Abstract
Eosinophilic esophagitis (EoE) is a chronic inflammatory disorder of the esophagus that is compounded by both genetic predisposition and aberrant responses to environmental antigens, particularly those that are food-derived. Data have indicated a unique transcriptional response in vivo that defines EoE and which is partially attributable to the Th2 cytokine interleukin 13 (IL-13). Moreover, a number of genetic risk variants in pro-inflammatory and epithelial cell genes associate with EoE susceptibility, demonstrating novel heritable mechanisms that contribute to disease risk. Here, we discuss recent advances in our understanding of the intrinsic (genetic) and extrinsic (environmental) components that illustrate the complex nature of EoE.
Keywords: Eosinophilic esophagitis, genetics, candidate gene, genome-wide association, polymorphism
The phenomenon of esophageal eosinophilia can be traced through the published literature as far back as 19621. Since these early reports, much progress has been made at the molecular and clinical levels to tease apart the intricacies that distinguish EoE from other inflammatory disorders including gastroesophageal reflux disease (GERD). As the prevalence of these diseases has increased since the late 1990s, the need for improved diagnoses, both from a therapeutic and research standpoint, has arisen. Moreover, the high degree of overlap in presenting symptoms in EoE and GERD have been problematic clinically and have necessitated the establishment of diagnostic criteria to differentiate the two diseases.
In 2007, the First International Gastrointestinal Research Symposium (FIGERS) published initial guidelines for the clinical diagnosis of EoE based on the symptomology and histology of the disease2. More recently, updated consensus recommendations from a panel of allergists, pathologists, and gastroenterologists have been stated (see Liacouras, et al. in this issue of J Allergy Clin Immunol). These recommendations emphasize that EoE is a chronic, antigen-driven clinicohistopathological disorder that is etiologically and epidemiologically distinct from GERD. Clinically, EoE is characterized by a spectrum of symptoms indicative of esophageal dysfunction. Pathologically, one or more esophageal mucosal biopsy specimens show eosinophil predominant inflammation in excess of 15 intraepithelial eosinophils per high-powered field (HPF). The disease is isolated to the esophagus and other causes of esophageal eosinophilia should be excluded. The peak age of EoE diagnosis occurs within the first three years of life3, most likely resulting from antigen hypersensitivity as solid foods are introduced, although diagnoses in adults is also common. Disease remission typically occurs with treatment, which may include dietary exclusion and/or topical corticosteroids4. The symptomology of EoE, if untreated, follows a progressive trend according to patient age. In pediatric patients, these symptoms begin as difficulty feeding and vomiting and can result in failure to thrive5, 6. In adolescent and adult patients, abdominal pain, dysphagia, and food impaction are the chief presentations of disease6, 7. Endoscopic examination has identified common esophageal abnormalities associated with EoE such as linear furrowing with loss of vascularity, ring-like structures, and the presence of white exudate on the esophageal epithelium2, 8. Histologically, the esophageal epithelium exhibits extensive basal layer hyperplasia with papillae elongation and fibrosis within the lamina propria and accumulation of eosinophils, B lymphocytes9, and CD4+ and CD8+T lymphocytes10, regulatory T cells11, and mast cells12–14.
In addition to being resistant to acid neutralization therapy, another distinguishing feature of EoE is the high rate of concurrent atopy. Studies have indicated a prominent role for food allergies in EoE with published frequencies ranging from 46–79% within the EoE population3, 5, 15, 16. In comparison to the estimated 22% of peanut allergic individuals who develop tolerance later in life17, only a very small percentage (< 10%) of EoE patients develop tolerance to food antigens as defined by sustained disease remission15, demonstrating the chronic nature of EoE. The most effective therapies currently used to manage EoE are food antigen avoidance or swallowed glucocorticoid treatment18. While these treatments can reduce or eliminate disease symptoms, relapse commonly occurs following re-introduction of allergens or discontinuation of treatment, suggesting food antigen hypersensitivity is a fundamental feature of EoE. A number of empirical and test-based food elimination trials have further implicated food hypersensitivity in the EoE population, however, the high variability and low predictive value of skin-prick tests and serum IgE measurements as demonstrated in these studies suggest that the clinical utility for standardized assessment for food-specific reactivity in EoE patients remains to be determined (see Chehade and Aceves16 for a thorough review of clinical food trials in EoE). Other atopic diseases such as atopic dermatitis (AD), asthma, and allergic rhinitis are also common in the EoE population5. Although these diseases are largely mediated by enhanced sensitivity to aeroallergens, the exacerbated Th2 inflammation and tissue remodeling that occurs within the affected tissues indicate shared mechanisms of disease with EoE.
Epidemiology
With the expansion of EoE cases reported worldwide, multiple studies have aimed to establish a baseline prevalence of EoE and determine whether disease incidence has increased. From 2000–2003, the estimated prevalence of EoE in a pediatric population was approximately 4 in 10,000 with an incidence rate of 0.9–1.3 in 10,000 new cases per year 5. A similar prevalence (~2 cases per 10,000) and incidence rate (1.4 per 100,000) during a 16-year period were observed in an adult Swiss cohort19. A study of 1000 esophageal biopsies from a randomized Swedish cohort showed a disease prevalence of 0.4% as defined using an esophageal eosinophil level of > 20/hpf20. However, in a retrospective study examining esophageal biopsies from 666 pediatric patients diagnosed with esophagitis from 1982–1999, data suggest that while the prevalence of EoE was increasing, the incidence of disease was relatively stable despite the marked increase in esophagogastroduodenoscopies during that time period21. Using the current recommended eosinophil threshold for EoE diagnosis (> 15/hpf), 198 of these patients had sufficient eosinophils levels with many having histologic indication of basal layer expansion and lamina propria fibrosis to indicate a retrospective diagnosis of EoE21. These findings suggest that enhanced disease recognition, rather than a true increase in disease incidence, underlies the emergence of EoE within the last decade. Notably, dysphagia was significantly associated with retrospective EoE cases, and the ancestry and gender of these cases were similar to those currently reported, with the majority being Caucasians (81%) and male (72%)21.
An interesting questionnaire-based study on the geographic distribution of EoE across the U.S. has indicated higher disease prevalence in urbanized areas, with a higher concentration of EoE observed in the northeastern states. Here, the estimated prevalence of EoE in the entire U.S. was 52 per 100,00022. A similar trend of a higher EoE prevalence in urban areas was shown to be independent of race, indicating that environment has an equally important contribution as genetics to EoE risk23. Certainly the high incidence of asthma among urban populations as demonstrated by multiple groups has garnered significant attention24 and given credence to the general hypothesis that increased exposure to aeroallergens is a predisposing factor. The findings by Spergel et al. and Franciosi et al., which define these EoE “hotzones” within urban settings, implicate a similar impact of socioeconomic factors in EoE susceptibility.
