Abstract
The Unfolded Protein Response (UPR) is an ensemble of signal transduction pathways that respond to perturbations in the oxidative, pro-folding environment of the endoplasmic reticulum. During the past decade, ongoing research implicated these pathways in maintaining homeostasis of cells and organisms exposed to various stresses. Herein, we highlight recent findings regarding the functional role of the UPR in both normal and pathophysiological processes.
The lumen of the endoplasmic reticulum (ER) provides an oxidative compartment wherein proteins destined for secretion or insertion into cellular membranes are co-translationally modified with sugar moieties and folded. Stresses that compromise the ER environment impair maturation resulting the accumulation of mis-folded proteins and activation of a stress response termed the Unfolded Protein Response (UPR)1,2. While chemicals that inhibit N-linked glycosylation (tunicamycin) or deplete ER calcium (thapsigargin) are frequently used to impair protein folding within the ER, environmental stresses that reduce carbon source availability (glucose), and oxygen, which occurs under pathogenic conditions such as cancer and viral infection, also have a direct impact on secretory homeostasis and thereby trigger the UPR3,4. The UPR is characterized by increased transcription of genes encoding ER molecular chaperones including BiP/GRP78 and GRP94, protein disulfide isomerase (PDI), and the pro-apoptotic transcription factor CHOP (C/EBP homologous protein), which is also known as growth arrest and DNA damage gene-153 (GADD153)5. The induction of ER chaperones is in turn coordinated with a marked decrease in the rate of overall protein synthesis and with cell cycle arrest. Inhibition of protein synthesis serves to lower the overall rate of protein traffic into the ER, but the fact that this process is counterbalanced by an increased synthesis of ER chaperones highlights the specificity of the UPR. This mini-review will focus on advances in our understanding of the role of the UPR in determining cell fate in response to micro-environmental stresses to which cells must respond.
Signal Transduction from the ER
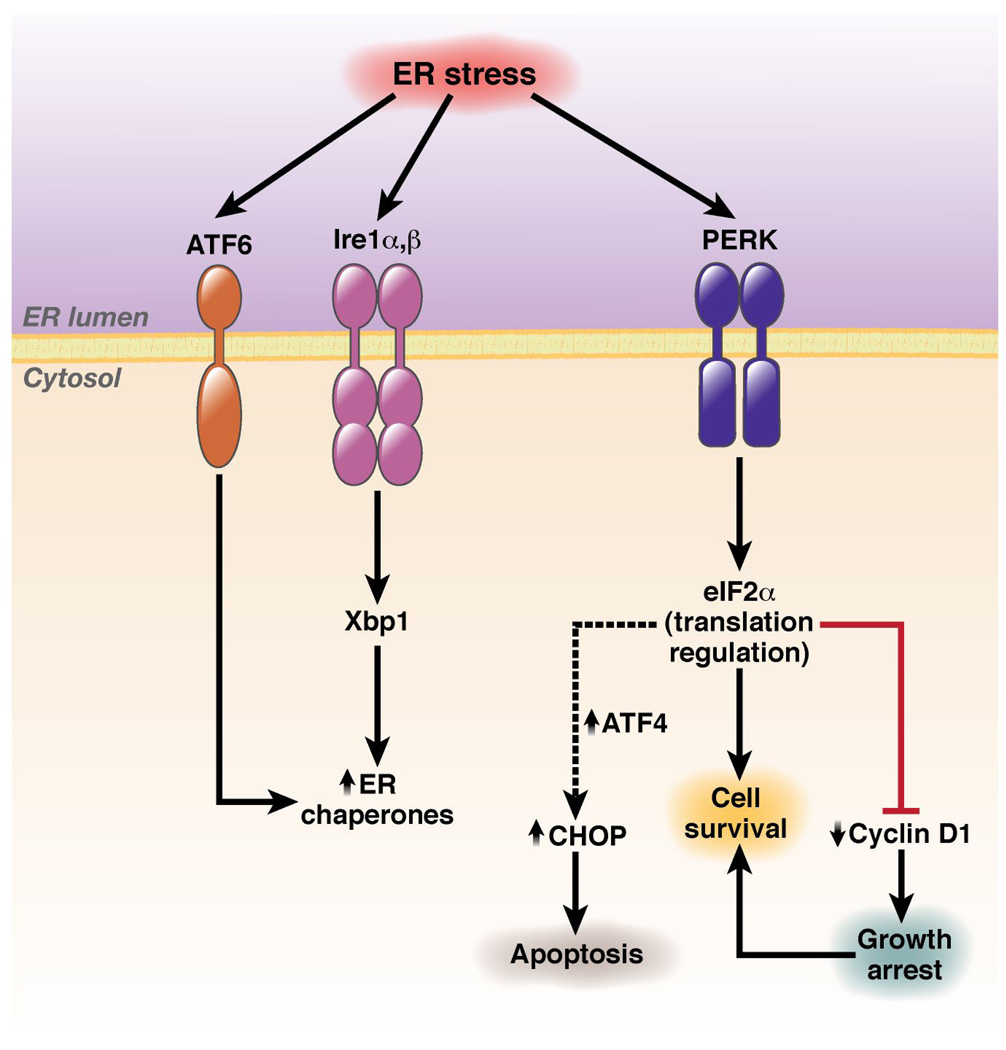
