Abstract
Sirtuins (SIRT1 7), the mammalian homologues of the Sir2 gene in yeast, have emerging roles in age-related diseases, such as cardiac hypertrophy, diabetes, obesity, and cancer. However, the role of several sirtuin family members, including SIRT1 and SIRT3, in cancer has been controversial. The aim of this review is to explore and discuss the seemingly dichotomous role of SIRT3 in cancer biology with particular emphasis on its potential role as a tumor promoter and tumor suppressor. This review will also discuss the potential role of SIRT3 as a novel therapeutic target to treat cancer.
Keywords: Sirtuin-3, SIRT3, Cancer, Tumor promoter, Tumor Suppressor
1. Introduction
Cancer is a leading cause of death worldwide, and the second cause of death in the United States after heart disease [1]. Despite advances in technology and improved therapeutic approaches to treat this devastating disease, shortcomings remain, especially in treating aggressive and metastatic disease and in predicting individual responses to treatment. These observations underscore the complexity of this disease and the need for personalized cancer therapy to increase the efficacy of treatment in individual cancer patients [2]. Therefore, discovering new pathways that regulate cancer processes is crucial to developing better approaches for cancer prevention and treatment.
The study of sirtuins (SIRT1–7) in cancer tumorigenesis and therapy is an exciting and promising new area in cancer research [3,4]. SIRT3 is of particular interest. It regulates both cell death and survival, and therefore, a controversy has emerged in the literature about its role as a tumor promoter and/or tumor suppressor. In this review, we will explore the controversy to provide the reader with a better understanding of the factors that contribute to this issue, and to discuss future directions and the possibility of using SIRT3 as a novel therapeutic target to treat cancer.
2. Sirtuins: an overview
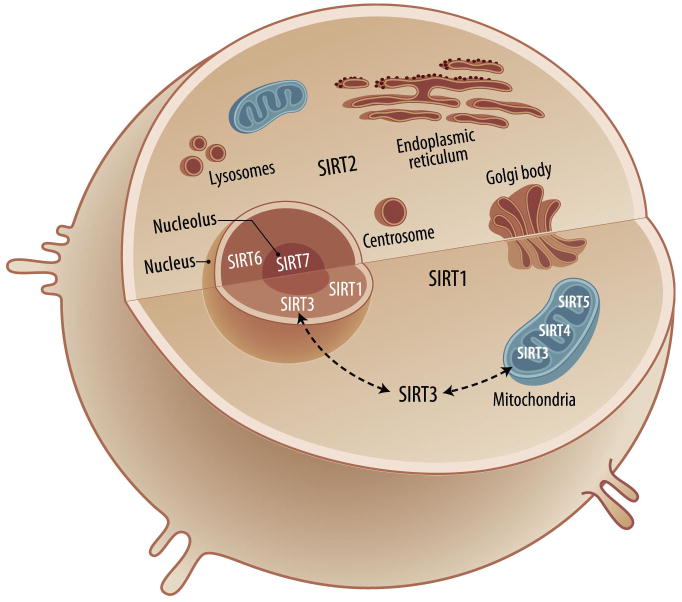
Sirtuins (SIRT1–7) are the mammalian homologues of the silent information regulator 2 (Sir2) first discovered in Saccharomyces cerevisiae as an NAD+-dependent histone deacetylase (HDAC). They are classified as class III HDACs: they require NAD+ as a cofactor to exert their biological function. They contain an evolutionarily conserved core domain, which is essential for their activity as NAD-dependent deacetylases or ADP-ribosyltransferases [5,6]. Sirtuin biology is complex, and sirtuins are widely expressed in normal tissues [7]. They are involved in a myriad of cellular and tissue functions, such as regulating oxidative stress, repairing DNA, increasing genomic stability, and affecting cell survival, apoptosis, development, metabolism, aging and longevity [3,4]. Some sirtuins are located in different cellular compartments (Fig. 1). Those in the same compartment, such as the mitochondrial SIRT3, 4, and 5, have different sequences and thus unique and diverse cellular functions and can interact with different targets [4,5,6].
Figure 1. Sirtuins subcellular localization.
SIRT1 is predominantly located in the nucleus, and also in the cytosol. SIRT2 is localized in the cytosol. SIRT3, SIRT4, and SIRT5 are mitochondrial proteins, but SIRT3 may also be found in the nucleus and cytosol under different cellular events. SIRT6 and SIRT7 are localized in the nucleus and nucleolus, respectively.
SIRT1 is the best-characterized member of the mammalian sirtuins. It is located predominately in the nucleus and modulates cellular stress and survival by deacetylating p53 [8,9], FOXO, and Ku70 [10,11], thus promoting tumorigenesis. SIRT1 is thought to have a role in skin, colon, breast and lung cancer, via one or more of these mentioned targets [12,13,14,15,16]. It also regulates vascular endothelial homeostasis, thereby controlling angiogenesis and vascular function, [17]. Thus, it is likely crucial in regulating cell survival, and its functions may contribute to cancer tumorigenesis.
On the other hand, SIRT1 might be a tumor suppressor [18,19,20]. For example, SIRT1 mutant mice possess an impaired DNA repair response, genomic instability, and increased incidence of tumorigenesis. Moreover, SIRT1 levels were lower in breast cancer and hepatic cell carcinoma than in normal controls [19]. These studies highlight the discrepancy in the literature about the biological functions of SIRT1 and underscore the complexity of sirtuin biology (See review by Deng et al. [21]).
SIRT2 is found in the cytosol, where it colocalizes with microtubules and deacetylates α-tubulin [22]. It controls cell-cycle progression [23] and is downregulated in human gliomas, suggesting a tumor suppressor role in brain cancer [24].
The gene for the nuclear protein SIRT6 is located on chromosome 19p13.3; a region frequently affected by chromosomal alterations in acute leukemia [25]. In addition, SIRT6-deficient mice possess an aging-like phenotype and genomic instability [26,27].
SIRT7, which is localized in the nucleolus and functions as a positive regulator of RNA polymerase I-mediated transcription, is required for cell proliferation and survival [28]. It is located on chromosome 17q25.3; a region frequently associated with chromosomal alterations in leukemias and lymphomas [29]. SIRT7 is also upregulated in breast and thyroid cancers [30,31,32].
The remaining three sirtuins, SIRT3, SIRT4, and SIRT5, are mitochondrial sirtuins [7,33]. Although SIRT4 lacks deacetylation activity, it has weak ADP-ribosyltransferase activity [34,35] and plays an important role in insulin regulation [36]. SIRT4 knockout mice are viable, fertile, and display no phenotype abnormalities, compared to wild-type littermates, but show increased levels of insulin secretion [34]. In contrast to SIRT1 and SIRT3, SIRT4 activity is downregulated by calorie restriction (CR) [34]. SIRT5 has less deacetylase activity than SIRT1-3 [37] and remains the least-characterized sirtuin. SIRT5 is located on chromosome 6p23, an area linked to numerous abnormalities associated with malignant diseases, such as acute myeloid leukemia [38]. In contrast to SIRT4- and SIRT5-deficient mice, SIRT3-deficient mice show greater mitochondrial hyperacetylation than wild-type mice, suggesting that SIRT3 is a key mitochondrial deacetylase [39].
