Abstract
The hallmark of chronic lymphocytic leukemia (CLL) is the relentless accumulation of mature lymphocytes, mostly due to their decreased apoptosis. CD74 was recently shown to serve as a survival receptor on CLL cells. In this study, we show that stimulation of CD74 with its natural ligand, migration inhibitory factor, initiates a signaling cascade that results in upregulation of TAp63, which directly regulates CLL survival. In addition, TAp63 expression elevates the expression of the integrin VLA-4, particularly during the advanced stage of the disease. Blocking of CD74, TAp63, or VLA-4 inhibits the in vivo homing of CLL cells to the bone marrow (BM). Thus, CD74 and its target genes TAp63 and VLA-4 facilitate migration of CLL cells back to the BM, where they interact with the supportive BM environment that rescues them from apoptosis. These results could form the basis of novel therapeutic strategies aimed at blocking homing of CLL cells in their return to the BM and attenuating their survival.
Chronic lymphocytic leukemia (CLL) is characterized by the progressive accumulation of small, mature CD5+ lymphocytes in the peripheral blood, lymphoid organs, and bone marrow (BM). The hallmark of the disease is decreased apoptosis, resulting in accumulation of these malignant cells (1). In addition, a strong relationship between BM infiltration patterns and clinical stages of the disease is observed, showing that the BM becomes filled with CLL cells as disease advances (2, 3).
We have recently shown that overexpression of CD74 is an important survival mechanism in CLL, starting from the very early disease stages (4). CD74 is a type II integral membrane protein that was originally thought to function mainly as an MHC class II chaperone (5). A small proportion of CD74 is modified by the addition of chondroitin sulfate (CD74-CS), and this form of CD74 is expressed on the surface of immune cells. In addition to its chaperone activity, surface CD74 also functions as a survival receptor (6–8). It was shown previously that macrophage migration inhibitory factor (MIF) binds to the CD74 extracellular domain, a process that results in the initiation of a signaling pathway in a CD44-dependent manner (8–10).
Our recent study showed that activation of CD74 by MIF on CLL cells initiates a signaling cascade that can be observed from the very early stages of the disease. This pathway induces NF-κB activation, resulting in the secretion of IL-8, which in turn promotes cell survival. Blocking of this pathway by the mAb hLL1 (milatuzumab) (11) leads to decreased cell survival (4).
The TAp63 form of p63 was found to regulate normal B cell survival in a CD74-dependent manner (12). The p63 gene exhibits high sequence and structural homology to p53 (13). The p63 gene contains two transcriptional start sites that enable the generation of transcripts containing (TAp63) or lacking (ΔNp63) the N-terminal transactivation domain. p63 plays a role in developmental regulation of limbs, skin, most epithelial tissues, and epidermal differentiation (14, 15). Furthermore, p63 was recently shown to have a vital role in cellular adhesion and survival in basal cells of the mammary gland and in other stratified epithelial tissues (16).
In the current study, we wished to determine whether p63 isoforms regulate CLL survival, migration, and homing. We show in this study that MIF-induced CD74 activation initiates a signaling cascade that results in upregulation of TAp63, which directly regulates CLL survival. In addition, elevated TAp63 expression upregulates cell surface expression of the VLA-4 integrin, resulting in augmented migration and homing of advanced CLL to the BM.
Materials and Methods
Patient population
B lymphocytes taken from the peripheral blood of patients with CLL who satisfied diagnostic and immunophenotypic criteria for CLL at various disease stages were provided as described previously, in accordance with the protocol approved by the Institutional Review Board of the Kaplan Medical Center (Rehovot, Israel) (17). The diagnosis of CLL was based on standard criteria, and patients were staged according to the Rai staging system (18). The characteristics of the patients are summarized in Table I. In our experiments, early cells refers to stages 0–II, and advanced cells refers to stage III and IV (according to the Rai staging system) (19).
Table I.
