Abstract
Motivation: The intensification of DNA sequencing will increasingly unveil uncharacterized species with potential alternative genetic codes. A total of 0.65% of the DNA sequences currently in Genbank encode their proteins with a variant genetic code, and these exceptions occur in many unrelated taxa.
Results: We introduce FACIL (Fast and Accurate genetic Code Inference and Logo), a fast and reliable tool to evaluate nucleic acid sequences for their genetic code that detects alternative codes even in species distantly related to known organisms. To illustrate this, we apply FACIL to a set of mitochondrial genomic contigs of Globobulimina pseudospinescens. This foraminifer does not have any sequenced close relative in the databases, yet we infer its alternative genetic code with high confidence values. Results are intuitively visualized in a Genetic Code Logo.
Availability and implementation: FACIL is available as a web-based service at http://www.cmbi.ru.nl/FACIL/ and as a stand-alone program.
Contact: dutilh@cmbi.ru.nl.
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The recent increases in read lengths have established next-generation DNA sequencing as a mature technique, with the first machines capable of single molecule sequencing currently being shipped to researchers. In most studies, the researchers' interests lie beyond translation: the focus is on proteins that are encoded in the DNA, and their function. Most analyses consider the translation between DNA and protein a trivial exercise. After all the genetic code or codon table, i.e. the ‘dictionary’ that translates codons (nucleic acid triplets) into amino acids (AAs), is largely universal and unambiguous (Koonin and Novozhilov, 2009). However, exceptions in the code of bacteria (Bove, 1993), eukaryotic nuclei (Helftenbein, 1985; Meyer et al., 1991), organelles (Barrell et al., 1979) and their associated viruses (Shackelton and Holmes, 2008) have been reported and, given the increasing phylogenetic breadth of sequenced taxa (Wu et al., 2009), many more such findings may be anticipated. A quick survey of the DNA sequences currently in Genbank shows that a total of 0.65% is annotated as being alternatively translated. If a novel sequence uses a non-canonical code, open-reading frames may be different than anticipated due to the reassignment of stop codons and alternative translations of coding codons. This affects both protein sequence and function prediction, so these considerations demand an easy way to assess the genetic code used on a sequenced fragment or assembled contig.
Non-canonical codes are generally identified by inspecting an alignment of the codons on the DNA against homologous protein sequences identified by, e.g. BlastX. The program Gendecoder (Abascal et al., 2006) automates this process, but it focuses on metazoan mitochondria and requires an annotated Genbank file as input. DNA sequencing increasingly yields fragments and assembled contigs that contain sufficient information for reliable genetic code prediction, but performing BlastX searches before knowing the correct translation table is untenable with the rate of DNA sequencing accelerating faster than CPU power. Moreover, the alignment may introduce errors that need to be addressed, preferably by an automated and validated approach.
2 METHODS
2.1 Training data
We used 5866 annotated DNA sequences to construct the training dataset: 3269 bacterial, 176 archaeal and 2421 organellar genomes (Supplementary Table 3), representing all such genomes available on July 13, 2010 in the Entrez genome database. From these genomes, we composed a training set by randomly selecting 1000 regions each of length 100, 200, 300,…, 1000, 2000, 3000,…, 10 000, 20 000, 30 000,…, 100 000, 200 000, 300 000,…, 1 000 000 nt, i.e. a total of 37 000 fragments. We made sure these genomic regions did not overlap to avoid redundant training data. The complete set of training data is available from the FACIL (Fast and Accurate genetic Code Inference and Logo) web site: http://www.cmbi.ru.nl/FACIL/input/complete_training_table.txt.gz.
2.2 Random Forest analysis
Random forest (RF) is a non-parametric classification algorithm that uses many classification trees in parallel (Breiman, 2001). It uses a random subset of the cases in the training dataset and the remainder of the cases for testing and calculating the accuracy scores. The randomForest R package version 2.11.0 was used with 100 trees (using 1000 trees gave almost the same results, see tab in Supplementary Table 1) and default parameters (63% of codons used for training, 37% for testing, square root of the number of variables to train individual trees). In FACIL, the RFs assess the correctness of the homology-based predictions: the response variables of RF1 and RF2 were ‘stop codon’ or ‘coding codon’, the response of RF3 was either ‘correct AA translation’ or ‘incorrect AA translation’.