3. SIRT3 subcellular localization
Determining SIRT3’s subcellular localization is important for finding its targets and substrates, explaining its cellular functions, and identifying important signaling cascades that may involve it. Human SIRT3 is expressed as a full-length 44-kD protein that is targeted to the mitochondria by its N-terminal localization sequence [40]. In the mitochondria, SIRT3 is cleaved via the mitochondrial matrix processing peptidase (MPP) to a short 28-kD protein, which is important for SIRT3 enzymatic activity [40,41]. Others reported that both forms for SIRT3 are enzymatically active [42]. Although most studies support a mitochondrial localization for SIRT3 [7,39,40,41,43,44,45,46,47], others suggest that SIRT3 might be present in the nucleus [42,47,48]. In addition, Sundaresan et al. reported that, although the long form of SIRT3 is found in the nucleus, cytoplasm, and mitochondria, the short form is extensively localized in the mitochondria, and during cellular stress, levels of both forms are increased in the nucleus and mitochondria of cardiomyocytes [49]. Despite this controversy, one can conclude that SIRT3 exerts a major role in the mitochondria and might also have a role in other cellular compartments [50].
4. SIRT3 and cell survival
Mitochondria contain large numbers of key molecules that regulate cell survival, death, and metabolic pathways and help to control the balance between health and disease [4,51,52]. For example, SIRT3 is critical for maintaining mitochondrial integrity and function. Along with SIRT4, and SIRT5, SIRT3 is a mitochondrial sirtuin [33], and SIRT3−/− mice manifest hyperacetylated mitochondrial proteins, impaired fatty-acid oxidation, and reduced levels of ATP [39,53,54]. In an early report supporting a prosurvival role for SIRT3 in vivo, rodents fasted for 48 hours had increased levels of the NAD+ biosynthetic enzyme Nampt in their mitochondria. The activity of Nampt, a stress and nutrient-responsive protein involved in maintaining cell viability, is regulated by SIRT3 [55]. In addition, under genotoxic stress, mitochondrial SIRT3 and SIRT4 were required to protect against genotoxic cell death in human embryonic kidney (HEK293) and fibrosarcoma cell lines [55].
With its central role in mitochondrial biology, SIRT3 contributes to cell survival by modulating oxidative stress pathways. Benigni et al. demonstrated that knockout of the angiotensin II type 1 receptor, a gene responsible for promoting high blood pressure and various pathological conditions, such as heart, kidney and brain diseases, promoted longevity in mice [56]. This receptor knockout was associated with increased numbers of mitochondria, attenuation of oxidative stress, and upregulation of Nampt and SIRT3 levels. SIRT3 protects cardiomyocytes and HeLa cells from genotoxic and oxidative stress-mediated cell death. By binding to and deacetylating Ku70, SIRT3 augments Ku70-Bax interactions, prevents Bax translocation to the mitochondria, and prevents apoptosis during stress-mediated conditions [49]. In addition, SIRT3 protects the heart from cardiac hypertrophy, at least in part, by attenuating reactive oxygen species (ROS) [57] and/or by regulating the mitochondrial permeability transition pore (mPTP) via deacetylating Cyclophilin-D [58]. SIRT3 utilizes exogenous NAD+ to block cardiac hypertrophy by activating the LKB1-AMP kinase pathway [59] (See review by Pillai et. al. [60]). Under CR conditions, SIRT3 deacetylates and activates superoxide dismutase 2 (SOD2), thus protecting cells from ROS-mediated cell damage [61]. SIRT3 also mediates deacetylation of the evolutionarily conserved lysine 122 needed for the activity of manganese superoxide dismutase (MnSOD) in response to oxidative stress, thus protecting cells from stress-mediated damage [62]. Furthermore, in neurons, SIRT3 acts as prosurvival factor, thus protecting neurons from excitotoxic injury such as N-methyl-D-aspartate (NMDA)-induced neuronal death [63].
SIRT3 also exerts a prosurvival role in multiple cancer pathways. The tumor suppressor, p53 was recently identified as a new target for SIRT3 deacetylation in bladder cancer [64]. SIRT3 rescued p53-induced growth arrest in human bladder tumor–derived EJ-p53 cells, supporting a prosurvival role for SIRT3 [64]. Ashraf et al. reported that increased transcriptional levels of SIRT3 were associated with lymph node–positive breast cancer, and SIRT3 expression was significantly higher in these samples than normal breast biopsies [30]. We recently reported that SIRT3 levels were significantly higher in oral squamous cell carcinoma (OSCC) cell lines and human OSCC tissue microarray samples (TMAs) than in normal controls [65]. Furthermore, SIRT3 downregulation inhibited cell growth and proliferation and increased the sensitivity of OSCC cells to radiation and chemotherapy treatments. To further demonstrate the role of SIRT3 in oral cancer carcinogenesis in vivo, we used a floor-of-mouth oral cancer murine model that mimics human OSCC [66,67] to study the effect of SIRT3 downregulation on OSCC tumor growth in immunodeficient mice. Downregulating SIRT3 reduced tumor burden in vivo, implicating a prosurvival role for SIRT3 in oral cancer [65].
Anoikis, apoptotic cell death triggered by loss of extracellular matrix (ECM) contacts, is dysregulated in many chronic debilitating and fatal diseases, including cancer, and resistance to anoikis contributes to the development and progression of cancer [68,69,70]. We recently reported that anoikis activates a CD95/Fas-mediated signaling pathway regulated by receptor interacting protein (RIP), a kinase that shuttles between CD95/Fas-mediated cell death and integrin/FAK-mediated survival pathways in oral squamous cell carcinoma (OSCC) cells [69]. Interestingly, we found that, as OSCC cells become resistant to anoikis, their SIRT3 expression increases and their RIP expression decreases. These cells exhibit a greater tumor burden in vivo. Additionally, SIRT3 is highly expressed in OSCC tissues and cells, where its expression pattern is opposite to that of RIP expression (Kapila lab; unpublished data). Thus, these observations suggest that SIRT3 may play a role in mediating anoikis resistance and tumorigenesis.
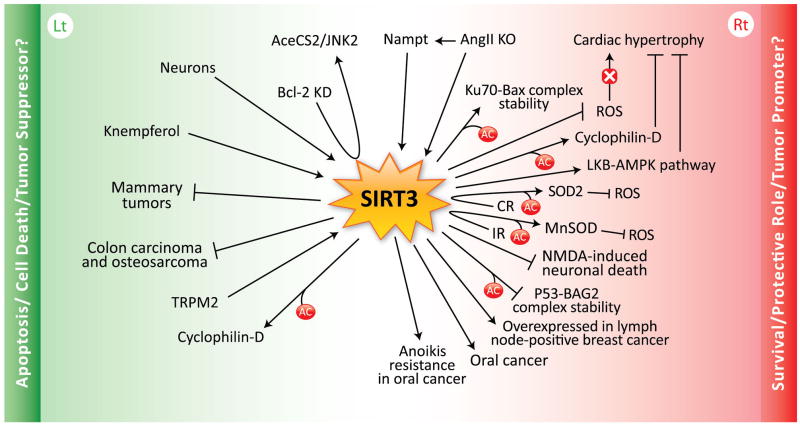
In summary, SIRT3 promotes survival and protects several cell types from cellular damage by maintaining mitochondrial integrity and functions and by enhancing their resistance to stress-mediated cell death. Similarly, SIRT3 overexpression in cancer cells promotes survival signals while suppressing apoptotic signals, thereby enhancing tumorigenesis (Fig. 2).