Genetic Heritability
An underlying genetic predisposition to EoE has been proposed by multiple groups that show a disproportionate prevalence of disease in Caucasians and males, and within families of affected individuals15, 23. For instance, data over a 14-year period demonstrated that 90% of the EoE patients were Caucasian and 75% were male15. Reports of a familial occurrence of EoE and esophageal dilatation in 6.8% and 9.7% of EoE patients, respectively, suggest the prevalence of EoE and associated esophageal dysfunction is high among related individuals5. Furthermore, the increased risk of EoE among siblings is dramatic when compared to other disorders. For instance, the estimated sibling recurrence risk among siblings of EoE patients (λs = ~ 80) is markedly higher compared to that of other atopic diseases with familial inheritance patterns such as asthma (λs = ~ 2)26. However, despite this strong familial inheritance, comparison of familial to sporadic cases of EoE showed no difference in esophageal pathology (with the exception of linear furrowing) and gene expression profiles25. Nonetheless, genetic predisposition and family history likely have a significant role in EoE susceptibility and thus detailed family histories are paramount when encountering these patients.
Transcriptome Analysis
A major step toward the molecular mapping of EoE was achieved when gene expression profiling of patient esophageal biopsies showed a remarkable transcript signature that distinguishes EoE from normal controls and patients with chronic esophagitis27. Altered expression of approximately 574 genes comprise this EoE “transcriptome”, which exhibits a high level of conservation among patient gender, age, and atopic history and strongly correlates with esophageal eosinophil levels. The most highly induced gene in the esophagus of EoE patients is the eosinophil chemoattractant eotaxin-3 (CCL26), which was overexpressed 53-fold in EoE esophageal biopsies compared to normal esophageal biopsies in this study27. Eotaxin-3 belongs to the eotaxin family (eotaxins 1–3) of CC chemokines and, through its receptor CCR3, activates downstream G protein signaling to drive eosinophil chemotaxis and activation. Of the eotaxins, only CCL26 is upregulated in EoE, and its expression correlates with eosinophil (and mast cell) levels within esophageal biopsies, indicating a specific contribution in the disease27. Notably, levels of CCL26 transcript in a single biopsy are highly sensitive (89%) in distinguishing EoE from control populations28 despite the histological “patchiness” of EoE across multiple biopsy specimens. In fact, histological examination of at least three biopsies is required to achieve similar diagnostic sensitivity2, 4. Immunofluorescence and in situ hybridization studies on esophageal biopsies identify the esophageal epithelium as the main source of eotaxin-3 production27. In vivo models of EoE further illustrate the crucial role of eotaxin-3 in disease as mice deficient in the eotaxin receptor Ccr3 are protected from esophageal eosinophilia following allergen challenge27. Steroid therapy, in particular swallowed glucocorticoids, effectively normalizes as much as 98% of the EoE transcriptome29, including CCL26, indicating the dynamic nature and reversibility of the gene dysregulation.
In addition to eotaxin-3, a number of immune cell-specific genes exhibit differential expression levels in EoE. For instance, immunoglobulin genes and genes involved in antibody class switching are elevated, reflecting the increase in the esophageal B cell population in EoE9. Mast cell-specific genes, specifically carboxypeptidase 3A (CPA3), high-affinity IgE receptor (FcεRI), and tryptase-α (TPSAB1), are abundantly represented in the EoE transcriptome, and mast cells are indeed a prominent inflammatory cell in the esophagus of EoE patients when specifically examined using anti-tryptase staining12, 27. Based on mast cell levels, a specific esophageal transcriptome is also identified in EoE patients, which only partially overlaps with the transcriptome defined by eosinophil levels alone12, indicating that mast cells and eosinophils are likely independently involved, at least in part. Significant increases in mast cell degranulation and mastocytosis within the epithelium, lamina propria, and smooth muscle layer12, 13, which can be ameliorated with steroid therapy12, further implicate these cells in the local inflammatory milieu within the esophagus.
A significant portion of the gene transcriptional changes associated with EoE occurs within the esophageal epithelium. These structural, non-immune cells can influence multiple aspects of disease phenotype, including inflammatory cell recruitment, tissue remodeling and hyperproliferation. The human esophageal epithelium is composed of non-keratinized, stratified squamous epithelia with a proliferating basal layer of one to three cells in depth and a differentiating suprabasal layer migrating towards the esophageal lumen30. Many of the histopathological features of the esophagus that are associated with EoE indicate gross defects in cell adherence as indicated by dilated intercellular spaces, expansion of the basal cell layer, and extracellular matrix deposition within the lamina propria. Studies have highlighted IL-13 as a critical signaling molecule capable of altering global gene expression of the esophageal epithelium. Ex vivo microarray analysis showed that treatment of biopsy-derived primary esophageal epithelial cells with IL-13, which is upregulated at the mRNA level in EoE, can largely recapitulate the EoE transcriptome29. This study also confirmed epithelial cells as the primary source of CCL26 in EoE, which was upregulated by an astounding 279-fold following IL-13 stimulation ex vivo29. Notably, esophageal epithelial cells derived from EoE and control individuals respond similarly to IL-13 as assessed by CCL26 production31.
Animal models have provided demonstrative data highlighting the robust pro-inflammatory action of IL-13 in an in vivo setting. Lung-specific overexpression of ll13 in mice induces an asthma-like phenotype in the absence of antigen challenge that is characterized by marked inflammatory cell infiltration into the lungs and enhanced airway mucus production32. However, this model also promotes inflammation within the esophagus, such as esophageal eosinophilia and tissue remodeling including fibrosis, angiogenesis, and epithelial hyperplasia33. The esophageal remodeling in this model occurs independent of eosinophilia and is inhibited by the type 2 IL-13 receptor (IL13Rα2)33. In summary, these findings implicate the esophageal epithelium as the pathogenic target of IL-13 signaling in EoE as demonstrated by the induction of pronounced histologic and molecular changes that occur in the presence of this potent Th2 cytokine.
The epidermal differentiation complex (EDC) on human chromosome 1q21 is a cluster of genes that regulates terminal differentiation and formation of the cornified envelope of the epithelium34. Despite the lack of a cornified layer in the esophagus, the EDC locus contains the highest density of dysregulated genes in the EoE transcriptome compared with all other loci in the genome31. Loss-of-function mutations in several EDC genes, including filaggrin (FLG), have been reported for various cutaneous disorders35–39. FLG, involucrin (IVL), and several small proline-rich repeat (SPRR) family members (2C, 2D, and 3) are expressed in esophageal epithelial cells but are downregulated in response to IL-13 ex vivo31, implicating a homeostatic role for the EDC in the esophageal epithelium.. Loss of FLG expression and subsequent defects in epidermal barrier function have been demonstrated in AD40, 41, which frequently co-occurs with EoE. However, no significant difference in FLG expression is observed between atopic and non-atopic EoE patients31, suggesting an alternative function for filaggrin in regulating the epithelial structure within the human esophagus.
