Abstract
Summary: Here we present Sequence Variant Analyzer (SVA), a software tool that assigns a predicted biological function to variants identified in next-generation sequencing studies and provides a browser to visualize the variants in their genomic contexts. SVA also provides for flexible interaction with software implementing variant association tests allowing users to consider both the bioinformatic annotation of identified variants and the strength of their associations with studied traits. We illustrate the annotation features of SVA using two simple examples of sequenced genomes that harbor Mendelian mutations.
Availability and implementation: Freely available on the web at http://www.svaproject.org.
Contact: d.ge@duke.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
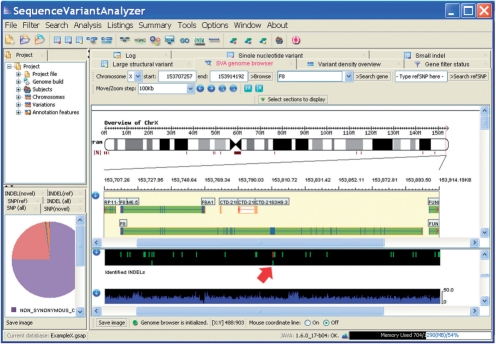
Recent advances in next-generation sequencing (NGS) technologies have made it possible to search through entire genomes for variants that influence traits of interest (Choi et al., 2009; Lupski et al., 2010; Ng et al., 2009; Roach et al., 2010). However, analyzing NGS data still requires addressing a number of considerable computational challenges. A variety of methods have been developed to firstly align the sequence reads (Langmead et al., 2009; Li and Durbin, 2009), and secondly to identify the genetic variants (Bentley et al., 2008; Chen et al., 2009; Hormozdiari et al., 2009; Li et al., 2009). In this work, we focus on the downstream annotation necessary for the analysis of variants and provide simple means of visualizing variants in their genomic context using a built-in genome browser (Fig. 1). One fundamental difference from the previous approach of using genome-wide association studies (GWAS) (McCarthy et al., 2008), is that many of the variants identified in NGS will be newly discovered. A second fundamental difference is that unlike GWAS data, sequence data, at least in principle, provides complete information about the variation present in a genome. For both of these reasons, it is essential to develop a computational environment that can assess the likely functions of the variants observed, and whether or not they are present in existing variant databases. In most contexts, such functional assignments will need to be considered along with statistical association evidence in order to prioritize variants in terms of their likelihoods of influencing traits of interest. We also emphasize that in the early days of interpreting sequence data for complex traits, the sequence data itself will often not be sufficient to provide statistical confidence of association. Instead, the bioinformatic prioritization helps to point towards variants of interest that can be evaluated in much larger sample sets by direct genotyping. We have designed Sequence Variant Analyzer (SVA) to meet these challenges, and in this report, we use two examples to illustrate some of the annotation features.
Fig. 1.
A screenshot of SVA, highlighting a frameshift indel that is located on exon 14 of the Factor VIII gene and is the cause of type A hemophilia.
2 METHODS
SVA was developed using the Java programming language, using the NetBeans IDE (Oracle, Redwood Shores, CA, USA). The graphical user interface was developed using Java Swing. A standard edition of SVA is available by DVD ROM, or a downloadable package (http://www.svaproject.org/). Additionally, we provide a much smaller evaluation edition. SVA uses a customized indexing, storing and optimizing mechanism. Therefore, to run SVA, installation and optimization of database systems is not required. SVA is also multiple-core aware, although Input/Output (I/O) consumes the majority of the computational time. SVA is an elaboration of WGAViewer (Ge et al., 2008), which was designed for the interpretation of GWAS datasets.
A number of publicly available genomic and biological databases are used by SVA for the annotation of genomic variants. We used the NCBI RefSeq (Pruitt et al., 2007) as the reference genome in SVA. Both human reference genome builds 36 and 37 are currently supported. We used the Ensembl core and variation databases (Flicek et al., 2010) as the main source of annotation features for genes and variations. We also provide options to include user-specified annotation tracks, for example, gene annotation dataset or transcription factor binding sites downloadable from the UCSC genome browser. We used the HUGO Gene Nomenclature Committee (HGNC) database as the primary source for naming canonical genes. Gene functions were annotated by integrating information from the Ensembl core, the gene ontology (GO) (Ashburner et al., 2000), and the KEGG pathway databases (Kanehisa et al., 2010). We used RefSNP, HapMap (Frazer et al., 2007), the 1000 Genomes Project (Durbin et al., 2010), and the DGV databases (Iafrate et al., 2004) for checking the novelty of identified genetic variants. We will release compiled annotation databases when new reference genome builds become available. Within each reference genome build, users may update their gene annotation databases by directly downloading the annotation data files following our online instructions.