Figure 2. SIRT3 diverse cellular functions. (Rt) The role of SIRT3 in survival, cell protection, and tumor promotion.
Nicotinamide phosphoribosyltransferase (Nampt), a stress and nutrient-responsive gene, protects against genotoxic cell death via SIRT3 upregulation. Gene knockout (KO) of Ang II promotes longevity in mice either directly or indirectly through Nampt or SIRT3 upregulation. SIRT3 deacetylates Ku70, augmenting Ku70-Bax interaction, thus attenuating apoptosis and promoting cell survival in cardiomyocytes. In the heart, SIRT3 also prevents cardiac hypertrophy by attenuating reactive oxygen species (ROS), deacetylating cyclophilin-D, and activating the anti-hypertrophic LKB1-AMP kinase signaling pathway. Calorie restriction (CR) prevents aging and age-related diseases by augmenting SIRT3 levels and functions in the cell, at least in part, by deacetylating and activating superoxide dismutase 2 (SOD2), thus protecting the cell from ROS-induced cell death. In addition, ionizing radiation (IR) in normal cells may enhance SIRT3-deacetylated-MnSOD activation, therefore, again protecting the cell from ROS-induced cell death. Although it is still controversial, in neurons SIRT3 seems to protect cells from excitotoxic injury such as N-methyl-D-aspartate (NMDA)-induced neuronal death. SIRT3 deaceylates p53, attenuating p53-BAG2 complex stability (BAG2; BCL2-associated athanogene 2), thus decreasing apoptosis. Lymph node–positive breast cancer is associated with increased levels of SIRT3. SIRT3 is also overexpressed in oral cancer and in anoikis-resistant oral squamous cell carcinoma (OSCC) cells, thus promoting OSCC cell survival and preventing anoikis-mediated cell death.. (Lt) The role of SIRT3 in apoptosis, cell death, and tumor suppression. In colorectal carcinoma, Bcl-2 knockdown (KD) was associated with SIRT3 upregulation and apoptosis by deacetylating AceCS2 and switching on the JNK2 signaling pathway. In neurons, low potassium (LK)-induced apoptosis is mediated by SIRT3. In leukemia, the treatment with Kaempferol, a flavonoid that auto-oxidizes and generates ROS, induces apoptosis by SIRT3 and Bax upregulation, thus switching on caspase-3 cascades and apoptosis. SIRT3 is downregulated in human breast cancer cells compared to normal controls, and SIRT3−/− mice developed mammary tumors over a 24-month period. In human colon carcinoma and osteosarcoma cells, SIRT3 also works as a tumor suppressor by suppressing ROS and HIF1-α. In HEK-293 cells, the transient receptor potential melastatin-related channel 2 (TRPM2), a nonselective cation channel induces cell death in response to oxidative stress via SIRT3. Unlike non-transformed cells, in some cancer cells, SIRT3 deacetylates cyclophilin-D, inducing the dissociation of hexokinase II/VDAC complex in the mitochondria, thus activating apoptosis.
5. SIRT3, apoptosis, and cell death
In contrast, other reports support a proapoptotic role for SIRT3. SIRT3 induces growth arrest and apoptosis in several colorectal carcinoma and osteosarcoma cells and in non-cancer human cell lines, such as retinal epithelial and lung fibroblast cells [71]. This action is mediated, in part, by SIRT3 modulation of the JNK2 signaling pathway in these cell lines [71]. Interestingly, the same group reported earlier that SIRT1 and JNK2 function as constitutive suppressors of apoptosis in colorectal carcinoma [72]. Thus, SIRT1 and SIRT3 have opposite roles in colorectal carcinoma. A similar observation was made in neurons [73], however, the role of SIRT3 in neurons is still controversial [63]. In leukemia cell lines, treatment with Kaempferol, a flavonoid that auto-oxidizes and generate ROS, induces apoptosis via increasing Bax and SIRT3 levels and activating caspase-3 cascades [74].
Recently, SIRT3 was reported to suppress tumors. Kim et al. implanted SIRT3−/− mouse embryonic fibroblasts (MEFs) expressing Myc/Ras into the hind limbs of nude mice. After 3 weeks, these mice developed tumors, but mice implanted with SIRT3+/+ Myc/Ras, SIRT3−/− Myc, or SIRT3−/− Ras MEFs did not. Importantly, SIRT3 knockout MEFs did not undergo spontaneous immortalization or possess a tumorigenic phenotype, unless they became immortalized by the action of Myc or Ras. This transformation-permissive phenotype was mediated by increased levels of ROS, chromosomal instability, and altered intracellular metabolism. Some SIRT3−/− mice developed mammary tumors over the 24-month observation period. In addition, SIRT3 expression was found to be decreased in commercially obtained TMAs of human breast cancer samples and in other cancers (glioblastoma, prostate, head and neck, and others), based on a review of gene expression data from other sources [75]. These findings suggest that SIRT3 is a tumor suppressor. In agreement with this report, others found that, in breast cancer patients, SIRT3 levels were lower or undetectable in most of the samples than in normal individuals, and specifically breast and ovarian cancers were frequently associated with focal deletion of the SIRT3 gene. Additionally, tumors lacking SIRT3 (SIRT3-KO-MEFs transformed with Ras and E1a oncogenes) grew faster and were bigger than transformed SIRT3-WT tumors in a xenograft model. This downregulation was associated with upregulation of hypoxia inducible factor-1 α (HIF1-α) targeted genes [76]. Similarly, Bell et al. also demonstrated a tumor suppressor role for SIRT3 in human colon carcinoma and osteosarcoma cells, via the ability of SIRT3 to negatively regulate ROS and HIF1-α [77].
In HEK-293 cells, the transient receptor potential melastatin-related channel 2 (TRPM2), a nonselective cation channel, confers susceptibility to cell death in response to oxidative stress. This cell death was reduced by treating with the general sirtuin chemical inhibitor, NAM, and with selective downregulation of both SIRT3 and SIRT2 [78], thus supporting a proapoptotic role for these sirtuins in the context of TRPM2 and oxidative stress. SIRT3 also deacetylates cyclophilin D, a protein required for hexokinase II binding to voltage-dependent anion channels (VDACs) to maintain mitochondrial integrity. Thus, in some cancer cells, SIRT3 induces hexokinase II to dissociate from the mitochondria and activate apoptosis. However, in non-transformed cells, activation of SIRT3 may prevent necrotic cell death [79]. From these findings, SIRT3 seems to function as a proapoptotic signal in some cancer and non-cancer cell lines, and it may prevent a transformation-permissive phenotype in certain normal cells, thus guarding the cell as a tumor suppressor (Fig. 2).
