Dear editor,
We have developed a method that achieves a faster and more thorough ablation of cellular proteins by integrating RNAi and protein knockout techniques (Figure 1A). While this combination approach is generally applicable to any target protein, it is ideal for those that cannot be sufficiently eradicated by RNAi and for long-lived proteins that endure well after nascent protein biosynthesis has been halted.
Figure 1.
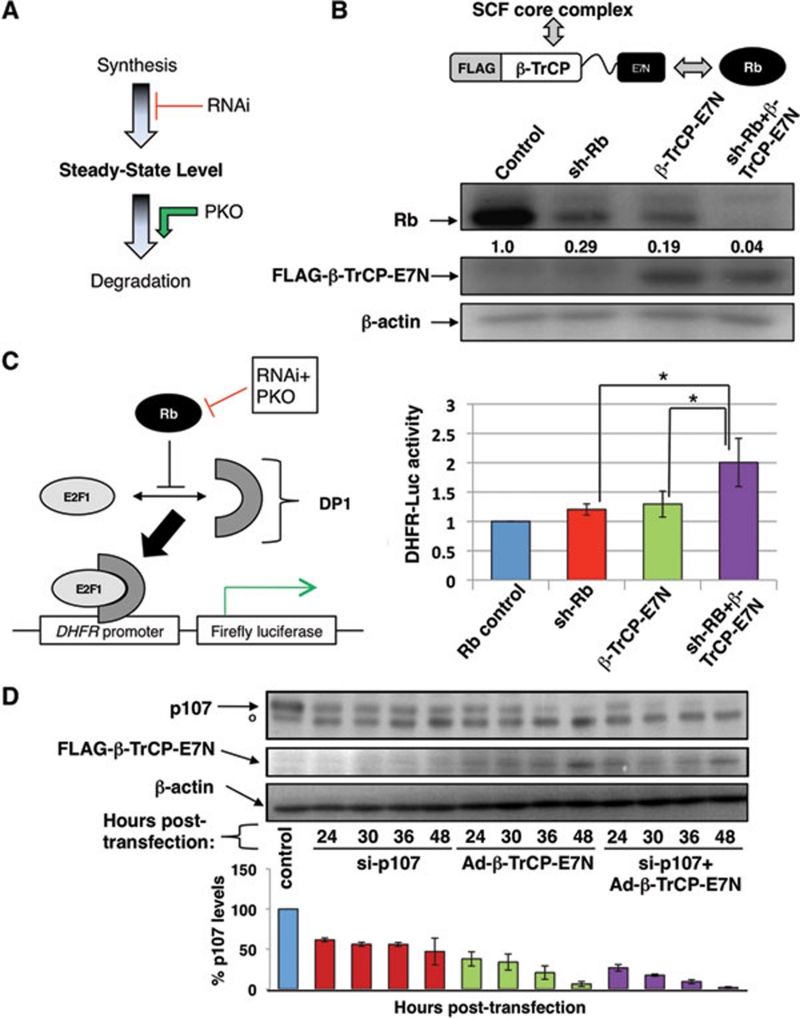
Combining RNAi and PKO to achieve maximal and rapid protein ablation. (A) Schematic diagram of the combination approach, which entails both a reduction in nascent protein biosynthesis via RNAi as well as enhanced post-translational degradation via PKO to ablate overall steady-state protein expression levels. (B) (Top) β-TrCP-E7N chimeric E3 ligase contains the β-TrCP F-box motif to facilitate interaction with the SCF core E3 ubiquitin ligase complex and the E7N peptide, which binds Rb (as well as pocket proteins p107 and p130). (Bottom) SAOS-2 cells were transfected by nucleofection (Amaxa) with pCDNA3-Rb-HA, and either pMSCV-LMP-sh-Rb, pCDNA3-β-TrCP-E7N or both. Control cells were transfected with pMSCV-LMP. All samples were selected with puromycin 24 h post-transfection, then analyzed by immunoblotting with antibodies against Rb, FLAG or β-actin. (C) (Left) Rb-mediated inhibition of E2F1, which binds the DHFR promoter along with its cofactor DP-1, reduces transcription of the DHFR-luciferase reporter. (Right) Dual-luciferase assay (Promega) of SAOS-2 cells transfected with pCDNA3-Rb-HA, pCDNA-E2F1, pCMV-DP1, DHFR-luc, pRL-Tk (Renilla) and the indicated knockdown constructs. Cells were lysed after 24 h of puromycin selection and the ratio of Firefly/Renilla (F/R) luciferase signal was measured in triplicate. Measurements were normalized to control and the graph indicates average (of three experiments) fold difference in F/R ratio. The symbol * indicates P-value < 0.05. (D) (Top) C33A cells were transfected (Invitrogen) with anti-RBL1 siRNA oligonucleotides (Thermo Scientific), then infected with adenovirus bearing the Ad1-β-TrCP-E7N construct. °Non-specific species. (Bottom) Average p107 expression levels were obtained from three separate experiments and normalized to p107 levels in C33A cells that were mock transfected and infected.
RNAi has been harnessed to specifically target transcripts from a single gene and subsequently reduce the steady-state level of its translated protein product. As such, it has become an indispensible and widely used tool in reducing target protein levels in order to assess their biological activity. However, in some cases, RNAi does not sufficiently ablate protein expression to create an intracellular environment that simulates complete inactivation of the target protein. Another factor to consider is the stability of the target protein, which dictates the rate and, therefore, the efficiency of its depletion. Even in cases of total disruption of nascent protein production, residual expression of long-lived target proteins may distort or obscure the assessment of the protein ablation phenotype.