2.3 Globobulimina pseudospinescens sequencing and assembly
Approximately 10 000 single-cell G.pseudospinescens organisms were isolated by hand from Gullmar Fjord sediment (Risgaard-Petersen et al., 2006). After washing, total DNA was extracted using the QIAamp DNA Micro Kit and sequenced by Illumina Genome Analyser II. Using Edena (Hernandez et al., 2008), 9 950 730 32 nt reads were assembled with parameters m=16 and M=16, which yielded the highest N50 value (N50=170). The raw data and assembly are available from the Gene Expression Omnibus (GEO) under accession number GSE26664. The total DNA of a eukaryote may contain up to three different translation codes (De Grey, 2005): nuclear, mitochondrial and plastid (if the organism is photosynthetic, but this is not the case for G.pseudospinescens). To avoid mixing these signals, the user can choose to feed individual contigs to FACIL, but this might lead to a bad genetic code prediction due to shortage of data. Thus, we selected those contigs that were likely derived from the G.pseudospinescens mitochondrial genome as follows. The 8456 assembled contigs were queried by BlastX version 2.2.22+ (Camacho et al., 2009) against all proteins encoded by completely sequenced mitochondria, downloaded from NCBI organelle genome resources (http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=2759) on July 28, 2010. Importantly, we used the standard genetic code for this BlastX search in order not to impose a bias in the genetic code on the contigs and our results. The 150 contigs with a high-scoring BlastX hit (E-value ≤0.01) were considered to be of mitochondrial origin (average length 223 nt, median length 191 nt). These sequences are available as ‘example’ input data on the FACIL web server. They contain fragments of mitochondrial genes like cytochrome B and several ATP synthase, cytochrome-c oxidase and NADH dehydrogenase subunits. We found no evidence for multiple copies (e.g. a nuclear and a mitochondrially encoded copy) of the encoded genes after a BlastN search (E-value ≤0.01) of the contigs against themselves.
3 RESULTS
3.1 Homology-based prediction
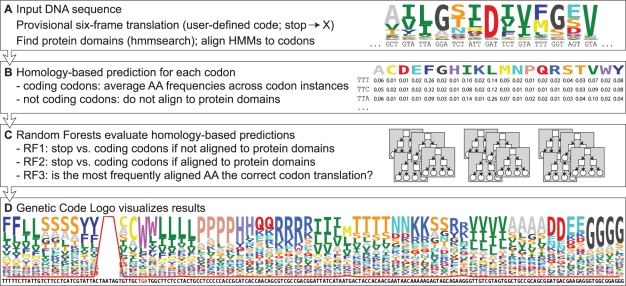
We present FACIL, a method to predict and evaluate the coding of every codon for any nucleic acid sequence, without requiring a priori annotation of proteins. First, FACIL queries all Pfam-fs protein domain hidden markov model (HMMs) [local alignment models (Finn et al., 2010)] against a provisional six-frame translation of the DNA. All known variant codes differ by at most a few codons, so a provisional translation can help to align the AAs in the protein domains to the codons in the DNA. By default, our provisional translation uses the standard code, but by iterating FACIL, using the newly identified codes as input, it is in principle possible to find more distant codes. Stop codons are translated as X to enable the initial alignment of all sites by hmmsearch (HMMER 3.0, http://hmmer.org; default parameters and Pfam trusted score cutoff; Fig. 1a). This sensitive profile-based homology search algorithm allows FACIL to identify homologous regions even if codons are consistently mis-translated. Thus, potentially unique codes can be identified even for organisms that are taxonomically divergent from known species, provided that homologous domains are found. For each codon, FACIL examines which AAs are most frequently associated to it among the aligned protein domains, taking into account the frequency distribution of AAs per position as defined by the domain HMMs. Because we use protein domains as the search unit and the HMMER 3.0 hmmsearch algorithm is fast, FACIL is extremely fast and insensitive to fragmented DNA, frameshifts due to, e.g. sequencing or assembly errors, introns and split gene sequences, and does not require gene annotation.
Fig. 1.
Outline of the FACIL algorithm, see text for details. The Genetic Code Logo visualizes the results, including the reliability of alternative genetic code predictions. The example shows the predicted code for G.pseudospinescens mitochondrial fragments, generated by entering the ‘example’ input data on the FACIL web server. The logo shows the 64 codons from left to right (predicted alternatives in red), each with a stack of AAs. The stack height indicates the percentage of RF3 trees supporting the predicted translation, the letter sizes indicate the scaled AA alignment scores and the red line is the percentage of RF1 or RF2 trees that predict a stop codon.