6. SIRT3, metabolism, and cancer
Metabolism is important in cancer development and prevention [80]. Cancer cells are metabolically active and need ATP to maintain their growth, proliferation, and survival [81,82]. Thus, understanding how specific regulators of metabolism are altered in cancer will be very helpful for developing therapies. Furthermore, cancer cells shift their mode of ATP/energy production from oxidative phosphorylation to glycolysis. This is called the “Warburg effect” [83,84]. However, cancer cells can switch between glycolysis and fatty acid oxidation, depending on the environment and substrate availability. This suggests that targeting one metabolic pathway may not be sufficient as a treatment strategy, since this may lead to resistance and more aggressive cancer phenotypes [85,86]
Mitochondria are important determinants of energy regulation, metabolic homeostasis, and cellular lifespan [52,87,88], and in the mitochondria, SIRT3 regulates numerous metabolic processes, such as fatty-acid oxidation, oxidative phosphorylation, and the TCA cycle. These observations implicate SIRT3 as a key regulator of cancer processes. Several studies have highlighted the role of SIRT3 in metabolism and homeostasis and revealed new targets and substrates for SIRT3-dependent deacetylation [33,89]. However, few linked SIRT3-regulated metabolism to cancer. Those that addressed SIRT3 as a tumor suppressor demonstrated that SIRT3−/− mice have depleted levels of ATP: about 50% less in the heart, liver and kidney, and increased ROS production than in wildtype mice [54,75]. Indeed, increased ROS levels promote mutagenesis and genomic instability [90], as was the presumed case in SIRT3−/− mice [75]. In addition, SIRT3 critically regulates the Warburg effect. Thus, SIRT3 mediates metabolic destabilization of HIF1-α, a factor that regulates the metabolic shift to glycolysis in cancer cells, and its upregulation is associated with tumorigenesis [76,77].
SIRT3 regulates mitochondrial energy homeostasis. More specifically, it maintains ATP basal levels by regulating mitochondrial electron transport by deacetylating the 39-kD protein NDUFA9 [54] and succinate dehydrogenase [91]. Moreover, ATP synthase (ATP5A) and the chaperone protein HSP70, which protect against oxidative stress, are targets for SIRT3 [92]. SIRT3 also regulates fatty-acid oxidation by deacetylating long-chain acyl coenzyme A dehydrogenase (LCAD), thereby augmenting its enzymatic activity. Mice lacking SIRT3 have hyperacetylated LCAD and fatty-acid oxidation disorders during fasting, including reduced ATP levels, hypoglycemia, and cold intolerance [53]. SIRT3 downregulation in the hepatocyte cell line, HepG2, results in dysfunction in the electron transfer chain, reduction of mitochondrial membrane potential, and increased levels of ROS [47]. The first discovered target of SIRT3-mediated deacetylation was acetyl-CoA synthetase 2 (AceCS2). SIRT3 deacetylates and activates AceCS2, an enzyme important in converting acetate to acetyl-CoA in the presence of ATP and CoA, an enzyme required for TCA (Krebs) cycle initiation [93,94,95]. Since SIRT3 also modulates several enzymes of the TCA cycle, including isocitrate dehydrogenase 2 (IDH2), this is yet another mechanism by which SIRT3 helps to modulate energy production from carbohydrates, fats, and proteins [96]. In addition, under CR conditions, SIRT3 seems to modulate IDH2 function and protect the cell from oxidative stress-induced cell death [97]. Interestingly, mutations of IDH2 are associated with some cancer types, such as gliomas and acute myeloid leukemias; however, the use of IDH2 as a potential therapeutic target is still controversial [98,99,100].
Shi et al. demonstrated that SIRT3 regulates thermogenesis by modulating mitochondrial proteins, such as peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1α) and the uncoupling protein 1 (UCP1) [44]. On the other hand, SIRT3 is a target for PGC-1α, whereby ROS levels are regulated in the cell [101]. PGC-1α itself has important metabolic roles, including regulating adaptive thermogenesis, gluconeogenesis, mitochondrial biogenesis, respiration, and it protects cells against ROS generation and damage. PGC-1α binds to the SIRT3 promoter, enhancing its expression and downstream signaling pathways, including the activation of anti-oxidants SOD2 and catalase [101].
Moreover, SIRT3 levels are lower in obese mice than in normal littermates, thus implicating a role for SIRT3 in controlling obesity [44]. Additionally, ob/ob mice treated with leptin, a key hormone in regulating fat metabolism and energy expenditure, showed increased levels of SIRT3, further supporting a role for SIRT3 in controlling obesity [102]. SIRT3 regulates ketone body production during fasting via deacetylating the mitochondrial protein 3-hydroxy-3-methylglu-taryl CoA synthase 2 (HMGCS2) [103], providing further evidence of the important roles of SIRT3 in regulating cellular metabolism.
In aggregate, although few studies have linked SIRT3-mediated metabolism directly to cancer, all these studies support a role for SIRT3 as a critical regulator of metabolism in the mitochondria, which in turn, participates in cancer development or prevention. Additionally, age-related diseases share common risk factors and perhaps even redundancy in their mechanisms of pathology [104]. Therefore, determining the role of SIRT3 in those diseases may help better define its potential role in their etiology and in the development of novel therapeutics.
7. Is SIRT3 a tumor promoter or suppressor?
Cancer cells possess six common traits including self-sufficiency in growth signals, insensitivity to antigrowth signals, evading apoptosis, sustained angiogenesis, limitless replicative potential, and tissue invasion and metastasis [105]. In addition, emerging hallmarks and enabling characteristics in cancer cells include dysregulation of cellular energy and avoidance of immune distraction, and the consequences of genomic instability and tumor-promoting inflammation are factors that contribute to creating a tumorigenic microenvironment, thus further facilitating and supporting the unique features of cancer cells phenotype [82]. The fact that SIRT3 can regulate most of these cancer processes, implicates SIRT3 as a novel potential therapeutic target to treat cancer. However, the discrepancy in the literature regarding the role of SIRT3 in cancer complicates how best to develop SIRT3 as a potential target for cancer therapy. It may be most appropriate to consider the cell-dependent context, the background of each cell line tested, and the influence of different dysregulated pathways in these cell lines, as in the case for SIRT1 in colon cancer [14].
The finding that SIRT3 is overexpressed in metabolically active tissues, such as the heart, where SIRT3 protects against genomic and stress-mediated apoptosis, at least in part, via ROS reduction and increases in Ku70-Bax interactions [49,57], is one mechanism by which cancer cells that overexpress SIRT3 similarly resist cell death. This was indeed demonstrated by the same group in the HeLa cervical cancer cell line [49].