Previously, we demonstrated that knockdown of the retinoblastoma protein (Rb), a protein that possesses a relatively long half-life of 9 h, can be achieved at the post-translational level through directly accelerated ubiquitin-mediated proteolysis 1, 2. Briefly, we constructed a chimeric E3 ligase that is able to recruit Rb to the SCFβTrCP (Skp1, Cul1 and F-box-containing βTrCP substrate receptor) E3 ligase complex 3, and facilitate Rb polyubiquitination and its subsequent proteolysis (Figure 1B, top). In doing so, we demonstrated that it is possible to engineer RING family ubiquitin ligases (such as the SCF complex) to recruit, ubiquitinate and induce proteolysis of specific intracellular proteins using a technique we designated “protein knockout” (PKO) 4, 5, 6.
Here, we posit that the maximum level of protein ablation can be achieved in the shortest period of time by combining the post-translational PKO system with transcript-targeting RNAi (Figure 1A). To test this two-pronged or double knockdown method, we utilized an anti-RB1 shRNA construct (a kind gift of Michael Reed) as well as the β-TrCP-E7N construct to target ectopically expressed Rb in SAOS-2 cells 7. As shown in Figure 1B (bottom), either sh-Rb or β-TrCP-E7N alone was able to reduce Rb expression levels to 29% and 19% (relative to control levels), respectively, by 48 h. Introduction of both sh-Rb and PKO constructs resulted in nearly total elimination of Rb expression (yielding only 4% Rb expression levels), surpassing the reduction seen in either shRNA or PKO construct alone (Figure 1B). Therefore, these results confirm that the combination approach is more effective than using either a post-transcriptional or post-translational technique alone.
We next compared the single and combined knockdown techniques in their ability to disrupt cellular Rb function. One established function of Rb is inhibiting E2F1 transcriptional activity. Here, we utilized a luciferase reporter gene construct driven by the E2F1-responsive DHFR promoter (DHFR-Luc) (Figure 1C, left) 8. SAOS-2 cells were transiently transfected with sh-Rb and/or β-TrCP-E7N together with the DHFR-Luc reporter. Expression of both constructs yielded 2-fold higher E2F1 reporter activity than control, while single knockdown samples only displayed a relatively modest increase (20-30%) (Figure 1C, right). Thus, from a functional perspective, the effects of integrated RNAi and PKO also significantly exceed those of RNAi or PKO alone.
To assess the kinetic rate of target depletion by the combined RNAi and PKO technique, and also to demonstrate the versatility of the technique, we next targeted endogenous p107 protein. C33A cells were transfected with anti-RBL1 siRNA oligonucleotides to target p107 transcript, and then infected with adenovirus bearing the β-TrCP-E7N construct. Relative to the untreated control, the siRNA-transfected samples showed a 39% reduction in p107 expression at 24 h and reached 53% reduction by 48 h (Figure 1D). Ad1-β-TrCP-E7N-infected samples showed more marked p107 depletion than the siRNA-transfected samples with 62% and 94% reduction at 24 and 48 h, respectively. Strikingly, the siRNA+Ad1-β-TrCP-E7N samples achieved the greatest level of p107 knockdown at all time points, with 73% reduction in p107 levels at 24 h and 98% at 48 h (Figure 1D). Thus, the combination approach ablates p107 expression more rapidly and with higher efficiency than RNAi or PKO alone.
In summary, we have proved the concept that combining the powerful techniques of RNAi and ubiquitin ligase-mediated protein knockout can maximize target protein ablation and is anticipated to significantly improve our ability to examine cellular protein function. We propose that double knockdown is a valuable and novel method that is particularly relevant for proteins that are not responsive to RNAi-mediated knockdown and for analyses that require the most rapid and thorough target protein ablation possible.
Acknowledgments
We thank Jennifer Lee for editing the manuscript and Jianxuan Zhang for technical advice, Zhi-Xiong Jim Xiao, Shao Ning Yang, Michael Reed and Ari Melnick for reagents and technical support. This work is supported in part by the Irma T Hirschl Career Scientist Award and the National Institute of Health grant (CA098210) to PZ. JH is supported by a Ruth L Kirschstein National Service Award (NRSA) Institutional Research Training Grant (T32 GM008539).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Materials and Methods
References
- Zhou P, Bogacki R, McReynolds L, Howley PM. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins. Mol Cell. 2000;6:751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zheng N., Zhou P. Exploring the functional complexity of cellular proteins by protein knockout. Proc Natl Acad Sci. 2003;100:14127–14132. doi: 10.1073/pnas.2233012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou P. Ectopic targeting of substrates to the ubiquitin pathway. Methods Enzymol. 2005;399:823–833. doi: 10.1016/S0076-6879(05)99053-8. [DOI] [PubMed] [Google Scholar]
- Zhou P. Targeted protein degradation. Curr Opin Chem Biol. 2005;9:51–55. doi: 10.1016/j.cbpa.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Zagorski WA, Knudsen ES, Reed MF. Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res. 2007;67:8264–8273. doi: 10.1158/0008-5472.CAN-06-4753. [DOI] [PubMed] [Google Scholar]
- Sdek P, Ying H, Zheng H, et al. The central acidic domain of MDM2 is critical in inhibition of retinoblastoma-mediated suppression of E2F and cell growth. J Biol Chem. 2004;279:53317–53322. doi: 10.1074/jbc.M406062200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods