Abstract
Summary: Scientists, educators and the general public often need to know times of divergence between species. But they rarely can locate that information because it is buried in the scientific literature, usually in a format that is inaccessible to text search engines. We have developed a public knowledgebase that enables data-driven access to the collection of peer-reviewed publications in molecular evolution and phylogenetics that have reported estimates of time of divergence between species. Users can query the TimeTree resource by providing two names of organisms (common or scientific) that can correspond to species or groups of species. The current TimeTree web resource (TimeTree2) contains timetrees reported from molecular clock analyses in 910 published studies and 17 341 species that span the diversity of life. TimeTree2 interprets complex and hierarchical data from these studies for each user query, which can be launched using an iPhone application, in addition to the website. Published time estimates are now readily accessible to the scientific community, K–12 and college educators, and the general public, without requiring knowledge of evolutionary nomenclature.
Availability: TimeTree2 is accessible from the URL http://www.timetree.org, with an iPhone app available from iTunes (http://itunes.apple.com/us/app/timetree/id372842500?mt=8) and a YouTube tutorial (http://www.youtube.com/watch?v=CxmshZQciwo).
Contact: sbh1@psu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
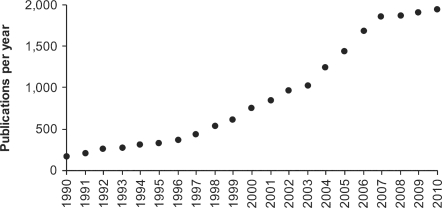
Molecular clocks have emerged as an important tool for timing the origin and divergence of species. This is because a vast majority of species and their ancestors have left little or no traces in the fossil record. Furthermore, the addressing of many biological questions (e.g. origin of life and eukaryotes, colonization of land by plants and fungi, and radiation of mammals) requires the knowledge of time of divergence (Avise 2009; Hedges and Kumar, 2009; Kumar, 2005). The increasing utility of molecular clocks is reflected in the increasing numbers of publications each year that report species divergence times using molecular data (Fig. 1). This has been driven by the development of methods that accommodate rate variation in estimating divergence times and their confidence intervals as well as the ease of sequence data generation owing to advances in sequence technologies (Battistuzzi et al., 2010; Drummond et al., 2006; Hedges and Kumar, 2009).
Fig. 1.
Growth in the number of publications per year reporting molecular clock based estimates of species divergence times. Potentially relevant publications (>19 000) were retrieved from Google Scholar (March, 2011) by using the query: (“molecular clock” OR “divergence time” OR “divergence times” OR “molecular time” OR “molecular divergence” OR “molecular date” OR “molecular dates” OR “divergence dates”) NOT (circadian OR disorder OR periodicity). A randomly selected subset of 20 publications was examined for each year to generate the estimated numbers of relevant publications. We expect these numbers to be minimums because our search strings will not produce all relevant articles.
Scientists and non-scientists frequently need to know when two species diverged in geologic time. Beyond that basic information, many wish to know if there is a consensus in the expert scientific community regarding the average or range of published times, and relevant publications. Answers cannot be generated by text mining the corpus of published literature (e.g. Google search). This is because the primary information on species and divergence times is in evolutionary trees that (i) appear in graphical formats (images) and (ii) contain hierarchical information among species, neither of which is accessible in text searches.
For this reason, we have developed a professional resource in which divergence times reported in the form of phylogenetic trees scaled to time (timetrees), in tables, and in the regular text in 910 studies have been collected in a relational database of hierarchical (tree) data. These data, gathered by visual inspection of articles and by author-submitted data files, encompass 17 341 species among all major groups of life and include expert reports from a consortium of 102 scientists (http://www.timetree.org/consortium.php) in addition to individual studies. This new release advances our earlier prototype resource, which contained data from 70 studies with taxonomic scope limited to tetrapods (amphibians, reptiles, mammals and birds) (Hedges et al., 2006). In addition to database expansion in terms of studies and species diversity, we have also developed a completely new application for Apple's iPhone (and compatible devices) in order to make the knowledgebase available to a greater diversity of potential users and for easier and ready access beyond the classical website view alone.