SIRT3 is overexpressed to a greater extent in several human oral cancer cells and tissues than in normal controls, and SIRT3 downregulation in these cells inhibited OSCC cell growth and proliferation and enhanced radio- and chemo-therapeutic drug cytotoxicity. These observations suggest that these cells rely on SIRT3 signaling for survival. In addition, SIRT3 downregulation in OSCC cells in vivo reduced tumor burden in mice [65], further underscoring the critical role of SIRT3 in promoting survival and tumorigenesis in OSCC. Moreover, under suspension conditions, oral cancer cells aggregate to become anoikis resistant, maintaining their survival signals to escape suspension-induced cell death. One of those survival signals seems to be mediated by increased SIRT3 expression levels (Kapila lab, unpublished data).
Nampt protection against cell death, which was dependent on SIRT3 expression in fibrosarcoma cells [55], and the ability of SIRT3 overexpression to rescue p53-induced cell growth arrest in human bladder cancer [64], also demonstrates a prosurvival role for SIRT3 in these tumors. Overexpression of SIRT3 in lymph node-positive breast cancer, implicates a role for SIRT3 in advanced stages of breast cancer [30]. Together, these findings support a prosurvival role for SIRT3 in cancer, and the concept that SIRT3 functions as a tumor promoter in these tumors.
In contrast to the findings of Ashraf et al. [30], others showed that SIRT3 levels were lower in human breast cancer samples than in normal controls, and SIRT3 levels were further downregulated in advanced stages of breast cancer, supporting an opposite role for SIRT3 in breast cancer [75,76]. Interestingly, Kim et al. showed that MEFs from SIRT3 knockout mice did not become immortalized spontaneously. Instead, they required at least one oncogenic hit with either Myc or Ras to become immortalized in vitro and both Myc and Ras to develop tumors in vivo [75]. This suggests that environmental or genetic factors control how SIRT3 functions in cells. In addition, this group found that seven out of twenty SIRT3 knockout mice developed mammary tumors over a 24-month period, while none of the SIRT3 wildtype mice developed tumors [75]. However, after abrogating the function of an important cellular regulator such as SIRT3, 24 months is a reasonable time for genomic alterations and mutations to accumulate, which are critical initiating factors in the multistep process of cancer development [106,107]. Additionally, since SIRT3 is a critical regulator of the Warburg effect in cancer cells, downregulation of SIRT3, such as in breast cancer, would be associated with increased levels of ROS and a shift in metabolism toward glycolysis via the upregulation of HIF1-α and its targeted genes [76]. Moreover, in human colon carcinoma and osteosarcoma cells, SIRT3 also suppresses, ROS, HIF1-α and its targeted genes. Thus, colon carcinoma cells with stable knockdown of SIRT3 demonstrated enhanced tumorigenesis in a xenograft model, and augmented HIF1-α protein stability and transcriptional activity compared to controls [77].
In contrast, tumors with high levels of SIRT3 as part of their genomic and signaling dysregulation could take advantage of this overexpression to sustain their survival signals by downregulating ROS levels and maintaining high amounts of ATP/energy sufficient for their cancer cell machinery. In this regard, reduced levels of ROS were associated with higher levels of antiapoptotic mitochondrial proteins, such as Bcl-2 and Bcl-xL in OSCC cells [108]. Interestingly, ROS differentially regulates apoptosis and malignant transformation in several cancer types by regulating Bcl-2 [109,110]. Thus, superoxide plays a proapoptotic role by downregulating and degrading Bcl-2 proteins by ubiquitination, whereas nitric oxide (NO)-mediated S-nitrosylation of Bcl-2, abrogates its ubiquitination and subsequent proteosomal degradation [109,110]. In addition, Bcl-2 family proteins have been linked to resistance to cancer therapy in B-cell lymphomas and oral cancer [111,112,113]. It would be interesting to study the role of SIRT3 in modulating Bcl-2 family proteins in these cancer cell types.
Interestingly, SIRT1 is overexpressed in drug-resistant cancer cells, including neuroblastoma, osteosarcoma, mammary, and ovarian carcinomas [114]. Similarly we found that two OSCC cell lines (UM-SCC-1 and UM-SCC-17B) that are highly resistant to radiation and cisplatin treatment [115,116] become sensitive to low doses of both treatments only when SIRT3 levels were downregulated. This suggests a role for SIRT3 in resistance-mediated mechanisms in oral cancer. This resistance could be mediated by regulating Bcl-2 family proteins or related signaling cascades via SIRT3-ROS modulation. These mechanisms are currently under investigation in our laboratory.
Additionally, depending on SIRT3 levels, current evidence suggests that SIRT3 controls mitochondrial ROS directly or indirectly in the cell [60,89], thereby dictating the fate of a given cell type. Therefore, a cell either undergoes damage and assumes an environmentally-permissive neoplastic phenotype [62,75,76,117] or a protective/stress-mediated resistance and undergoes cell survival [49,57,61,62,97].
Interestingly, contrasting findings highlight the dichotomy of SIRT3’s role in cancer processes in different cancer cell types. We found in oral cancer and others found in fibrosarcoma, cervical cancer, and bladder cancer that SIRT3 was required to protect these tumors from stress-mediated cell death by various stimuli [55,57,64,65]. Others showed that SIRT3 was required to suppress tumorigenesis, and to induce stress-mediated cell death in tumors, including colorectal carcinoma, osteosarcoma, leukemia, and breast cancer [71,74,75,76,77]. Furthermore, the recent findings that SIRT3 might deacetylate cyclophilin-D, resulting in either enhancing apoptosis in transformed cells such as HeLa cells [79] or promoting survival and protecting against age-related cardiac hypertrophy [58], clearly show that SIRT3 may function differently depending on cell type. Even in normal cells such as neurons, SIRT3’s role also seems to be controversial [63,73].
Thus, SIRT3’s function varies in different normal and tumor tissues and may be cell- and tumor-type specific. Its role must not be generalized, but should be examined in each cancer type separately to determine whether it functions as a tumor promoter or suppressor. More importantly, the genetic or epigenetic alterations that underlie cancer initiation and progression may differ from individual to individual even within the same cancer type, resulting in a dysregulation of different signaling cascades that may or may not depend on SIRT3. These issues complicate our ability to predict the importance of SIRT3 in a given carcinogenic event. Therefore, looking at the bigger picture by screening cancer patients to analyze their genomic, epigenomic, proteomic, and metabolomic profile will help discover the dysregulated pathways that lead to a given disease, and thereby help maximize and personalize the therapeutic approaches for each patient.