It is important to note that 2% of the EoE transcriptome is not reversible following disease remission induced by swallowed glucocorticoids29. Interestingly, these transcripts include genes that are involved in regulating homeostatic and pathogenic responses in the epithelium, such as cadherin-like 26 (CDH26), uroplakin 1B (UPK1B), periostin (POSTN), and desmoglein-1 (DSG1)29. DSG1 is a transmembrane desmosomal cadherin component of desmosomes and facilitates the calcium-dependent homotypic interactions between adjacent cells that impart both structure and mechanical strength to the epithelia. Expression of DSG1 is decreased in both glucocorticoid-treated and untreated EoE patients (77% and 87%, respectively) compared to normal controls. DSG1 is of particular importance as it is the target of multiple inherited and acquired cutaneous disorders. Pemphigus foliaceus and pemphigus vulgaris are autoimmune diseases in which autoantibodies targeting DSG1 decrease cellular adhesion, resulting in epidermal blistering42. Notably, epithelial microabcesses exhibiting pronounced eosinophilic inflammation that can be associated with pemphigoid disorders have also been demonstrated within the esophagus, such as in pemphigus vegetans43. Furthermore, multiple heterozygous mutations in the extracellular domain coding region of DSG1 have been linked with striate palmoplantar keratoderma (SPPK), a disease characterized by epidermal thickening on the palms and soles44. Collectively, these findings substantiate the significance of alterations in DSG1 in a spectrum of human diseases; it is tempting to speculate that tissue-specific decreases in DSG1 may be pathogenic and partially responsible for the tissue-specific inflammation in EoE.
Periostin (POSTN) is another key molecule that demonstrates steroid resistance in EoE. Periostin, which functions as a cell adhesion molecule that regulates extracellular matrix deposition45, 46, is dramatically upregulated in EoE by approximately 52-fold and while glucocorticoid therapy can reduce a significant portion this overexpression, POSTN remains elevated in glucocorticoid-treated patients (~2-fold)47. Periostin is expressed in the basal epithelium and papillae47 of the esophagus, suggesting a contributing role for the increased lamina propria fibrosis. Indeed, TGF-β, a pro-fibrotic stimulus that is expressed by eosinophils and mast cells in EoE patient biopsies13, 48, can induce a dramatic upregulation of POSTN expression in primary esophageal fibroblasts, supporting this potential mechanism for tissue fibrosis observed in EoE47, 49. Moreover, periostin can enhance eosinophil adhesion in vitro and Postn-deficient mice are protected from allergen-induced eosinophilia in the lung and esophagus47. Interestingly, periostin upregulation in bronchial epithelial cells enhances TGF-β-induced collagen synthesis50. As periostin also enhances cross-linking of collagen fibrils through upregulating the cleavage of mature, active lysyl oxidase51, these cumulative data suggest a positive feedback loop in which periostin has a central role in promoting the fibrotic responses in multiple inflammatory conditions.
In summary, esophageal transcript profiling has defined an EoE specific-transcript signature that is composed of dysregulated gene networks involved in Th2 inflammation and epithelial cell responses. These studies demonstrate that IL-13 is a central mediator and link between the immunological and histological changes that are germane to EoE, largely through its effects on the esophageal epithelium. Given the well-documented role of IL-13 in other atopic diseases such as asthma and AD, it is reasonable to speculate that IL-13 production in response to inhaled or absorbed antigens can also predispose individuals to other Th2 comorbidities such as EoE.
Genetic Variants and Disease Susceptibility
The number of studies investigating genetic variants associated with EoE are few compared to other more common and more widely-recognized atopic diseases such as AD and asthma. Regardless, there have been significant strides in uncovering EoE risk variants in a relatively short period of time14, 27, 52, 53 due in part to the technological advances in genotyping single-nucleotide polymorphisms (SNPs) in large case-control cohorts. In all, there have been four candidate gene studies, which tested for polymorphisms in genes with a published (or suspected) functional role in EoE, and one genome-wide association study (GWAS), used to identify EoE risk variants across the entire genome in an unbiased fashion (Table 1).
Table 1.
Genetic risk variants in EE
| Chr | SNP1 | Alleles2 | Gene/gene locus | SNP location3 | Study Design | P-value and Odds Ratio | Summary | Reference |
|---|---|---|---|---|---|---|---|---|
| 7q11 | rs2302009 | T>G | Eotaxin-3 (CCL26) | 3′ URT | Candidate gene study (117 cases, 225 controls) | P = 0.001, OR = 4.55 | Significance in a case-control association was also replicated by transmission disequilibrium testing in a trios cohort. | 25 |
| 19q13 | - (C-509T) | C>T | TGF-β1 (TGFB1) | promoter | Candidate gene study (20 cases) |
P = 0.02 for response status P = 0.01 for TGF-β1+ cells |
CC genotype correlated with therapy response; T allele associated with increased numbers of TGF-B1+ cells in lamina propria | 13 |
| 1q21 | rs61816761 (2282del4) | CAGT>- | Filaggrin (FLG) | exon | Candidate gene study (365 cases, 164 controls) | P = 0.018, OR = 4.89 | Loss-of-function mutation in FLG associated with EE | 29 |
| 5q22 | rs10062929 | C>A | TSLP | intron | Large – scale candidate gene study (257 cases, 342 controls) | meta-P = 3.16 × 10−6, OR = 0.36–0.45 | TSLP SNPs associated with EE risk independent of atopy | 52 |
| Xp22/Yp11 | rs36133495 | G>T | TSLP receptor (CRLF2) | exon | Candidate gene study (199 cases, 78 controls) | P = 0.039, OR = 2.05 | Ala>Val amino-acid change in TSLP receptor associated with male EE patients | 52 |
| 5q22 | rs3806932 rs7723819 |
A>G G>A |
TSLP WDR36 |
near gene near gene |
Genome-wide association study (351 cases, 3,104 controls) | meta-P = 3.19 × 10−9, OR = 0.54–0.73 meta-P = 7.67 × 10−9, OR = 0.55–0.71 |
Minor (protective) G allele correlated with decreased esophageal TSLP expression | 64 |
dbSNP Build 131 “rs” identifier given when appropriate.
major allele>minor allele.
SNP location in relation to the gene/gene locus.
Abbreviations used: Chr, chromosome; OR, odds ratio
Candidate Gene Studies
Blanchard et al. identified the first EoE risk variant in a likely candidate, CCL26. The CCL26 single-nucleotide polymorphism (rs2302009) was shown to be highly associated with disease risk (P = 0.001) with an odds ratio = 4.55 in a case-control cohort27. Transmission disequilibrium testing, which measures the transmission of a disease allele from unaffected, heterozygous parents to an affected offspring, confirmed the association of rs2302009 with EoE was not due to ancestral differences in the case-control analysis27. Additional studies have also linked this SNP to increased serum IgE levels and asthma susceptibility54. However, the observed association between rs2302009 and EoE was independent of atopic status, indicating a direct link with EoE susceptibility. Although rs2302009 is located within the 3′ untranslated region of the CCL26 transcript and could potentially affect mRNA stability, a functional effect of this SNP in either asthma or EoE has yet to be described.