3.2 RF-based evaluation of homology-based prediction
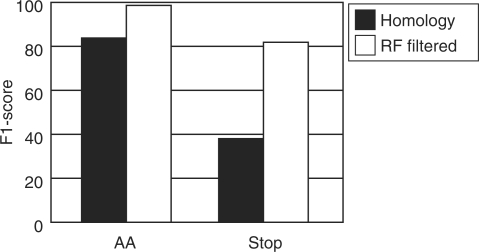
The homology-based prediction creates a matrix of 64 codons by 20 AAs for each DNA molecule (Fig. 1b), where we consider the AA that most frequently aligns to a codon within the protein domains as its most likely translation. The 83.3% of the AAs thus predicted are correct (Fig. 2), but stop codons (that do not align to the protein domains) may be over-predicted for short sequences with strong codon bias, causing a low precision of 24.0% for stop codons. Also, AAs with similar properties may align to a codon with almost equal frequencies, due to neutral evolution at the protein level (see ATA in Fig. 1d). All in all, this leads to relatively low precision and sensitivity scores (see Supplementary Table 2: precision: 83.3 and 24.0%, sensitivity: 83.3 and 89.6%, for AAs and stop codons, respectively). We expected that these errors can be identified by inspecting the variables relating to the homology-based prediction (Table 1). To quantify the reliability of the predictions and assess which parameters are important to achieve a reliable prediction, we implemented a RF approach (Fig. 1c). RF is a non-parametric classification algorithm capable of integrating many variables, yet difficult to overtrain due to the use of many classification trees in parallel that each are trained with a subset of the training data (Breiman, 2001). RFs can even capture subclasses in the training data: clusters of instances with a specific variable importance.
Fig. 2.
F1-score for predicting coding (AA) and stop codons by homology alone and after RF filtering. Values are based on the predictions for all codons from the random fragments of bacterial, archaeal and organellar genomes (see Supplementary Table 2).
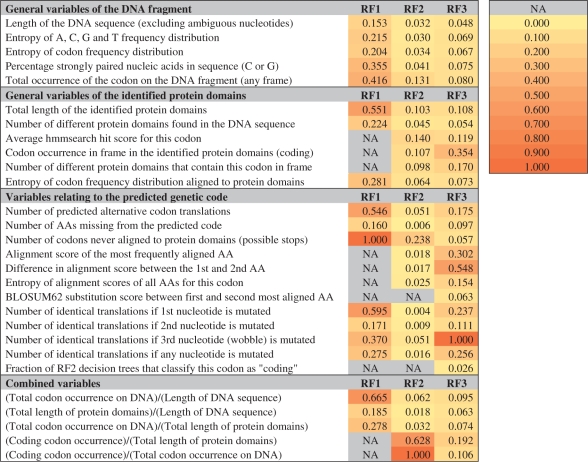
Table 1.
Variables used in each RF and their normalized importance (calculated as MeanDecreaseGini/max MeanDecreaseGini; see Supplementary Table 1)
 |
NA, not applicable.
For every DNA sequence, we evaluate a range of variables for each of the 64 codons to estimate the confidence of the homology-based prediction (Table 1). First, we include general variables of the DNA fragment including sequence length and the total occurrence of the codon in the sequence. Second, we include variables of the identified protein domains like the average hmmsearch hit score. Third, we include variables relating to the predicted genetic code (e.g. the number of AAs missing from the predicted genetic code, the number of codons never aligned to protein domains), variables that represent the confidence of the homology-based prediction [e.g. the similarity between the two top-scoring AAs as defined by their BLOSUM62 substitution score (Henikoff and Henikoff, 1992)] and variables that relate to the robustness of the predicted genetic code (number of single mutation codons translated to the same AA). Finally, we include several combined parameters, including the fraction of codons occurring in frame within the protein domains over their occurrence in the entire DNA sequence.
We trained three RFs of 100 trees, each specialized to answer a specific question. RF1 (91.03% accuracy) and RF2 (99.95% accuracy) were designed to discern stops from coding codons among those codons that do not and do align to protein domains, respectively. RF3 (95.08% accuracy) predicts whether the AA that most frequently aligns to a codon is indeed its correct translation. The assessment of the homology-based predictions by these RFs increased the precision and sensitivity scores (see Supplementary Table 2: precision: 97.1 and 99.3%, sensitivity: 88.1 and 75.8%, for AAs and stop codons, respectively). The decrease in sensitivity for stop codons is mainly due to the many training cases where not all codons are present in the protein domain alignments, e.g. for short input sequences. Thus, RF1 is strict in accepting them as true stop codons. We recommend to be critical of potentially novel alternative ‘rare coding codons’, especially when analyzing short input sequences. Note that these are cases where FACIL will not predict an AA translation, as the codon is not aligned to any protein domain.