8. Sirtuins as potential therapeutic targets for cancer
Several studies have implicated sirtuins as novel therapeutic targets for many age-related diseases, including cancer, but how sirtuins are involved in cancer is still not clear and controversial. In this review, we want to highlight the discrepancies in the literature about the roles of sirtuins in cancer, especially those of SIRT1 and SIRT3. A clear understanding of how individual sirtuins are involved in different cancer types is important for assessing their potential in possible therapies. Sirtuins seem to be involved in tumorigenesis, and thus, sirtuin inhibitors/modifiers might have therapeutic benefit. Several inhibitors and activators of sirtuins have been tested in different cancer cell lines, but few have been tested in vivo [51]. The sirtuin inhibitors, sirtinol and splitomicin, induced senescence-like growth arrest in breast and lung cancers [118]. NAM, another sirtuin inhibitor, induced apoptosis in lung cancer [8]. We demonstrated that sirtinol and NAM inhibited cell growth and proliferation and induced apoptosis in oral cancer cells [65]. Treatment of B-cell lymphoma cells with cambinol, a SIRT1 inhibitor, inhibited tumor cell growth and induced apoptosis in vitro, and reduced tumor size compared to controls in vivo [119]. Moreover, cambinol sensitized lung cancer cells to the DNA-damaging agent etoposide, thus inducing cell death and etoposide-induced cell-cycle arrest [119] (See review by Balcerczyk et al. [120]).
Resveratrol, a polyphenol phytoalexin and natural component found in the skin of red grapes and red wine, works as an activator of sirtuins and possesses diverse natural therapeutic benefits, including cardiac protection, anti-inflammatory and anti-carcinogenic effects, preventing obesity, and promoting longevity [121,122]. These therapeutic benefits seem to work, at least in part, by activating SIRT1 and SIRT3, although it is not yet clear whether these effects are mediated by direct or indirect mechanisms [121,122,123,124]. Interestingly, resveratrol modulates both survival and death signals, depending on the administered dose in vivo [125]. At low doses (2.5 or 5 mg/kg for 14 days in rats), resveratrol provided cardiac protection and lower levels of apoptosis than controls. In contrast, at high doses (25 or 50 mg/kg), resveratrol hindered cardiac function and promoted apoptosis of cadiomyocytes. The former effect was mediated by augmenting survival signaling pathways, including p-Akt, NFκB, and Bcl-2 activation. The later was mediated by switching on the death program by repressing the same pathways [125]. Moreover, resveratrol is the most studied sirtuin activator in cancer prevention. Many studies have shown resveratrol to be a natural anticarcinogenic agent, modulating different stages of cancer, including initiation, promotion, and progression in neuroblastoma, hepatoma, breast, lung, pancreatic, and prostate cancers in vitro or in vivo [126,127]. However, many of these studies yielded contradictory results even in the same tumor type. We reasoned that these discrepancies could be due to the different experimental approaches used to examine resveratrol. For instance, some data were collected from mice and others from rats. Some studies used animal carcinogenesis models with different genetic backgrounds, others used different doses of resveratrol, and yet others used different time frames for drug administration. Resolving these discrepancies would be very helpful. In addition, these drugs are all generalized inhibitors or activators of several sirtuin family members. Thus, some redundancy or even opposing actions of some sirtuin functions may be expected. Furthermore, different tumors have different genetic backgrounds that differ from one person to another. This diversity may explain why one patient responds well to a specific treatment but another patient with the same type of cancer does not.
The rapidly evolving era of personalized medicine holds great promise for the future of cancer therapy. For example, the use of RNA interference (RNAi) as a specifically targeted therapeutic approach may be useful especially in combination with conventional treatments. Currently, studies using RNAi are still in early stages of clinical trials. RNAi has been tested in different types of cancer, such as lung, advanced liver, and chronic myeloid leukemia. We recently used an OSCC floor-of-mouth model in which mice treated with shRNA-modified-OSCC cells to reduce SIRT3 levels had a lower tumor burden than controls [65]. This demonstrates the usefulness of targeted gene knockdown as a potential therapeutic approach (See review by Phalon et al. [128]).
Class-I and II HDAC inhibitors have been tested in phase-I and II clinical trials with or without conventional chemotherapeutic drugs. The agents were well tolerated with low toxicity and yielded promising results. Some of the agents used include phenyl acetate, suberoylanilide hydroxamic acid, and Trichostatin to treat patients with solid tumors, hematologic malignancies, and advanced leukemias [129]. To our knowledge, there are no published reports on clinical trials using class-III HDAC inhibitors of sirtuins to treat cancer. However, class-III HDAC activators of sirtuins such as resveratrol are currently in early stages of clinical trials, and have been tested for safety and potential treatment of age-related diseases such diabetes, neurodegenerative disorders and cancer (See references [130,131] about ongoing and published clinical trials).
9. Conclusions
We are just beginning to appreciate the role of sirtuins in treating age-related diseases, such as cancer. However, the current controversy regarding the role of SIRT3 in cancer, emphasizes the importance of examining this area further. Data support that SIRT3 can function as a tumor promoter or suppressor, depending on the cell- and tumor-type and the presence of different stress or cell death stimuli. Also, genetic and environmental factors underlying cancer initiation and development in each patient may contribute to this discrepancy. Thus, it is important to carefully examine the role of SIRT3 in each tumor type separately and in both in vitro and in vivo settings. Understanding the mechanistic differences in tumor types will enhance our knowledge of the complex biology of sirtuins in cancer and, ultimately, help us develop better therapeutic approaches. Therefore, screening cancer patients for genomic, proteomic, and metabolomic abnormalities and dysregulated signaling cascades to highlight important alterations might help identify useful targets for personalized cancer therapy and more successful cancer treatment.
Acknowledgments
This work is supported by NIH grant RO1DE014429 and R56DE014429 to YK.