TGF-β, an eosinophil and mast cell-derived mediator of fibrotic tissue responses, has been implicated in the same pathogenic process in EoE48. Moreover, TGF-β1 has recently been shown to stimulate esophageal smooth muscle contractility and potentially contribute to esophageal dysmotility in EoE13. A SNP within the TGFB1 promoter (C-509T) that associated with asthma susceptibility55, 56 was shown to create a binding site for the transcription factor YY1 that subsequently enhanced promoter activity56. In a small cohort of 20 EoE patients, homozygotes for the minor T allele of C-509T exhibited increased TGF-β1-positive lamina propria cells14. Conversely, the major C allele of C-509T was a positive prognostic indicator for therapeutic responses in EoE14. Determining the association of this and other TGFB1 SNPs in a larger disease cohort will be vital to assess the full genetic contribution of TGFB1 in EoE.
Polymorphisms in epithelial-specific genes have also been associated with EoE susceptibility. First, a loss-of-function SNP in FLG (2282del4) that was previously linked with AD susceptibility38 also associates with EoE risk; similar to the CCL26 SNP, this association is specific to EoE as atopy was found not to be a confounding factor31. A second and larger candidate gene study examined 736 SNPs in 52 genes known to be involved in epithelial cell structure or inflammatory responses. Here, an EoE cohort of 170 patients was genotyped using a custom SNP chip and compared to similarly genotyped controls with various atopic histories53. Importantly, SNPs in thymic stromal lymphopoietin (TSLP), a cytokine recently described as a “master regulator” of Th2 responses57, were shown to associate with EoE independent of patient atopic status. TSLP is derived primarily from epithelial cells in response to cytokines58, noxious substances59 and mechanical stress60 and exerts its effects on nearly every cell type involved in Th2 inflammation including eosinophils61 and mast cells62. For instance, TSLP activates dendritic cells to adopt a Th2 priming phenotype through the secretion of the chemokines TARC, MDC, and eotaxin-2 and OX40L expression, which activates Th2 cytokine production by naïve CD4+ T cells63–65. Thus, it is remarkable that TSLP critically regulates the exact processes involved in allergen sensitization that underscore the EoE phenotype. This study also identified an association between male EoE patients and a non-synonymous SNP in the TSLP receptor (CRLF2)53, which, given the male predilection for EoE, presents an intriguing scenario as CRLF2 is encoded on pseudoautosomal region 1 of the X and Y chromosomes66.
EoE GWAS
A broader, unbiased GWAS approach was undertaken to identify SNPs associated with EoE susceptibility. Here, two relatively large cohorts of EoE cases and normal controls were genotyped for 550,000 SNPs across the genome52. While only one locus on chromosome 5q22 was genome-wide significant following multiple testing correction, this region contains the genes encoding for TSLP and WD repeat domain 36 (WDR36). Esophageal expression of TSLP but not WDR36 is increased in EoE, and the protective minor allele for the most significantly EoE-associated SNP on 5q22 (rs3806932), which lies upstream of the TSLP locus, correlates with decreased TSLP expression in the esophagus. Notably, rs3806932 is in linkage disequilibrium (LD) with rs380693352, 67, suggesting that these two SNPs are inherited together more often than would be expected by chance. Data recently implicated rs3806933 in altering the binding of the transcription factor activator protein-1 to the TSLP promoter with a modest increase in promoter activity67. The other genome-wide significant SNP on 5q22 is upstream of the WDR36 gene, located approximately 14 kb away from TSLP, and lies within the same linkage block as rs380693252. WDR36 is critically involved in ribosomal RNA processing68 and is co-regulated with IL2 in activated T lymphocytes69. Moreover, SNPs in the region of WDR36 have been associated with peripheral blood eosinophilia70 as well as glaucoma susceptibility71. Thus, while TSLP appears to be the likely disease candidate on the 5q22 locus, the role of WDR36 warrants further investigation.
GWASs on other more-common gastrointestinal inflammatory diseases such as Crohn’s disease72, ulcerative colitis73, and celiac disease74–76 have successfully identified numerous disease risk variants aided in part by the well-developed patient cohorts being historically investigated for these diseases. For example, meta-analyses across large, independent case-control cohorts (often in excess of 10,000 combined patients) along with further refinement of the human genome polymorphism map have yielded sufficient sample sizes to detect significant disease associations with common variants that have relatively low effect sizes. The low sample size of the current EoE GWAS (251 EoE cases in total) not only emphasizes the magnitude of the 5q22 SNP associations but also suggests that there are likely additional EoE risk variants to be uncovered as further EoE cohorts are subjected to genome-wide genotyping and similar meta-analyses are performed (in essence, boosting the statistical power). A hint at what SNPs or gene loci may hold potential significance in these future studies can be gained by investigation into those that failed to reach the statistical threshold for significance from the previous GWAS52. For instance, SNPs in STAT6, the major signaling molecule downstream of IL-13 signaling, and in the aforementioned DSG1, which is largely resistant to steroid-dependent regulation in EoE, are highly associated with EoE but under the genome-wide significance threshold52. Given the robust immune component of EoE and the overexpression of HLA-DR in the esophagus of EoE patients77 it is quite surprising there was a lack of an association between EoE and polymorphisms in the HLA locus previously linked to other gastrointestinal diseases such as celiac disease75, 78.
Conclusions and Future Directions
In just over ten years since the recognition of EoE as a distinct inflammatory disorder, the rapid progress towards characterizing the disease on multiple fronts has underscored its complexity. We now have insight into the natural history of the EoE, its strong association with specific ethnicities and genders, the genetic and environmental factors involved, and the molecular pathogenesis of the disease (Fig. 1). Moreover, the burst of data illustrating EoE risk variants in CCL26, TGFB1, TSLP and CRLF2, and FLG provide insight into the upstream mechanisms that regulate the expression of genes that are operational (and likely synergistic) in multiple aspects of EoE pathogenesis (Fig. 2). For instance, perturbations in the TSLP signaling pathway as a result of variants either increasing TSLP levels or altering receptor function can amplify innate inflammatory responses to food antigens. Moreover, prolonged CCL26 expression may further enhance eosinophil recruitment and TGF-β1 secretion to exacerbate tissue remodeling. Variants affecting FLG expression may disrupt normal esophageal barrier function and result in increased antigen exposure and affect overall tissue integrity. Despite these advances, much work remains in terms of identifying true casual variants and determining their mechanistic function in these pathways. A major initiative currently underway is to expand upon the current genome-wide-associated polymorphisms by increasing the number of genotyped EoE patients; this will undoubtedly greatly expand the number of genetic loci linked with EoE risk. Moreover, deep sequencing efforts and extensive fine-mapping of the established EoE susceptibility loci such as TSLP could identify rare and/or casual variants that affect gene transcription.