Ire1 isoforms (α, ubiquitously expressed α and tissue restricted β), are composed of a luminal domain that senses stress, a single transmembrane domain, and a cytosolic tail that contains both a protein kinase domain and an RNase domain6, 7. Ire1 triggers increased expression of numerous ER chaperones through activation of the X-box binding protein 1 (Xbp1) transcription factor (Figure 1). Accumulation of Xbp1 is mediated by the RNase function of Ire1, which mediates an alternative splicing event that generates an alternative Xbp1 mRNA that is more efficiently translated8, 9. PERK, another ER transmembrane protein kinase activated in a manner analogous to the Ire110, catalyzes serine 51 phosphorylation of eIF2α resulting in down-regulation of general protein synthesis11, 12. While PERK is one of four distinct eIF2α protein kinases, which includes the heme-regulated kinase (HRI), the interferon-inducible, RNA-dependent protein kinase (PKR) and GCN2, only PERK function is required for cellular response to ER stress. The third signaling components are the transmembrane transcription factors ATF6α/β. While normally tethered to the ER, upon stress, ATF6 migrates to the trans-Golgi, where it is processed by S1P and S2P proteases to release the N-terminal DNA-binding transcription factor domain. The latter regulates transcription of a subset of Xbp1 responsive genes due to their related DNA binding specificities13.
Figure 1. UPR signal transduction.
ER stress signals induce UPR via promoting the activation of homologous transmembrane protein kinases, Ire1a/b, and PERK and a transmembrane transcription factor, ATF6. These regulators collectively transduce signals resulting in the upregulation of ER chaperones and CHOP while simultaneously down-regulating cellular protein synthesis and including that of the growth promoter cyclin D1 thereby inducing growth arrest.
Activation of both Ire1 and PERK requires dimerization via sequences within luminal domains14. Dimerization is antagonized by the ER chaperone BiP, which physically inhibits association. Increased levels of unfolded proteins in the ER lumen titrate BiP thereby permitting dimerization and activation of PERK/Ire1 molecules. ATF6 is also regulated by BiP, unlike Ire/PERK, BiP inhibits movement of ATF6 to the Golgi and thereby determines access to activating protease activity. Thus, the chaperone to misfolded protein ratio is considered a key determinant in UPR status.