Abbreviations
- NAD+
Nicotinamide adenine dinucleotide
- NAM
Nicotinamide
- CR
Calorie Restriction
- OSCC
Oral Squamous Cell Carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Minino AM, Xu J, Kochanek KD, Tejada-Vera B. Death in the United States, 2007. NCHS Data Brief. 2009:1–8. [PubMed] [Google Scholar]
- 2.Workman P, de Bono J. Targeted therapeutics for cancer treatment: major progress towards personalised molecular medicine. Curr Opin Pharmacol. 2008;8:359–362. doi: 10.1016/j.coph.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 4.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 6.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 7.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 9.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 10.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 11.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 12.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 13.Ota H, Akishita M, Eto M, Iijima K, Kaneki M, Ouchi Y. Sirt1 modulates premature senescence-like phenotype in human endothelial cells. J Mol Cell Cardiol. 2007;43:571–579. doi: 10.1016/j.yjmcc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Stunkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Salto-Tellez M, Ni B, Entzeroth M, Wood J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Sun D, Li F, Tian L, Li C, Li L, Lin R, Wang S. Downregulation of Sirt1 by antisense oligonucleotides induces apoptosis and enhances radiation sensitization in A549 lung cancer cells. Lung Cancer. 2007;58:21–29. doi: 10.1016/j.lungcan.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Hida Y, Kubo Y, Murao K, Arase S. Strong expression of a longevity-related protein, SIRT1, in Bowen’s disease. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- 17.Potente M, Dimmeler S. Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle. 2008;7:2117–2122. doi: 10.4161/cc.7.14.6267. [DOI] [PubMed] [Google Scholar]
- 18.Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 19.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 23.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, Oshimura M. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Mahlknecht U, Ho AD, Voelter-Mahlknecht S. Chromosomal organization and fluorescence in situ hybridization of the human Sirtuin 6 gene. Int J Oncol. 2006;28:447–456. [PubMed] [Google Scholar]
- 26.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 27.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voelter-Mahlknecht S, Letzel S, Mahlknecht U. Fluorescence in situ hybridization and chromosomal organization of the human Sirtuin 7 gene. Int J Oncol. 2006;28:899–908. [PubMed] [Google Scholar]
- 30.Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95:1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frye R. “SIRT8” expressed in thyroid cancer is actually SIRT7. Br J Cancer. 2002;87:1479. doi: 10.1038/sj.bjc.6600635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Nigris F, Cerutti J, Morelli C, Califano D, Chiariotti L, Viglietto G, Santelli G, Fusco A. Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. Br J Cancer. 2002;86:917–923. doi: 10.1038/sj.bjc.6600156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. 2010;1804:1645–1651. doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 34.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 35.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 36.Argmann C, Auwerx J. Insulin secretion: SIRT4 gets in on the act. Cell. 2006;126:837–839. doi: 10.1016/j.cell.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 37.Verdin E, Dequiedt F, Fischle W, Frye R, Marshall B, North B. Measurement of mammalian histone deacetylase activity. Methods Enzymol. 2004;377:180–196. doi: 10.1016/S0076-6879(03)77010-4. [DOI] [PubMed] [Google Scholar]
- 38.Mahlknecht U, Ho AD, Letzel S, Voelter-Mahlknecht S. Assignment of the NAD-dependent deacetylase sirtuin 5 gene (SIRT5) to human chromosome band 6p23 by in situ hybridization. Cytogenet Genome Res. 2006;112:208–212. doi: 10.1159/000089872. [DOI] [PubMed] [Google Scholar]
- 39.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper HM, Huang JY, Verdin E, Spelbrink JN. A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLoS ONE. 2009;4:e4986. doi: 10.1371/journal.pone.0004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 45.Cooper HM, Spelbrink JN. The human Sirt3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 46.Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 2009;18:514–525. doi: 10.1002/pro.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP, Jr, Leclerc J, Nguyen P, Gius D, Sack MN. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem. 2010;110:238–247. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–179. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 49.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku-70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?--functional localization of an NAD+-dependent protein deacetylase. Biochem J. 2008;411:e11–13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Lanza IR, Nair KS. Mitochondrial function as a determinant of life span. Pflugers Arch. 2010;459:277–289. doi: 10.1007/s00424-009-0724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-Sensitive Mitochondrial NAD(+) Levels Dictate Cell Survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P, Morigi M, Coffman TM, Remuzzi G. Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Invest. 2009;119:524–530. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, Gupta MP. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285:3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pillai VB, Sundaresan NR, Jeevanandam V, Gupta MP. Mitochondrial SIRT3 and heart disease. Cardiovasc Res. 2010;88:250–256. doi: 10.1093/cvr/cvq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 62.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SH, Lu HF, Alano CC. Neuronal Sirt3 Protects against Excitotoxic Injury in Mouse Cortical Neuron Culture. PloS one. 2011;6:e14731. doi: 10.1371/journal.pone.0014731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-Induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS ONE. 2010;5:e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D’Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henson B, Li F, Coatney DD, Carey TE, Mitra RS, Kirkwood KL, D’Silva NJ. An orthotopic floor-of-mouth model for locoregional growth and spread of human squamous cell carcinoma. J Oral Pathol Med. 2007;36:363–370. doi: 10.1111/j.1600-0714.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- 67.Wolter KG, Wang SJ, Henson BS, Wang S, Griffith KA, Kumar B, Chen J, Carey TE, Bradford CR, D’Silva NJ. (−)-gossypol inhibits growth and promotes apoptosis of human head and neck squamous cell carcinoma in vivo. Neoplasia. 2006;8:163–172. doi: 10.1593/neo.05691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakamoto S, Kyprianou N. Targeting anoikis resistance in prostate cancer metastasis. Mol Aspects Med. 2010;31:205–214. doi: 10.1016/j.mam.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamarajan P, Bunek J, Lin Y, Nunez G, Kapila YL. Receptor-interacting protein shuttles between cell death and survival signaling pathways. Mol Biol Cell. 2010;21:481–488. doi: 10.1091/mbc.E09-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coates JM, Galante JM, Bold RJ. Cancer therapy beyond apoptosis: autophagy and anoikis as mechanisms of cell death. J Surg Res. 2010;164:301–308. doi: 10.1016/j.jss.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 71.Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- 72.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 73.Pfister JA, Ma C, Morrison BE, D’Mello SR. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS ONE. 2008;3:e4090. doi: 10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, Fini M, Russo MA. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem. 2009 doi: 10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- 75.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism during Stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1alpha Destabilization. Cancer cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011 doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud AL, Scharenberg AM, Denu JM. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J Biol Chem. 2006;281:14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- 79.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Resendis-Antonio O, Checa A, Encarnacion S. Modeling core metabolism in cancer cells: surveying the topology underlying the warburg effect. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madhok BM, Yeluri S, Perry SL, Hughes TA, Jayne DG. Targeting Glucose Metabolism: An Emerging Concept for Anticancer Therapy. Am J Clin Oncol. 2010 doi: 10.1097/COC.0b013e3181e84dec. [DOI] [PubMed] [Google Scholar]