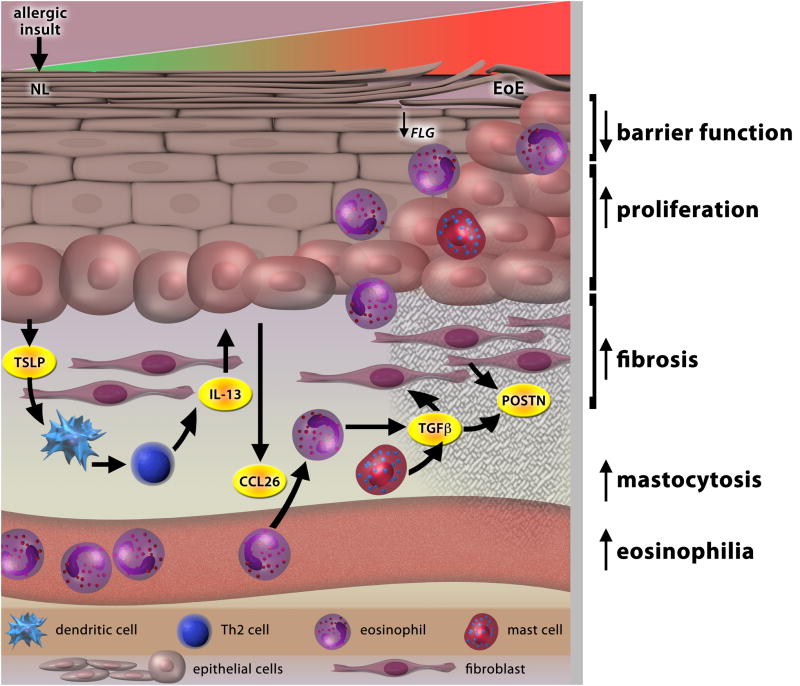
Figure 1. The molecular pathogenesis of EoE.
An allergic insult by either food antigens or aeroallergens initiates the transition of the esophagus from a normal (NL) to EoE phenotype through the production of TSLP by the esophageal epithelium. TSLP-activated DCs induce a robust Th2 response and enhanced IL-13, which in turn mediates marked dysregulation of gene expression (the EoE transcriptome). Enhanced eotaxin-3 (CCL26) secretion by the esophageal epithelium promotes eosinophil migration from the blood into the tissue. Eosinophil- and mast cell-derived TGF-β, along with IL-13, act on fibroblasts within the lamina propria to secrete periostin (POSTN) and stimulate the fibrotic response. Loss of FLG expression, partially due to IL-13 and/or genetic variants, may further enhance or even predispose EoE patients to antigen exposure and exacerbate Th2 inflammation.
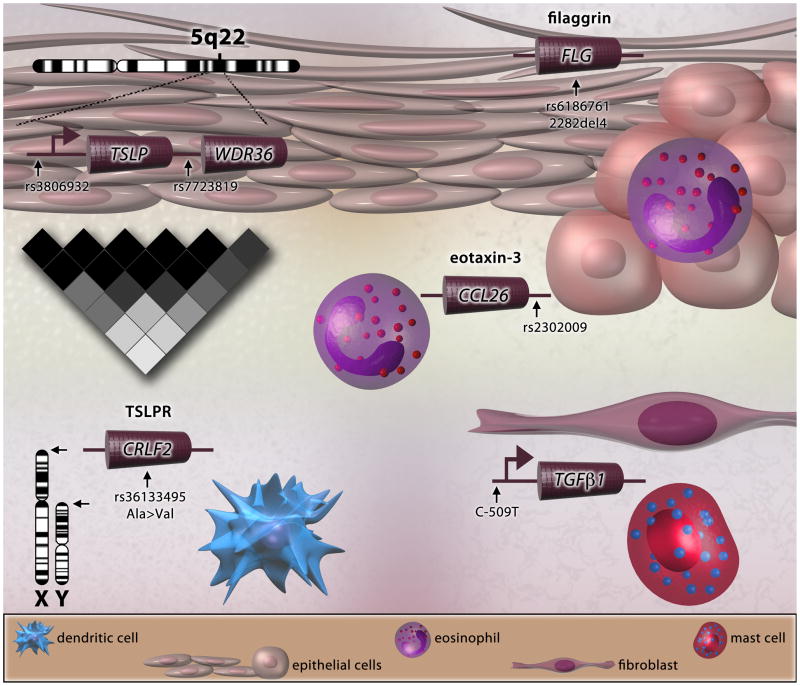
Figure 2. Genetic risk variants in EoE.
EoE risk variants near TSLP and in the TSLP receptor (TSLPR) gene (CRLF2) highlight a potential role for the TSLP pathway in EoE. SNPs in other key genes such as CCL26, TGFB1 and FLG can affect multiple aspects of EoE pathogenesis, including eosinophil chemotaxis, fibrosis and smooth muscle dysfunction, and decreased esophageal barrier function, respectively.
An additional area to be explored in EoE heritability will be the role of epigenetics, which can be defined as the study of heritable changes in gene expression that are not associated with DNA sequence variations which can include non-coding RNAs, histone modifications (acetylation and methylation), and DNA methylation81. Importantly, as these genomic alterations can be influenced by external stimuli such as diet and drugs, epigenetics can provide insight into the complex interactions between environmental exposures and disease-associated genes. The profiling of global epigenetic changes in large disease cohorts has already yielded promising results for cancer, cardiovascular disease, obesity81, and asthma82. As EoE is also influenced by environmental antigen exposure, the uncovering of an EoE epigenome through microRNA arrays, DNA methylation profiling, and chromatin immunoprecipitation-sequencing (ChIP-seq) technologies will provide a critical link to the global gene transcriptional changes already known to occur in EoE. Recent data have already indicated that IL-13 can elicit acetylation changes to histone H3 at the CCL26 promoter in esophageal epithelial cells, implicating epigenetic modifications represent a novel mechanism of gene regulation in EoE83.
The pivotal role of the esophageal epithelium in EoE and the transcriptional changes that occur within different stratified layers of the epithelium provide potential opportunities for non-invasive biomarkers for EoE. Laser-capture microscopy allows for the isolation of specific cell types from minute sections of tissues that can subsequently be subjected to microarray or mass spectrometry analysis. Such techniques have already be used to define the transcriptomes79 and proteomes80 of the various esophageal layers in Barrett’s esophagus. Identification of an EoE-specific transcript profile specific to the suprabasal epithelium may yield diagnostic targets from the skin or oral mucosa samples.