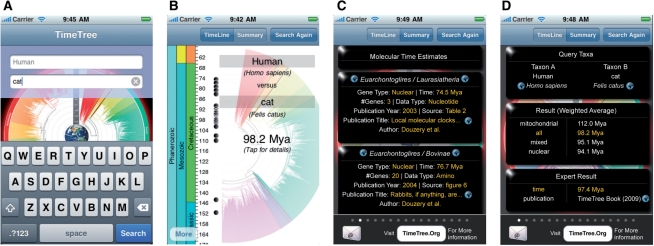
To use TimeTree2 for the iPhone, a user opens the app and is presented with a form for entering the names of two organisms of interest (Fig. 2A). The user then taps the ‘search’ button to initialize the search. TimeTree first uses a synonym list (from NCBI) to translate from many common names and any ambiguities among the query names are resolved by presenting the user with a choice between possible matches. Then, the two most-inclusive groups containing the query organisms are identified, and all published studies included in the TimeTree repository are scanned for taxa pairs across groups to find all published times of divergence for the evolutionary split (Hedges et al., 2006).
Fig. 2.
(A) User interface to provide common or scientific names to query for divergence times. (B) Time estimate along with a geologic timescale. (C) Details on individual studies represented by filled circles in the previous view. (D) Summary of divergence times, including the expert result.
The results of a query return the divergence time (average weighted by number of genes) for the organisms of interest (Fig. 2B). Times from individual studies are marked with a dot along a geological timescale to show the diversity of time estimates relating to the user query (Fig. 2B). By tapping on a marker, a user can navigate to more detailed information for the published study, including the names of taxa used in the study, gene type, divergence time, number of genes, data type (nucleotide or amino acid), author, publication title and source of information within the article (e.g. the specific table or figure) (Fig. 2C).
A Summary page is also available to the users (Fig. 2D), which shows more detailed information on the user-query, including the expert time estimates, if applicable, from The Timetree of Life (Hedges and Kumar, 2009). As an added feature, users can conveniently send query results via email. This feature also enables scientists to generate alternative summary statistics if they desire.
In summary, TimeTree resource has now matured to contain a substantial number of published studies, with studies being added at a steady pace over time. After release of TimeTree2, the rate of queries increased more than 10-fold, now surpassing 20 000 per month, with 60% coming from iPhones. This demonstrates that scientists at large and the general public are interested in the knowledge of species divergence times and use it when it is served in an accessible tool. Furthermore, providing access to scientific resources on new media (e.g. iPhone) decreases latent barriers and amplifies the usage of scientific information, as the iPhone app is currently being downloaded >1400 times each month. TimeTree already is being used by K–12 educators (Metzger, 2011) and we anticipate increased use in the education sector as the knowledgebase expands and graphical user interface is refined.
ACKNOWLEDGEMENTS
We are greatly indebted to L. N. Viswanathan for writing the iPhone app to our specifications and designs. We also thank members of the TimeTree group for their efforts in database management (L.-W. Wu), data acquisition and input (M. Heinicke, M. Hippenstiel, Y. Plavnik, L.-W. Wu), and web design and graphics (M. Owens, W. Parkhurst, M. Powell, L.-W. Wu). Thanks are also due to the members of the TimeTree Consortium for their support and invaluable advice.
Funding: Research grants from the NASA Astrobiology Institute (S.B.H.), US National Science Foundation (S.B.H., S.K.), Science Foundation of Arizona (S.K.), and the US National Institutes of Health (S.K.).
Conflict of Interest: none declared.
REFERENCES
- Avise J. Timetrees: beyond cladograms, phenograms, and phylograms. In: Hedges S.B., Kumar S., editors. The Timetree of Life. New York: Oxford University Press; 2009. pp. 19–15. [Google Scholar]
- Battistuzzi F.U., et al. Performance of relaxed-clock methods in estimating evolutionary divergence times and their credibility intervals. Mol. Biol. Evol. 2010;27:1289–1300. doi: 10.1093/molbev/msq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., et al. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B., et al. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hedges S.B., Kumar S. Discovering the timetree of life. In: Hedges S.B., Kumar S., editors. The Timetree of Life. New York: Oxford University Press; 2009. pp. 3–18. [Google Scholar]
- Hedges S.B., Kumar S. The Timetree of Life. New York: Oxford University Press; 2009. [Google Scholar]
- Kumar S. Molecular clocks: four decades of evolution. Nat. Rev. Genet. 2005;6:654–662. doi: 10.1038/nrg1659. [DOI] [PubMed] [Google Scholar]
- Metzger K.J. Helping students conceptualize species divergence events using the online tool “TimeTree: the timescale of life”. Am. Biol. Teach. 2011;73:106–108. [Google Scholar]