- 82.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 83.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferreira LM. Cancer metabolism: The Warburg effect today. Exp Mol Pathol. 2010;89:372–380. doi: 10.1016/j.yexmp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 85.Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9:230–234. doi: 10.1038/sj.pcan.4500879. [DOI] [PubMed] [Google Scholar]
- 87.Tsujimoto Y. Apoptosis and necrosis: intracellular ATP level as a determinant for cell death modes. Cell Death Differ. 1997;4:429–434. doi: 10.1038/sj.cdd.4400262. [DOI] [PubMed] [Google Scholar]
- 88.Osiewacz HD. Role of mitochondria in aging and age-related disease. Exp Gerontol. 2010;45:465. doi: 10.1016/j.exger.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 89.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tudek B, Winczura A, Janik J, Siomek A, Foksinski M, Olinski R. Involvement of oxidatively damaged DNA and repair in cancer development and aging. Am J Transl Res. 2010;2:254–284. [PMC free article] [PubMed] [Google Scholar]
- 91.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM, Che CM, Leung PT, Wang Y. Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics. 2009;9:2444–2456. doi: 10.1002/pmic.200800738. [DOI] [PubMed] [Google Scholar]
- 93.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.North BJ, Sinclair DA. Sirtuins: a conserved key unlocking AceCS activity. Trends Biochem Sci. 2007;32:1–4. doi: 10.1016/j.tibs.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 97.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss under Caloric Restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 99.Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, Wiemels JL, Nelson HH, Karagas MR, Wrensch MR, Kelsey KT, Wiencke JK. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. Journal of the National Cancer Institute. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu J, Zuo J, Xu Q, Wang X, Wang Z, Zhou D. Isocitrate dehydrogenase mutations may be a protective mechanism in glioma patients. Medical hypotheses. 2011;76:602–603. doi: 10.1016/j.mehy.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 101.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang W, Della-Fera MA, Hartzell DL, Hausman D, Baile CA. Adipose tissue gene expression profiles in ob/ob mice treated with leptin. Life Sci. 2008;83:35–42. doi: 10.1016/j.lfs.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 103.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 Deacetylates Mitochondrial 3-Hydroxy-3-Methylglutaryl CoA Synthase 2 and Regulates Ketone Body Production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Probst-Hensch NM. Chronic age-related diseases share risk factors: do they share pathophysiological mechanisms and why does that matter? Swiss Med Wkly. 2010;140:w13072. doi: 10.4414/smw.2010.13072. [DOI] [PubMed] [Google Scholar]
- 105.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 106.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 107.Foulds L. The natural history of cancer. J Chronic Dis. 1958;8:2–37. doi: 10.1016/0021-9681(58)90039-0. [DOI] [PubMed] [Google Scholar]
- 108.Li D, Ueta E, Kimura T, Yamamoto T, Osaki T. Reactive oxygen species (ROS) control the expression of Bcl-2 family proteins by regulating their phosphorylation and ubiquitination. Cancer Sci. 2004;95:644–650. doi: 10.1111/j.1349-7006.2004.tb03323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Azad N, Iyer A, Vallyathan V, Wang L, Castranova V, Stehlik C, Rojanasakul Y. Role of oxidative/nitrosative stress-mediated Bcl-2 regulation in apoptosis and malignant transformation. Ann N Y Acad Sci. 2010;1203:1–6. doi: 10.1111/j.1749-6632.2010.05608.x. [DOI] [PubMed] [Google Scholar]
- 110.Azad N, Iyer AK, Wang L, Lu Y, Medan D, Castranova V, Rojanasakul Y. Nitric oxide-mediated bcl-2 stabilization potentiates malignant transformation of human lung epithelial cells. Am J Respir Cell Mol Biol. 2010;42:578–585. doi: 10.1165/rcmb.2009-0094OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bauer JA, Kumar B, Cordell KG, Prince ME, Tran HH, Wolf GT, Chepeha DB, Teknos TN, Wang S, Eisbruch A, Tsien CI, Urba SG, Worden FP, Lee J, Griffith KA, Taylor JM, D’Silva N, Wang SJ, Wolter KG, Henson B, Fisher SG, Carey TE, Bradford CR. Targeting apoptosis to overcome cisplatin resistance: a translational study in head and neck cancer. Int J Radiat Oncol Biol Phys. 2007;69:S106–108. doi: 10.1016/j.ijrobp.2007.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stolz C, Hess G, Hahnel PS, Grabellus F, Hoffarth S, Schmid KW, Schuler M. Targeting Bcl-2 family proteins modulates the sensitivity of B-cell lymphoma to rituximab-induced apoptosis. Blood. 2008;112:3312–3321. doi: 10.1182/blood-2007-11-124487. [DOI] [PubMed] [Google Scholar]
- 113.Duan H, Heckman CA, Boxer LM. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol Cell Biol. 2005;25:1608–1619. doi: 10.1128/MCB.25.5.1608-1619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chu F, Chou PM, Zheng X, Mirkin BL, Rebbaa A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005;65:10183–10187. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- 115.Grenman R, Burk D, Virolainen E, Wagner JG, Lichter AS, Carey TE. Radiosensitivity of head and neck cancer cells in vitro. A 96-well plate clonogenic cell assay for squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 1988;114:427–431. doi: 10.1001/archotol.1988.01860160071024. [DOI] [PubMed] [Google Scholar]
- 116.Carey TE, Van Dyke DL, Worsham MJ, Bradford CR, Babu VR, Schwartz DR, Hsu S, Baker SR. Characterization of human laryngeal primary and metastatic squamous cell carcinoma cell lines UM-SCC-17A and UM-SCC-17B. Cancer Res. 1989;49:6098–6107. [PubMed] [Google Scholar]
- 117.Oberley LW. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed Pharmacother. 2005;59:143–148. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 118.Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y, Kaneki M. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 119.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 120.Balcerczyk A, Pirola L. Therapeutic potential of activators and inhibitors of sirtuins. BioFactors. 2010;36:383–393. doi: 10.1002/biof.112. [DOI] [PubMed] [Google Scholar]
- 121.Signorelli P, Ghidoni R. Resveratrol as an anticancer nutrient: molecular basis, open questions and promises. J Nutr Biochem. 2005;16:449–466. doi: 10.1016/j.jnutbio.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 122.Das DK, Mukherjee S, Ray D. Resveratrol and red wine, healthy heart and longevity. Heart Fail Rev. 2010;15:467–477. doi: 10.1007/s10741-010-9163-9. [DOI] [PubMed] [Google Scholar]
- 123.Rayalam S, Yang JY, Ambati S, Della-Fera MA, Baile CA. Resveratrol induces apoptosis and inhibits adipogenesis in 3T3-L1 adipocytes. Phytother Res. 2008;22:1367–1371. doi: 10.1002/ptr.2503. [DOI] [PubMed] [Google Scholar]
- 124.Mukherjee S, Ray D, Lekli I, Bak I, Tosaki A, Das DK. Effects of Longevinex (modified resveratrol) on cardioprotection and its mechanisms of action. Can J Physiol Pharmacol. 2010;88:1017–1025. doi: 10.1139/y10-082. [DOI] [PubMed] [Google Scholar]
- 125.Dudley J, Das S, Mukherjee S, Das DK. Resveratrol, a unique phytoalexin present in red wine, delivers either survival signal or death signal to the ischemic myocardium depending on dose. J Nutr Biochem. 2009;20:443–452. doi: 10.1016/j.jnutbio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 126.Athar M, Back JH, Tang X, Kim KH, Kopelovich L, Bickers DR, Kim AL. Resveratrol: a review of preclinical studies for human cancer prevention. Toxicol Appl Pharmacol. 2007;224:274–283. doi: 10.1016/j.taap.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bishayee A. Cancer prevention and treatment with resveratrol: from rodent studies to clinical trials. Cancer prevention research. 2009;2:409–418. doi: 10.1158/1940-6207.CAPR-08-0160. [DOI] [PubMed] [Google Scholar]
- 128.Phalon C, Rao DD, Nemunaitis J. Potential use of RNA interference in cancer therapy. Expert Rev Mol Med. 2010;12:e26. doi: 10.1017/S1462399410001584. [DOI] [PubMed] [Google Scholar]
- 129.Acharya MR, Sparreboom A, Venitz J, Figg WD. Rational development of histone deacetylase inhibitors as anticancer agents: a review. Mol Pharmacol. 2005;68:917–932. doi: 10.1124/mol.105.014167. [DOI] [PubMed] [Google Scholar]
- 130.Patel KR, Scott E, Brown VA, Gescher AJ, Steward WP, Brown K. Clinical trials of resveratrol. Annals of the New York Academy of Sciences. 2011;1215:161–169. doi: 10.1111/j.1749-6632.2010.05853.x. [DOI] [PubMed] [Google Scholar]
- 131. [(Accessed April 25, 2011).]; http://www.clinicaltrials.gov/ct2/results?term=resveratrol.