3 RESULTS
The SVA tool utilizes a knowledgebase of 8.9 GB, which is compiled and compressed into DVD ROM or can be downloaded from the SVA website. The main annotation functionality is performed locally and does not require an active internet connection. This tool has two distinct modules that allow analysis of the genomes included in a given project: the annotation module is used to determine genomic context and to predict the potential biological function of the identified variants (for example, synonymous or nonsynonymous, essential splice, etc.) in each genome. The results can then be visualized in SVA's built-in genome browser (visualization module). The annotated variants can also be exported to a separate statistical tool, for example ATAV (http://www.duke.edu/~minhe/atav/), to assess their statistical associations with traits. Importantly, SVA is designed to conveniently connect its bioinformatic annotations with the statistical association tests so that users are allowed to consider them both and even suggestive association evidence may catch the users' attention. There are also a number of integrated bioinformatic listing functions in SVA designed to help prioritize genetic variants. In broad overview, therefore, SVA inputs a set of identified variants for each genome, annotates them for possible functionality, and permits the user a variety of ways to visualize and filter the resulting data with relationship to phenotypic information. The basic principles of these core analyses are fairly simple, but the strength of these analyses is their ability to be seamlessly run across multiple genomes to prioritize all variants and genes at once.
Two examples illustrate SVA's utility: To illustrate the simple annotation features of SVA we have analyzed the sequence data for subjects with two different Mendelian diseases. (1) Hemophilia A is an X-linked recessive disorder that is characterized by excessive bleeding due to improper clotting. Genetic mutations in the Factor VIII gene are known to cause Hemophilia A; therefore, it is possible to use this disease as a positive control. We annotated 10 case and 10 control genomes and found that the gene with the most cases harboring a rare mutation was Factor VIII (F8) (Pelak et al., 2010). An SVA genome browser view of one of the identified indels is shown in Figure 1. (2) Metachondromatosis (MC) is an autosomal dominant condition affecting bone growth. Our center performed a whole-genome sequencing of one MC patient, following a linkage analysis that implicated six candidate regions spanning a total of 42 MB. We took a three-step strategy: we first applied SVA's genomic region filter; we next applied SVA's novelty filters to identify variants that were absent in dbSNP and absent in all eight control genomes; and finally, we applied SVA's functional filters to identify protein-truncating variants. These filtering steps quickly identify an 11-bp frameshifting deletion on the PTPN11 gene as the cause, which was validated in a separate family (Sobreira et al., 2010). We note that for these simple cases no association tests are required (beyond rarity or absence in controls), whereas in the case of complex traits SVA would normally be used alongside a program like ATAV to facilitate the consideration of variants that are both annotated as functional and that show some degree of statistical association with trait values.
4 CONCLUSIONS AND DISCUSSION
The overriding philosophy of SVA is that the interpretation of whole-genome sequence data benefits from simultaneous consideration of multiple lines of evidence, in particular bioinformatic annotation of variant function and statistical genetic association with trait values. We note this philosophy contrasts with what emerged as best practice for GWAS in which all variants were implicitly considered equally likely a priori to show association with traits (McCarthy et al., 2008). While there are several genome browser and annotation programs available today that are suitable for different needs (Fiume et al., 2010; Robinson et al., 2011; Wang et al., 2010; SeattleSeq: http://gvs.gs.washington.edu/SeattleSeqAnnotation/), we are unaware of any that performs the integrated features of SVA including: variant annotation and filtering by function and/or calling QC, visualization in a built-in browser and convenient export of user selected variants for statistical association testing. These features allow users to interact directly with the full set of data that are relevant to making a judgment about which variants show the strongest combined evidence of influencing the trait of interest. We also note that the SVA framework is fully generalizable, and over time we expect a number of new annotation features to be incorporated which will take advantage of knowledge of the functional regions of the human genome emerging from the ENCODE project (Birney et al., 2007) and related activities. Additionally, in principle, SVA could be modified to perform annotations in other species. Future development plans include developing versions of SVA in highly used model organisms.
Funding: The Bill & Melinda Gates Foundation (grant 157412); National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology (grant AI067854); National Institute of Neurological Disorders and Stroke (grant RC2NS070344); National Institute of Mental Health (grant RC2MH089915).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Ashburner M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.R., et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. PNAS. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R.M., et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume M., et al. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–1944. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., et al. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A.J., et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J.R., et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. New England J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M., et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Ng S.B., et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2009;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K., et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., et al. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach J.C., et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N.L.M., et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., et al. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.