At the interface of cell cycle progression
Activation of UPR signaling triggers a rapid arrest in G1 phase of the cell cycle. G1 phase progression requires the activities of one or more of the D-type cyclins (D1, D2, D3) in association with either CDK4 or CDK6 followed by activation of the cyclin E- and A-dependent kinase CDK2. Cell cycle arrest in response to mitogen deprivation or anti-proliferative cytokines can be achieved through degradation of unstable cyclin subunits, by specific post-translational modifications of the CDK subunits, or via association of active cyclin-bound CDKs with polypeptide CDK inhibitors (CKIs)15. While mitogen withdrawal inhibits cyclin D gene expression and accelerates cyclin D1 proteolytic degradation16, UPR-dependent signal transduction intersects with the cell cycle via PERK-dependent inhibition of cyclin D protein synthesis and rather than accelerating cyclin D degradation17, 18. Research utilizing genetically defined cells revealed that G1 arrest triggered by ER stress requires PERK-dependent phosphorylation of eIF2α18. The resulting inhibition of cyclin D1 translation together with its rapid rate of proteolysis results in its nearly immediate elimination and loss of cyclin D1-dependent kinase activity19. The ensuing cell cycle arrest in non-tumorigenic cells is facilitated alsoby p53-dependent increases in the CKI, p21Cip1 20. ER stress-induced growth arrest likely provides a window of opportunity or checkpoint that prevents cells from continuing their cell division cycle under conditions in which the proper folding and assembly of proteins are significantly compromised. Ultimately, the failure of the UPR to re-establish proper homeostatic balance can trigger apoptosis.
UPR and Metabolism
A primary function of the ER is post-translational folding of proteins destined for membranes or for secretion and a key modification essential for proper folding is asparagine-linked glyclosylation. Thus, tissues whose primary function is secretion are particularly dependent upon UPR signaling for homeostasis. Not surprisingly, significant pathologies in such tissues are caused by inadequacies in PERK function. Children suffering from Wallcot-Rallison disease, a recessive disorder characterized by loss-of-function PERK mutations, develop type I diabetes, a result of reduced beta-cell proliferation and exocrine cell apoptosis21. This phenotype is faithfully recapitulated in conventional PERK knockout mouse model22, 23. In addition to the pancreas, plasma cells, a B-cell whose function is the secretion of functional antibodies depend also upon UPR components. Rather than PERK however, plasma cells depends upon the combined activities of ATF6 and Ire1-Xbp124, 25 that function to maintain a required repertoire of ER chaperones.
Given that increased secretory demand as well as cell division requires membrane expansion, it should not come as a surprise that UPR signaling contributes also to aspect of lipid biogenesis. Ire1-dependent activation of Xbp1 contributes not only to chaperone expression, but contributes to phospholipid biosynthesis via regulation choline cytidyltransferase, a rate-limiting enzyme in the CDP-choline pathway25. Synthesis of this enzyme in plasma cells appears to be synergistically regulated by Xbp1 and ATF624, 25. Additionally, Xbp1 also plays a key role in the production of liver-derived plasma lipids26.
UPR signaling contributes to lipid biogenesis in the context of mammary gland during pregnancy. Knockout of PERK in the murine mammary gland reduced free fatty acid production and lipid deposition in the milk of nursing mice27. Perk deletion correlated with a loss of sustained of expression of key lipogenic enzymes such as ATP citrate lyase, fatty acid synthase, stearyl-CoA desaturase-1. Expression of these enzymes is acutely dependent upon Sterol Regulatory Element-Binding Protein 1c (SREBP1c) a transmembrane transcription factor whose release from ER membranes requires targeted movement to the Golgi where it is processed by S1P and S2P proteases. PERK loss inhibits SREBP1c Golgi processing, thereby reducing expression of key Lipogenic enzymes27.
Viral Impact on the UPR
Replication of viruses in eukaryotic cells requires the robust synthesis of viral polypeptides that taxes the folding machinery. Accordingly, many viruses including herpes simplex virus (HSV) and hepatitis C virus (HCV) are associated with UPR activation. This rapidly expanding field is covered by several excellent recent reviews28, 29. Viruses are known to both induce UPR and produce the means of inhibiting these responses. The latter might be necessary in order to protect the host cells from ER stress-mediated death, to enable translation of viral proteins and ensuing virus production28–32. However, in reality, a number of examples demonstrate poorly understood specificity with regard to which UPR branch is activated by specific viruses.