In conclusion, it is remarkable how the genetic dissection of EoE susceptibility has uncovered key pathways that are now being considered for treatment strategies. For example, our findings identify new targets for antibody neutralization strategies (such as anti-IL-13) and specific cell types for directed therapy such as mast cells and epithelial cells, which also supports the clinical value of topical steroid therapy. Therefore, over the next ten years, further unraveling of the genetics and environmental factors that compound EoE holds great promise for the future development of novel and highly effective therapies.
Acknowledgments
This work was supported in part by NIH U19 AI070235, R01 DK076893, R37 AI1045898, the PHS Grant P30 DK0789392, Department of Defense, Food Allergy Project, the Buckeye Foundation and the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation. J.D.S. is supported by a T32 NIH training grant (HL091805). M.E.R. has proprietary interest in reslizumab, a drug being developed by Cephalon.
We would like to thank all of the participating families, patients, physicians, and nurses as well as members of the clinical research team (A. Ahrens, B. Buckmeier Butz, A. Ellison, A. Greenberg, A. Greenler, T. Grotjan, S. Jameson, E. Stucke, and M. Mingler) at the Cincinnati Center for Eosinophilic Disorders for assistance with patient enrollment, DNA preparation and/or database management. We are also grateful to S. Hottinger for her editorial assistance with this review.
Abbreviations used
- EoE
eosinophilic esophagitis
- hpf
high-powered field
- GERD
gastroesophageal reflux disease
- IL-13
interleukin 13
- EDC
epidermal differentiation complex
- TSLP
thymic stromal lymphopoietin
- CCL26
chemokine (C-C motif) ligand 26
- TGF-β1
transforming growth factor, beta 1
- HLA
human leukocyte antigen
- FLG
filaggrin
- DSG1
desmoglein-1
- POSTN
periostin
- WDR36
WD repeat domain 36
- SNP
single-nucleotide polymorphism
- GWAS
genome-wide association study
Glossary
- BASAL ZONE HYPERPLASIA (BZH), PAPILLARY ELONGATION
Histological findings in EoE that are caused by active basal cell proliferation and increased extension of the vascular papillae and subepithelial lamina propria into the epithelial space. Other typical histologic features of EoE include dilated intercellular spaces and lamina propria fibrosis
- CCR3 and EOTAXINS
C-C Chemokine Receptor-3 (CCR3) binds eotaxins. While eotaxins-1, −2 target eosinophils to the lung and lower gastrointestinal tract, eotaxin-3 is present only in humans and functions as a chemoattractant for esophageal eosinophils
- CHROMATIN IMMUNOPRECIPITATION
ChIP technology uses antibodies to precipitate a protein bound to DNA. The bound DNA sequence can be analyzed to look for, e.g. target sequences for transcription factors or histone associated regions of DNA
- EPIGENETICS
The study of changes in DNA configuration that allow for changes in gene expression independent of sequence changes. For example, DNA methylation can cause DNA closing, making specific genetic regions inaccessible to RNA polymerase, transcription factors thus silencing gene expression. Histone acetylation can allow DNA to open and can increase transcription. Creb binding protein (CBP, p300) is a histone acetylase implicated in EoE pathogenesis
- FILAGGRIN
INVOLUCRIN, Involucrin is present in the cytoplasm of keratinocytes and is cross linked to cell membrane proteins via transglutaminase. This allows the formation of a strong epithelial barrier and decreases skin invasion by microorganisms. The usual function of filaggrin is to function as a natural moisturizing factor. Loss of function in the filaggrin gene causes ichthyosis vulgaris and predisposes to eczema, asthma, and eosinophilic esophagitis
- GENOME WIDE ASSOCIATION STUDY (GWAS)
GWAS uses gene chip technology and bioinformatics to analyze the human genome for single nucleotide polymorphisms in diseased and non-diseased states. Haplotypes (in blocks) that vary between diseased and non-diseased subjects are considered to be associated with the disease state
- INTERLEUKIN 13
IL-13 is a Th2 cell-derived interleukin capable of inducing multiple aspects of eosinophil- associated tissue remodeling. IL-13 overexpression in target organs of transgenic animals is associated with pulmonary, esophageal, and cutaneous fibrosis as well as angiogenesis
- LAMBDA (λ)
The familial relative risk is defined by the statistic λ which assesses the risk of disease in an individual with a diseased first degree biological relative to the risk in the population at large. The larger the λ, the stronger the genetic effect is on the disease
- LINEAR FURROWING, WHITE EXUDATES
Typical endoscopic findings in EoE include esophageal lichenification, linear furrowing, pallor, and white exudates/plaques (histologically comprised of eosinophils), strictures, and concentric rings due to motility or fibrosis
- SINGLE NUCLEOTIDE POLYMORPHISM (SNP)
Genetic variant in single nucleotide that may be normally present in the population and associated with risk for certain complex polygenic diseases
- TARC
Thymus and activation regulated chemokine (TARC), also known as CCL17 induces migration of CCR4+ T cells to the skin in eczema patients and can be elevated in the periphery of some EoE patients
- TRANSFORMING GROWTH FACTOR- Beta (TGFβ)
TGFβ is produced by epithelial cells, and inflammatory cells including eosinophils, and mast cells and has pro-fibrotic effects. TGFβ1, 2, and 3 reside on distinct chromosomes but utilize the same signaling pathway and receptors
- TRANSCRIPTOMES
PROTEOMES, Gene expression microarray defines transcriptional differences among subjects with and without a disease state. Gene chip technology and bioinformatics are utilized to analyze the level of gene expression (referred to as the “transcriptome”) across the genome in diseased versus control populations. Proteomes are the protein expression pattern of an organism. Proteomic analysis can use techniques such as 2D gel analysis and peptide sequencing or antibody arrays to find both previously known and unknown proteins associated with a particular disease state
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schreiber MH. Granuloma of the esophagogastric junction with eosinophilic infiltration. Gastroenterology. 1962;43:206–11. [PubMed] [Google Scholar]
- 2.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–63. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Assa’ad AH, Putnam PE, Collins MH, Akers RM, Jameson SC, Kirby CL, et al. Pediatric patients with eosinophilic esophagitis: an 8-year follow-up. J Allergy Clin Immunol. 2007;119:731–8. doi: 10.1016/j.jaci.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 4.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood S, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011 doi: 10.1016/j.jaci.2011.02.040. In press. [DOI] [PubMed] [Google Scholar]
- 5.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–1. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 6.Noel RJ, Rothenberg ME. Eosinophilic esophagitis. Curr Opin Pediatr. 2005;17:690–4. doi: 10.1097/01.mop.0000184291.34654.be. [DOI] [PubMed] [Google Scholar]