We found different variables to be important to each of these questions. For codons that are never aligned to protein domains, the most important variable to distinguish true stop codons from rare coding codons (RF1) is how many of the 64 possible codons did not align to any protein domain. If the sequence contains many codons that never align to a protein domain, this is likely a result of a combination of the low number of identified protein domains, the short length or the low complexity of the query sequence, although individually, those parameters were less important to RF1. Among codons that do align to protein domains, coding codons can be distinguished from spuriously aligned stop codons (RF2) by their occurrence ratio in-frame within protein domains and in the entire sequence. This includes off-frame occurrence in the coding region, where it has been hypothesized that stop codons are abundant to terminate frame-shifted translation (Seligmann and Pollock, 2004). To determine if the AA with the highest alignment score is indeed the correct coding translation for a codon (RF3), the difference between the first and second best alignment score is an important variable. Interestingly, however, the most important variable in RF3 turns out to be a basic characteristic of the genetic code, i.e. the translation redundancy at the third nucleotide of the codon. The genetic code is characterized by a low impact of wobble base pairing of tRNAs at the third nucleotide and apparently, spuriously high-scoring AAs can be recognized as being in violation with this rule. Note that, e.g. the amount of protein-coding sequence falling into Pfam domains is not an important distinguishing variable in any of the RFs. Its major contribution is in RF1, which is in accordance with the most important RF1 variable, i.e. the number of codons that did not align to any protein domain.
To assess potentially conflicting variable importance between standard and alternative codons, we did an additional experiment where we trained the RFs for each of these groups separately (see tabs in Supplementary Table 1). While the variable importances for the RFs trained with only standard codons were very comparable with the complete set, the RFs trained with alternatively encoded codons gave a different picture. For alternative codons that are never aligned to protein domains, the most important variable to distinguish true stop codons from rare coding codons (RF1) is the percentage of strongly paired nucleic acids (GC content). This reflects the difficulty in predicting alternative genetic codes for genomes with a high GC-skew. For alternative codons that do align to protein domains, RF2 assigns the highest importance to the difference in alignment score between the first and second most frequently aligned AA. To determine if the AA assigned by homology is indeed the correct coding translation (RF3), the translation redundancy at the first nucleotide of the codon is the most important distinguishing variable. This analysis pinpoints some of the specific variables that are important for the prediction of alternative codons.
3.3 Genetic Code Logo and web server
The output of FACIL, i.e. a predicted translation for each codon along with confidence values based on the supporting fraction of decision trees in the RFs, is visualized in a Genetic Code Logo (Fig. 1d). We implemented FACIL into a web server (www.cmbi.ru.nl/FACIL/) that enables the user to easily obtain a code prediction with details and a Genetic Code Logo for any sequenced genome or set of contigs. This site also contains a downloadable stand-alone version of the software. Both the web server and the stand-alone version of FACIL take FASTA formatted DNA sequences as input and allow the user to specify the genetic code used for the provisional translation.
3.4 Mitochondrial genetic code of Globobulimina pseudospinescens
Alternative genetic codes are perhaps most abundant in mitochondrial genomes. To illustrate the use of our method, we set out to decipher the genetic code of the mitochondrial genome of the foraminifer G. pseudospinescens. Foraminifera belong to the Rhizaria, a kingdom with only very few protein sequences in the databases, none of which are derived from a mitochondrial genome. This means that no close relatives of this species are represented in the Pfam-fs protein domains used in FACIL. Moreover, the data are particularly challenging, as the genome sequence is highly fragmented and incomplete (150 contigs with an average length of 223 nt). Nevertheless, we obtain strong support that the G.pseudospinescens mitochondrial genome uses the ‘Protozoan Mitochondrial Code’ (NCBI translation table 4, see Fig. 1d and Supplementary Fig. 1), with all 62 high-confidence AA translations correctly classified by RF3, including the alternative translation of TGA into tryptophan (W). Both stop codons (TAA and TAG) were identified by RF1 (red line). BlastX searches and manual curation are consistent with these results (Supplementary Dataset 1 and Supplementary Table 4). Running the 8306 remaining contigs (average length 168 nt) through FACIL predicted the ‘Standard Code’ (NCBI translation table 1) as the most similar code, with 63 of the 64 codons predicted correctly. In the homology step, ATG was more often aligned to leucine (L) than to methionine (M; see Supplementary Fig. 2), but that translation was considered unreliable by RF3 and filtered out. TGA was identified as a stop codon. This analysis exemplifies the value of our method for the reliable discovery of code variants, even in fragmented DNA from taxonomically divergent organisms.
3.5 Performance
As explained above, a FACIL query consists of two main steps. The first is a homology search where the six-frame translation of the input sequences are queried for known protein domains by hmmsearch (Fig. 1a), the second is an evaluation of the alignment-based predictions by three specialized RFs (Fig. 1c). For the 150 G.pseudospinescens sequences (length ~223 nt) presented as an example, these steps take approximately 4 and 1 min, respectively, on our current web server (3 GHz, 32 GB memory). This brings the total run time for prediction of the genetic code to 5 min, only a fraction of the 50 min required for a BlastX search against the proteins in the NCBI Refseq protein database on the same machine (E-value ≤0.01; Supplementary Dataset 1). Indeed, the main performance gain of FACIL comes from the difference in database size that it queries. FACIL only needs to go through 9318 Pfam-fs profiles, whereas the BlastX-based analysis queries at least the Refseq database (9 004 816 proteins in the December 2010 version we used) and preferably even non-redundant protein database, especially for less well-characterized organisms. Moreover, the BlastX results need to be parsed by a custom script and simply selecting the most often aligned AA for each codon may lead to errors, e.g. the two stop codons are occasionally aligned in BlastX hits (Supplementary Table 4). The RFs in FACIL filter out these cases. For large-scale datasets, great improvement may be expected by running hmmsearch in parallel on a high-speed hardware accelerator.
4 DISCUSSION
Currently, there is no standard available for inference of the genetic code of an unannotated DNA sequence, and a range of ad hoc methods that lack quality control and reported reliability scores obscure this research area [the notable exception being Gendecoder (Abascal et al., 2006)]. With FACIL, we present an easy, fast and reliable tool to predict the genetic code for nucleic acid sequences that does not depend on any a priori gene annotation. FACIL detects alternative genetic codes even in species distantly related to known organisms.
Previously, genetic code prediction has explicitly [Gendecoder (Abascal et al., 2006)] or implicitly (BlastX searches) benefited from phylogenetic relatedness for reliable predictions. With FACIL, we chose to rely on general protein domains for two reasons. First, this eliminates the requirement to know the taxonomic placement of the organism from which the DNA was derived, which may in particular be difficult for early-branching organisms. Second, this greatly improves its speed (see Section 3.5). Nevertheless, future genetic code predictors may benefit from a more phylogenetically balanced selection of reference sequences, a proper model of evolution and a probabilistic phylogenetic method that considers the AAs at neighboring nodes of the tree and uses branch lengths to calculate the probability of AAs at an unknown state node.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the anonymous reviewers for their useful suggestions.
Funding: B.E.D. was supported by the Netherlands Organisation for Scientific Research (NWO) Horizon grant (050-71-058) and by NWO Veni grant (016.111.075). R.J. was supported through the Virgo consortium (BSIK 03012), an Innovative Cluster approved by the Netherlands Genomics Initiative. R.S. was supported by NWO Horizon grant (050-71-053). M.Sc. was supported by the Darwin Center for Biogeosciences (project 142.16.1091). M.St. was supported by European Research Council starting grant MASEM (242635). This work is part of the programme of BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO.
Conflict of Interest: none declared.
REFERENCES
- Abascal F., et al. GenDecoder: genetic code prediction for metazoan mitochondria. Nucleic Acids Res. 2006;34:W389–W393. doi: 10.1093/nar/gkl044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B.G., et al. A different genetic code in human mitochondria. Nature. 1979;282:189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Bove J.M. Molecular features of mollicutes. Clin. Infect. Dis. 1993;17(Suppl. 1):S10–S31. doi: 10.1093/clinids/17.supplement_1.s10. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Camacho C., et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grey A.D.N.J. Forces maintaining organellar genomes: is any as strong as genetic code disparity or hydrophobicity? Bioessays. 2005;27:436–446. doi: 10.1002/bies.20209. [DOI] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985;13:415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G. Amino-acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D., et al. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Novozhilov A.S. Origin and evolution of the genetic code: the universal enigma. IUBMB Life. 2009;61:99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F., et al. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc. Natl Acad. Sci. USA. 1991;88:3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risgaard-Petersen N., et al. Evidence for complete denitrification in a benthic foraminifer. Nature. 2006;443:93–96. doi: 10.1038/nature05070. [DOI] [PubMed] [Google Scholar]
- Seligmann H., Pollock D.D. The ambush hypothesis: hidden stop codons prevent off-frame gene reading. DNA Cell Biol. 2004;23:701–705. doi: 10.1089/dna.2004.23.701. [DOI] [PubMed] [Google Scholar]
- Shackelton L.A., Holmes E.C. The role of alternative genetic codes in viral evolution and emergence. J. Theor. Biol. 2008;254:128–134. doi: 10.1016/j.jtbi.2008.05.024. [DOI] [PubMed] [Google Scholar]
- Wu D., et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.