epigenetic modifications. Nature Biotechnology. 2010;10:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- [17].Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- [18].Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, Narayan D, Molinaro AM, Halaban R, Weissman SM. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Research. 2009;19:1462–1470. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei C. Dynamic changes in the human methylome during differentiation. Genome Research. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Law J, Jacobsen S. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Research. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird P, Jones P. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- [27].Jones P, Liang G. Rethinking how DNA methylation patterns are maintained. Nature Reviews Genetics. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- [29].Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- [30].Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- [31].Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones P. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Feng S, Cokus S, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen S. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- [34].Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA ethylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harrington MA, Jones PA, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci USA. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mancini DN, Singh SM, Archer TK, Rodenhiser DI. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- [38].Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg A. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg A. Reply to “Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells”. Nat Genet. 2010;42:5–6. doi: 10.1038/ng0110-4. [DOI] [PubMed] [Google Scholar]
- [40].Ball MP, Li J, Gao Y, Lee J, Leproust E, Park IH, Xie B, Daley GQ, Church G. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- [42].Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Research. 2010;10:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Down T, Rakyan V, Turner D, Flicek P, Li H, Kulesha E, Gräf S, Johnson N, Herrero J, Tomazou E, Thorne N, Bäckdahl L, Herberth M, Howe K, Jackson D, Miretti M, Marioni J, Birney E, Hubbard T, Durbin R, Tavaré S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Meth. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of epigenetic DNA modifications with a protein nanopore. Chemical communications (Cambridge, England) 2010 doi: 10.1039/c0cc02864a. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nature nanotechnology. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- [48].Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- [49].Schalkwyk L, Meaburn E, Smith R, Dempster E, Jeffries A, Davies M, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]