- 7.Atkins D, Kramer R, Capocelli K, Lovell M, Furuta GT. Eosinophilic esophagitis: the newest esophageal inflammatory disease. Nat Rev Gastroenterol Hepatol. 2009;6:267–78. doi: 10.1038/nrgastro.2009.45. [DOI] [PubMed] [Google Scholar]
- 8.Dellon ES, Gibbs WB, Fritchie KJ, Rubinas TC, Wilson LA, Woosley JT, et al. Clinical, endoscopic, and histologic findings distinguish eosinophilic esophagitis from gastroesophageal reflux disease. Clin Gastroenterol Hepatol. 2009;7:1305–13. doi: 10.1016/j.cgh.2009.08.030. quiz 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59:12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucendo AJ, De Rezende L, Comas C, Caballero T, Bellon T. Treatment with topical steroids downregulates IL-5, eotaxin-1/CCL11, and eotaxin-3/CCL26 gene expression in eosinophilic esophagitis. Am J Gastroenterol. 2008;103:2184–93. doi: 10.1111/j.1572-0241.2008.01937.x. [DOI] [PubMed] [Google Scholar]
- 11.Fuentebella J, Patel A, Nguyen T, Sanjanwala B, Berquist W, Kerner JA, et al. Increased number of regulatory T cells in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2010;51:283–9. doi: 10.1097/MPG.0b013e3181e0817b. [DOI] [PubMed] [Google Scholar]
- 12.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:140–9. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–204. e4. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 14.Aceves SS, Newbury RO, Chen D, Mueller J, Dohil R, Hoffman H, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65:109–16. doi: 10.1111/j.1398-9995.2009.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spergel JM, Brown-Whitehorn TF, Beausoleil JL, Franciosi J, Shuker M, Verma R, et al. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48:30–6. doi: 10.1097/MPG.0b013e3181788282. [DOI] [PubMed] [Google Scholar]
- 16.Chehade M, Aceves SS. Food allergy and eosinophilic esophagitis. Curr Opin Allergy Clin Immunol. 2010;10:231–7. doi: 10.1097/ACI.0b013e328338cbab. [DOI] [PubMed] [Google Scholar]
- 17.Skolnick HS, Conover-Walker MK, Koerner CB, Sampson HA, Burks W, Wood RA. The natural history of peanut allergy. J Allergy Clin Immunol. 2001;107:367–74. doi: 10.1067/mai.2001.112129. [DOI] [PubMed] [Google Scholar]
- 18.Brown-Whitehorn TF, Spergel JM. The link between allergies and eosinophilic esophagitis: implications for management strategies. Expert Rev Clin Immunol. 2010;6:101–9. doi: 10.1586/eci.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straumann A, Simon HU. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–9. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Ronkainen J, Talley NJ, Aro P, Storskrubb T, Johansson SE, Lind T, et al. Prevalence of oesophageal eosinophils and eosinophilic oesophagitis in adults: the population-based Kalixanda study. Gut. 2007;56:615–20. doi: 10.1136/gut.2006.107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBrosse CW, Collins MH, Buckmeier Butz BK, Allen CL, King EC, Assa’ad AH, et al. Identification, epidemiology, and chronicity of pediatric esophageal eosinophilia, 1982–1999. J Allergy Clin Immunol. 2010;126:112–9. doi: 10.1016/j.jaci.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spergel JM, Book WM, Mays E, Song L, Shah SS, Talley NJ, et al. Variation in Prevalence, Diagnostic Criteria, and Initial Management Options for Eosinophilic Gastrointestinal Diseases in the United States. J Pediatr Gastroenterol Nutr. 2010 doi: 10.1097/MPG.0b013e3181eb5a9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franciosi JP, Tam V, Liacouras CA, Spergel JM. A case-control study of sociodemographic and geographic characteristics of 335 children with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:415–9. doi: 10.1016/j.cgh.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Togias A, Fenton MJ, Gergen PJ, Rotrosen D, Fauci AS. Asthma in the inner city: the perspective of the National Institute of Allergy and Infectious Diseases. J Allergy Clin Immunol. 2010;125:540–4. doi: 10.1016/j.jaci.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 25.Collins MH, Blanchard C, Abonia JP, Kirby C, Akers R, Wang N, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6:621–9. doi: 10.1016/j.cgh.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118:1054–9. doi: 10.1016/j.jaci.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. e7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Odze RD. Pathology of eosinophilic esophagitis: what the clinician needs to know. Am J Gastroenterol. 2009;104:485–90. doi: 10.1038/ajg.2008.40. [DOI] [PubMed] [Google Scholar]
- 31.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–41. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–88. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo L, Fulkerson PC, Finkelman FD, Mingler M, Fischetti CA, Blanchard C, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol. 2010;185:660–9. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.South AP, Cabral A, Ives JH, James CH, Mirza G, Marenholz I, et al. Human epidermal differentiation complex in a single 2.5 Mbp long continuum of overlapping DNA cloned in bacteria integrating physical and transcript maps. J Invest Dermatol. 1999;112:910–8. doi: 10.1046/j.1523-1747.1999.00613.x. [DOI] [PubMed] [Google Scholar]
- 35.Kainu K, Kivinen K, Zucchelli M, Suomela S, Kere J, Inerot A, et al. Association of psoriasis to PGLYRP and SPRR genes at PSORS4 locus on 1q shows heterogeneity between Finnish, Swedish and Irish families. Exp Dermatol. 2009;18:109–15. doi: 10.1111/j.1600-0625.2008.00769.x. [DOI] [PubMed] [Google Scholar]
- 36.McLean WH, Palmer CN, Henderson J, Kabesch M, Weidinger S, Irvine AD. Filaggrin variants confer susceptibility to asthma. J Allergy Clin Immunol. 2008;121:1294–5. doi: 10.1016/j.jaci.2008.02.039. author reply 5–6. [DOI] [PubMed] [Google Scholar]
- 37.Weidinger S, Illig T, Baurecht H, Irvine AD, Rodriguez E, Diaz-Lacava A, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–9. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 39.Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–42. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 40.Fallon PG, Sasaki T, Sandilands A, Campbell LE, Saunders SP, Mangan NE, et al. A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming. Nat Genet. 2009;41:602–8. doi: 10.1038/ng.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Regan GM, Sandilands A, McLean WH, Irvine AD. Filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2009;124:R2–6. doi: 10.1016/j.jaci.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Kottke MD, Delva E, Kowalczyk AP. The desmosome: cell science lessons from human diseases. J Cell Sci. 2006;119:797–806. doi: 10.1242/jcs.02888. [DOI] [PubMed] [Google Scholar]
- 43.Ichimiya M, Nakano J, Muto M. Pemphigus vegetans involving the esophagus. J Dermatol. 1998;25:195–8. doi: 10.1111/j.1346-8138.1998.tb02379.x. [DOI] [PubMed] [Google Scholar]
- 44.Hunt DM, Rickman L, Whittock NV, Eady RA, Simrak D, Dopping-Hepenstal PJ, et al. Spectrum of dominant mutations in the desmosomal cadherin desmoglein 1, causing the skin disease striate palmoplantar keratoderma. Eur J Hum Genet. 2001;9:197–203. doi: 10.1038/sj.ejhg.5200605. [DOI] [PubMed] [Google Scholar]
- 45.Conway SJ, Molkentin JD. Periostin as a heterofunctional regulator of cardiac development and disease. Curr Genomics. 2008;9:548–55. doi: 10.2174/138920208786847917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snider P, Hinton RB, Moreno-Rodriguez RA, Wang J, Rogers R, Lindsley A, et al. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. 2008;102:752–60. doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–96. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119:206–12. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Li-Kim-Moy JP, Tobias V, Day AS, Leach S, Lemberg DA. Esophageal Subepithelial Fibrosis and Hyalinization Are Features of Eosinophilic Esophagitis. J Pediatr Gastroenterol Nutr. 2011 doi: 10.1097/MPG.0b013e3181ef37a1. [DOI] [PubMed] [Google Scholar]
- 50.Sidhu SS, Yuan S, Innes AL, Kerr S, Woodruff PG, Hou L, et al. Roles of epithelial cell-derived periostin in TGF-beta activation, collagen production, and collagen gel elasticity in asthma. Proc Natl Acad Sci U S A. 2010;107:14170–5. doi: 10.1073/pnas.1009426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maruhashi T, Kii I, Saito M, Kudo A. Interaction between periostin and BMP-1 promotes proteolytic activation of lysyl oxidase. J Biol Chem. 2010;285:13294–303. doi: 10.1074/jbc.M109.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:160–5. e3. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chae SC, Lee YC, Park YR, Shin JS, Song JH, Oh GJ, et al. Analysis of the polymorphisms in eotaxin gene family and their association with asthma, IgE, and eosinophil. Biochem Biophys Res Commun. 2004;320:131–7. doi: 10.1016/j.bbrc.2004.05.136. [DOI] [PubMed] [Google Scholar]
- 55.Ueda T, Niimi A, Matsumoto H, Takemura M, Yamaguchi M, Matsuoka H, et al. TGFB1 promoter polymorphism C-509T and pathophysiology of asthma. J Allergy Clin Immunol. 2008;121:659–64. doi: 10.1016/j.jaci.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med. 2004;169:214–9. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 57.Ziegler SF. The role of thymic stromal lymphopoietin (TSLP) in allergic disorders. Curr Opin Immunol. 2010;22:795–9. doi: 10.1016/j.coi.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogiatzi SI, Fernandez I, Bichet JC, Marloie-Provost MA, Volpe E, Sastre X, et al. Cutting Edge: Proinflammatory and Th2 cytokines synergize to induce thymic stromal lymphopoietin production by human skin keratinocytes. J Immunol. 2007;178:3373–7. doi: 10.4049/jimmunol.178.6.3373. [DOI] [PubMed] [Google Scholar]
- 59.Smelter DF, Sathish V, Thompson MA, Pabelick CM, Vassallo R, Prakash YS. Thymic stromal lymphopoietin in cigarette smoke-exposed human airway smooth muscle. J Immunol. 2010;185:3035–40. doi: 10.4049/jimmunol.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oyoshi MK, Larson RP, Ziegler SF, Geha RS. Mechanical injury polarizes skin dendritic cells to elicit a T(H)2 response by inducing cutaneous thymic stromal lymphopoietin expression. J Allergy Clin Immunol. 2010;126:976–84. 84, e1–5. doi: 10.1016/j.jaci.2010.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong CK, Hu S, Cheung PF, Lam CW. Thymic stromal lymphopoietin induces chemotactic and prosurvival effects in eosinophils: implications in allergic inflammation. Am J Respir Cell Mol Biol. 2010;43:305–15. doi: 10.1165/rcmb.2009-0168OC. [DOI] [PubMed] [Google Scholar]
- 62.Allakhverdi Z, Comeau MR, Jessup HK, Yoon BR, Brewer A, Chartier S, et al. Thymic stromal lymphopoietin is released by human epithelial cells in response to microbes, trauma, or inflammation and potently activates mast cells. J Exp Med. 2007;204:253–8. doi: 10.1084/jem.20062211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito T, Wang YH, Duramad O, Hori T, Delespesse GJ, Watanabe N, et al. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J Exp Med. 2005;202:1213–23. doi: 10.1084/jem.20051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–80. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- 65.Liu YJ. Thymic stromal lymphopoietin: master switch for allergic inflammation. J Exp Med. 2006;203:269–73. doi: 10.1084/jem.20051745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tonozuka Y, Fujio K, Sugiyama T, Nosaka T, Hirai M, Kitamura T. Molecular cloning of a human novel type I cytokine receptor related to delta1/TSLPR. Cytogenet Cell Genet. 2001;93:23–5. doi: 10.1159/000056941. [DOI] [PubMed] [Google Scholar]
- 67.Harada M, Hirota T, Jodo AI, Doi S, Kameda M, Fujita K, et al. Functional analysis of the thymic stromal lymphopoietin variants in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2009;40:368–74. doi: 10.1165/rcmb.2008-0041OC. [DOI] [PubMed] [Google Scholar]
- 68.Gallenberger M, Meinel DM, Kroeber M, Wegner M, Milkereit P, Bosl MR, et al. Lack of WDR36 leads to preimplantation embryonic lethality in mice and delays the formation of small subunit ribosomal RNA in human cells in vitro. Hum Mol Genet. 2011;20:422–35. doi: 10.1093/hmg/ddq478. [DOI] [PubMed] [Google Scholar]
- 69.Mao M, Biery MC, Kobayashi SV, Ward T, Schimmack G, Burchard J, et al. T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics. 2004;83:989–99. doi: 10.1016/j.ygeno.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 70.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 71.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14:725–33. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 72.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–5. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 74.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–9. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel AJ, Fuentebella J, Gernez Y, Nguyen T, Bass D, Berquist W, et al. Increased HLA-DR expression on tissue eosinophils in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2010;51:290–4. doi: 10.1097/MPG.0b013e3181e083e7. [DOI] [PubMed] [Google Scholar]
- 78.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40:710–2. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El-Serag HB, Nurgalieva ZZ, Mistretta TA, Finegold MJ, Souza R, Hilsenbeck S, et al. Gene expression in Barrett’s esophagus: laser capture versus whole tissue. Scand J Gastroenterol. 2009;44:787–95. doi: 10.1080/00365520902898127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stingl C, van Vilsteren FG, Guzel C, Ten Kate FJ, Visser M, Krishnadath KK, et al. Reproducibility of Protein Identification of Selected Cell Types in Barrett’s Esophagus Analyzed by Combining Laser-Capture Microdissection and Mass Spectrometry. J Proteome Res. 2011;10:288–98. doi: 10.1021/pr100709b. [DOI] [PubMed] [Google Scholar]
- 81.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010;126:453–65. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CBP-mediated histone 3 acetylation. J Biol Chem. 2011 doi: 10.1074/jbc.M110.210724. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]