The most obvious reasons for such selectivity is likely the very need to correct the protein synthesis-controlling and pro-apoptotic branches of the pathway in order to complete the replication. The rapid translation of viral proteins will pose an additional problem for the virus as it challenges the capacity of the host cell to properly fold key proteins. Accordingly, viruses have evolved to ameliorate specific UPR branches that control translation. For example, HSV was shown to express glycoprotein B that specifically inhibits PERK activation33. Furthermore, HSV is also generates the γ134.5 protein that is capable of maintaining the low levels of eIF2α phosphorylation through the phosphatase-mediated stimulation of eIF2α de-phosphorylation34. The latter mechanism is likely required to safeguard against activation of other kinases capable of phosphorylation of eIF2α independently of activated PERK (e.g. PKR)35. Yet, this kinase can be also inhibited by proteins produced by viruses of medical importance such as HCV36.
Conversely, the flavivirus West Nile virus strain Kunjin virus can specifically activate Ire1α but has a negligible effect on PERK37. In the latter case, synthesis of hydrophobic non-structural proteins inhibits the anti-viral effects of IFNα/β. HCV appears to utilize an analogous mechanism38. This relationship between induction/resolution of UPR and responsiveness of cells to anti-viral effects of type I IFN is significant as the UPR may negatively impact local immunity39. Although temporally detrimental for viral maturation, activation of PERK also has a side effect of feeding into the ligand-independent pathway that governs downregulation and degradation of IFNα/β receptor. This receptor on cell surface mediates all cellular responses to endogenous IFNα/β or pharmaceutical IFNα used to treat chronic viral infections such as hepatitis C and B40. The levels of this receptor are regulated by the ligand-inducible ubiquitination and degradation of the IFNAR1 chain41. Ubiquitination of IFNAR1 depends on IFN-induced IFNAR1 phosphorylation by protein kinase D242–46. This pathway negatively regulates IFNα/β signaling in cells previously exposed to IFNα/β and have likely generated a robust anti-viral response to these cytokines. There is also an alternative basal (ligand-independent) pathway that leads to downregulation of IFNAR1 from the surface of cells that have not been yet exposed to IFNα/β47. Because this pathway does not require ligand, it can impair the ability of a naïve cell to respond to its future encounters with IFNα/β.
Phosphorylation of IFNAR1 by Casein kinase 1α is key to ligand-independent turnover48, 49. Remarkably, the ability of this kinase to phosphorylate IFNAR1 and trigger its ubiquitination and degradation can be greatly increased in cells exposed to ER stress49. More importantly, these UPR-induced events required activity of PERK49. The functional outcome of this regulation is a tempered ability of already infected cell to react to its future encounters to IFNα/β and to mount an efficient anti-viral state49, 50. Intriguingly, whereas the induction of UPR by HCV infection in experimental models has been thoroughly documented39, a search for upregulated markers of UPR in liver biopsy samples from patients with chronic HCV was inconclusive51. The latter results could be explained by localized changes that are limited to rather small clusters of infected cells at a given time52. Future concerted efforts are required to investigate the role of UPR in hepatitis C pathogenesis and liver injury.
Apoptotic response to the UPR
In most pathophysiological conditions activation of the UPR has cytoprotective effects, a feature of reduced global protein synthesis with selective increased synthesis of proteins with cytoprotective roles, cells experiencing prolonged or acute ER stress undergo ER-initiated apoptosis. Indeed prolonged exposure of cells to tunicamycin or thapsigargin induce apoptosis53, 54. Conversely, cells with a compromised UPR, (e.g. cells deficient for PERK/eIF2α or IRE/XBP-1), are significantly more sensitive to ER-induced cell death than wild-type cells presumably due to the continuous accumulation of misfolded proteins in the ER, a process termed “proteotoxicity”55, 56. Although the mechanisms for ER-induced apoptosis are poorly understood, but there is strong evidence that Ca2+ release from ER stores and subsequent caspase activation, including caspase-3 and perhaps caspases 4 and 7 mediate programmed cell death under these conditions57.
Well known for their contribution to the intrinsic apoptotic pathway through the mitochondria, bcl-2 family members are localized also to the ER. ER localized bcl-2 may contribute to ER membrane permeability by maintaining the pro-death bak and bax in their inactive conformations58. ER stress induces oligomerization of bax and bak to their active states that can then induce an ER Ca+2 leak that triggers apoptosis. ER release of calcium is known to activate calpains, and induce cleavage of Bid, a BH-3 only bcl-2 family protein. Cleavage of Bid enhances its capacity to induce mitochondria membrane permeabilization, thereby leading to cytochrome c release and activation of downstream caspases.
UPR and Autophagy
Another emerging role for an ER-dependent pro-survival pathway is the involvement of ER and eIF2α phosphorylation in the autophagic pathway. Previous work59, 60 has established that ER stress can activate autophagy, a highly conserved lysosome-dependent mechanism for degrading intracellular constituents. It has been proposed that autophagy, as a pro-survival mechanism under short-term nutrient deprivation, can counteract apoptotic mechanisms. However, similar to UPR activation, prolonged or acute activation of autophagy can induce clonogenic cell death61. Interestingly, several agents that induce ER stress (e.g., proteasome inhibitors, thapsigargin, etc) induce autophagy. Extreme hypoxia can also induce autophagy, primarily via activation of ATF462, 63. These findings establish autophagy an important adaptive response cells use to survive ER stress, especially in the conditions of the solid tumor microenvironment.
Tumorigenic utilization of the UPR
Accumulating evidence demonstrates activation of the UPR in solid tumors. BiP/GRP78 accumulation in malignant human breast lesions has been observed frequently64. With regard to signal transduction from the ER, loss of Ire165, as well as its downstream target transcription factor XBP166, compromise tumor progression and inhibit neovascularization. Moreover, transgenic mice with directed expression of the XBP-1 spliced isoform (XBP-1s) develop bone lytic lesions and subendothelial Ig deposition - pathologic disorders reminiscent of multiple myeloma in humans67. PERK has also been implicated in facilitating tumor progression. Initial work demonstrated that murine fibroblasts derived from PERK null embryos and transformed with SV40 large/small T-antigen and Ki-RasV12 failed to grow in immune compromised mice as did human colorectal carcinoma cells expressing a dominant-negative form of PERK68. Subsequently, deletion of PERK in MMTV-Neu mice was found to modestly increased tumor latency, but more profoundly inhibit metastatic spread. Furthermore, deletion of PERK in established tumors, significantly reduced tumor progression69, 70.
In addition to involvement in tumor progression, recent work has also implicated UPR in Inflammatory Bowel Disease, response to of the GI tract to hypoxic insult increasing susceptibility to bacterial translocation and inflammatory response71–73. Adaptive signaling from the UPR has even been implicated in response to alcohol consumption74 emphasizing the role of this pathway as a sensor of both cellular and environmental stress.
Collectively, the current work supports the concept of developing inhibitors of UPR signaling as anti-neoplastic therapeutics and potentially for a variety of diseases that impact the secretory apparatus of the cell. However, caution must be utilized. Our understanding of the breadth of the UPR signaling with respect to cellular and organismal homeostasis remains limited. Efforts to dissect the molecular role of each component within cellular contexts provide new and exciting avenues of research, but also add to the complexity of downstream effects that must be considered when targeting pathways. As better animal models are generated to assess the function of the UPR in various physiological and pathological diseases, such as cancer, we will be in a position to better anticipate uses and outcomes of targeted therapies.
Acknowledgments
This work was supported by grants from the National Institutes of Health P01 CA104838 and a Leukemia & Lymphoma Scholar award (JAD); CA142425 (SYF); CA 094214 and CA 13962 (CK); and the Abramson Cancer Center (JAD, SYF, CK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim Biophys Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 5.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Online Only)
- 6.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 8.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 10.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 14.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 15.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 16.Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc Natl Acad Sci U S A. 1999;96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- 21.Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 22.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang WFD, Li Y, Iida K, McGrath B, Cavener DR. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 25.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0808517105. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DY, Lee J, Sugden B. The unfolded protein response and autophagy: herpesviruses rule! J Virol. 2009;83:1168–1172. doi: 10.1128/JVI.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 30.Wang T, Weinman SA. Causes and consequences of mitochondrial reactive oxygen species generation in hepatitis C. J Gastroenterol Hepatol. 2006;21 Suppl 3:S34–S37. doi: 10.1111/j.1440-1746.2006.04591.x. [DOI] [PubMed] [Google Scholar]
- 31.Waris G, Tardif KD, Siddiqui A. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem Pharmacol. 2002;64:1425–1430. doi: 10.1016/s0006-2952(02)01300-x. [DOI] [PubMed] [Google Scholar]
- 32.Schroder M, Kaufman RJ. Divergent roles of IRE1alpha and PERK in the unfolded protein response. Curr Mol Med. 2006;6:5–36. doi: 10.2174/156652406775574569. [DOI] [PubMed] [Google Scholar]
- 33.Mulvey M, Arias C, Mohr I. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol. 2007;81:3377–3390. doi: 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng G, Feng Z, He B. Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2alpha dephosphorylation by the gamma(1)34.5 protein. J Virol. 2005;79:1379–1388. doi: 10.1128/JVI.79.3.1379-1388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gale M, Jr, Katze MG. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 36.Gale MJ, Jr, Korth MJ, Katze MG. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol. 1998;10:157–162. doi: 10.1016/s0928-0197(98)00034-8. [DOI] [PubMed] [Google Scholar]
- 37.Ambrose RL, Mackenzie JM. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol. 2010 doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luquin E, Larrea E, Civeira MP, Prieto J, Aldabe R. HCV structural proteins interfere with interferon-alpha Jak/STAT signalling pathway. Antiviral Res. 2007;76:194–197. doi: 10.1016/j.antiviral.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor DR, Silberstein E. Innate immunity and hepatitis C virus: eluding the host cell defense. Front Biosci. 2009;14:4950–4961. doi: 10.2741/3579. [DOI] [PubMed] [Google Scholar]
- 41.Coccia EM, Uze G, Pellegrini S. Negative regulation of type I interferon signaling: facts and mechanisms. Cell Mol Biol (Noisy-le-grand) 2006;52:77–87. [PubMed] [Google Scholar]
- 42.Kumar KG, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279:46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- 43.Kumar KG, Tang W, Ravindranath AK, Clark WA, Croze E, Fuchs SY. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. Embo J. 2003;22:5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar KG, Varghese B, Banerjee A, Baker DP, Constantinescu SN, Pellegrini S, Fuchs SY. Basal ubiquitin-independent internalization of interferon alpha receptor is prevented by Tyk2-mediated masking of a linear endocytic motif. J Biol Chem. 2008;283:18566–18572. doi: 10.1074/jbc.M800991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marijanovic Z, Ragimbeau J, Kumar KG, Fuchs SY, Pellegrini S. TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem J. 2006;397:31–38. doi: 10.1042/BJ20060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng H, Qian J, Varghese B, Baker DP, Fuchs SY. Ligand-Stimulated Downregulation of the Interferon Alpha Receptor: Role of Protein Kinase D2. Mol Cell Biol. 2011 doi: 10.1128/MCB.01154-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Plotnikov A, Banerjee A, Suresh Kumar KG, Ragimbeau J, Marijanovic Z, Baker DP, Pellegrini S, Fuchs SY. Ligand-independent pathway that controls stability of interferon alpha receptor. Biochem Biophys Res Commun. 2008;367:388–393. doi: 10.1016/j.bbrc.2007.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Carvalho LP, Bhattachariya S, Carbone CJ, Kumar KG, Leu NA, Yau PM, Donald RG, Weiss MJ, Baker DP, McLaughlin KJ, Scott P, Fuchs SY. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and Type I interferon signaling. Mol Cell Biol. 2009;29:6401–6412. doi: 10.1128/MCB.00478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya S, HuangFu WC, Liu J, Veeranki S, Baker DP, Koumenis C, Diehl JA, Fuchs SY. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J Biol Chem. 2010;285:2318–2325. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McPherson S, Powell EE, Barrie HD, Clouston AD, McGuckin M, Jonsson JR. No evidence of the unfolded protein response in patients with chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2011;26:319–327. doi: 10.1111/j.1440-1746.2010.06368.x. [DOI] [PubMed] [Google Scholar]
- 52.Asselah T, Bieche I, Mansouri A, Laurendeau I, Cazals-Hatem D, Feldmann G, Bedossa P, Paradis V, Martinot-Peignoux M, Lebrec D, Guichard C, Ogier-Denis E, Vidaud M, Tellier Z, Soumelis V, Marcellin P, Moreau R. In vivo hepatic endoplasmic reticulum stress in patients with chronic hepatitis C. J Pathol. 2010;221:264–274. doi: 10.1002/path.2703. [DOI] [PubMed] [Google Scholar]
- 53.Lam M, Dubyak G, Chen L, Nunez G, Miesfeld RL, Distelhorst CW. Evidence that BCL-2 represses apoptosis by regulating endoplasmic reticulum-associated Ca2+ fluxes. Proc Natl Acad Sci U S A. 1994;91:6569–6573. doi: 10.1073/pnas.91.14.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Mechanism of caspase activation. J Biol Chem. 2001;276:33869–33874. doi: 10.1074/jbc.M102225200. [DOI] [PubMed] [Google Scholar]
- 55.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 56.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 57.Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, Arnold SM. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002;3:411–421. doi: 10.1038/nrm829. [DOI] [PubMed] [Google Scholar]
- 58.Zong WX, Li C, Hatzivassiliou G, Lindsten T, Yu QC, Yuan J, Thompson CB. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J Cell Biol. 2003;162:59–69. doi: 10.1083/jcb.200302084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 62.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 29:4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 64.Fernandez PM, Tabbara SO, Jacobs LK, Manning FC, Tsangaris TN, Schwartz AM, Kennedy KA, Patierno SR. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res Treat. 2000;59:15–26. doi: 10.1023/a:1006332011207. [DOI] [PubMed] [Google Scholar]
- 65.Drogat B, Auguste P, Nguyen DT, Bouchecareilh M, Pineau R, Nalbantoglu J, Kaufman RJ, Chevet E, Bikfalvi A, Moenner M. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res. 2007;67:6700–6707. doi: 10.1158/0008-5472.CAN-06-3235. [DOI] [PubMed] [Google Scholar]
- 66.Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, Le QT, Koong AC. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–5947. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 67.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, Munshi N, Horner J, Ivanova EV, Protopopov A, Anderson KC, Tonon G, DePinho RA. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. Embo J. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta S, McGrath B, Cavener DR. PERK regulates the proliferation and development of insulin-secreting beta-cell tumors in the endocrine pancreas of mice. PLoS ONE. 2009;4:e8008. doi: 10.1371/journal.pone.0008008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, Diehl JA. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29:3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grootjans J, Hodin CM, de Haan JJ, Derikx JP, Rouschop KM, Verheyen FK, van Dam RM, Dejong CH, Buurman WA, Lenaerts K. Level of activation of the unfolded protein response correlates with Paneth cell apoptosis in human small intestine exposed to ischemia/reperfusion. Gastroenterology. 140:529–539. doi: 10.1053/j.gastro.2010.10.040. e3. [DOI] [PubMed] [Google Scholar]
- 72.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaser A, Tomczak M, Blumberg RS. "ER stress(ed out)!": Paneth cells and ischemia-reperfusion injury of the small intestine. Gastroenterology. 140:393–396. doi: 10.1053/j.gastro.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lugea A, Tischler D, Nguyen J, Gong J, Gukovsky I, French SW, Gorelick FS, Pandol SJ. Adaptive unfolded protein response attenuates alcohol-induced pancreatic damage. Gastroenterology. 140:987–997. doi: 10.1053/j.gastro.2010.11.038. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]