Patient data
| Patient No. | Age (y) | Sex | Zap70 | Doubling Time | Previous Chemotherapy | RAI Stage | Modified RAI Risk (iwCLL) | In Our Study |
|---|---|---|---|---|---|---|---|---|
| 1 | 79 | M | Positive | 3 mo | No | II | Intermediate | Early |
| 2 | 50 | M | ND | 2 mo | FC/R | IV | High | Advanced |
| 3 | 72 | M | Negative | NA | COP/R | IV | High | Advanced |
| 4 | 62 | F | Positive | Not reached | No | 0 | Low | Early |
| 5 | 52 | M | Positive | 6 mo | No | 0 | Low | Early |
| 6 | 84 | F | Positive | 2 y | None | 0 | Low | Early |
| 7 | 76 | F | Positive | 3 mo | FC/R | IV | High | Advanced |
| 8 | 75 | F | ND | 12 mo | No | 0 | Low | Early |
| 9 | 59 | M | Negative | Not reached | No | I | Intermediate | Early |
| 10 | 82 | F | Positive | 7 mo | No | I | Intermediate | Early |
| 11 | 76 | F | Positive | 2 mo | L/P, FC/R | IV | High | Advanced |
| 12 | 75 | M | Negative | Not reached | No | 0 | Low | Early |
| 13 | 58 | F | Positive | NA | COP | IV | High | Advanced |
| 14 | 84 | M | Positive | 1.5 y | No | 0 | Low | Early |
| 15 | 59 | F | Positive | 3 mo | L/P | II | Intermediate | Early |
| 16 | 72 | F | Negative | 3 mo | COP | III | High | Advanced |
| 17 | 67 | F | Positive | Not reached | No | 0 | Low | Early |
| 18 | 74 | M | ND | 10 mo | No | II | Intermediate | Early |
| 19 | 60 | M | Positive | Not reached | No | 0 | Low | Early |
| 20 | 80 | F | Negative | 12 mo | No | 0/I | Intermediate | Early |
| 21 | 70 | M | Positive | NA | L/P | IV | High | Advanced |
| 22 | 86 | F | Positive | Not reached | No | II | Intermediate | Early |
| 23 | 72 | M | Negative | NA | FC | III | High | Advanced |
| 24 | 63 | M | Positive | NA | CHOP-like | IV | High | Advanced |
| 25 | 78 | M | Positive | 1 mo | FCR, L/P | IV | High | Advanced |
| 26 | 88 | M | ND | Newly diagnosed | No | IV | High | Advanced |
| 27 | 70 | M | Negative | Not reached | No | I | Intermediate | Early |
| 28 | 82 | M | Positive | Not reached | No | I | Intermediate | Early |
| 29 | 74 | F | Negative | Not reached | No | 0 | Low | Early |
| 30 | 81 | F | Positive | 2 mo | L/P | III | High | Advanced |
| 31 | 80 | M | positive | Not reached | No | I | Intermediate | Early |
| 32 | 80 | F | Positive | >2 y | L/P | IV | High | Advanced |
| 33 | 76 | M | Positive | NA | L/P | IV | High | Advanced |
| 34 | 65 | M | ND | Not reached | No | 0 | Low | Early |
| 35 | 49 | M | Positive | Not reached | No | 0 | Low | Early |
| 36 | 50 | F | ND | 8 mo | L/P | III | High | Advanced |
| 37 | 60 | M | Negative | 24 mo | No | II | Intermediate | Early |
| 38 | 70 | M | Negative | Not reached | No | I | Intermediate | Early |
| 39 | 56 | F | ND | 6 mo | L/P | I | Intermediate | Early |
| 40 | 75 | F | Positive | 12 mo | No | 0 | Low | Early |
| 41 | 72 | F | ND | Not reached | No | 0 | Low | Early |
| 42 | M | Positive | 3 mo | FCR | IV | High | Advanced | |
| 43 | F | Negative | Several y | No | I | Intermediate | Early | |
| 44 | F | Negative | Not reached | No | II | Intermediate | Early | |
| 45 | 77 | M | Positive | 3 mo | COP | III | High | Advanced |
| 46 | 80 | M | Positive | 5 mo | COP, FCR | IV | High | Advanced |
| 47 | 54 | M | Positive | 2 mo | FCR | II | Intermediate | Early |
| 48 | 78 | F | ND | Not reached | No | 0 | Low | Early |
| 49 | 34 | M | Negative | 12 mo | No | I | Intermediate | Early |
| 50 | 50 | F | Negative | Not reached | No | I | Intermediate | Early |
| 51 | 73 | F | Positive | 6 mo | L/P, CHOP | IV | High | Advanced |
| 52 | 77 | M | ND | Not reached | No | 0 | Low | Early |
| 53 | 55 | F | Negative | 6 mo | Newly diagnosed | IV | High | Advanced |
| 54 | 65 | M | Negative | 1 y | No | I | Intermediate | Early |
| 55 | 70 | F | Negative | Not reached | No | 0 | Low | Early |
| 56 | 76 | F | Negative | Not reached | No | 0 | Low | Early |
| 57 | 76 | F | Negative | Not reached | No | 0–I | Intermediate | Early |
| 58 | 72 | M | Positive | 2 mo | CHOP-like | IV | High | Advanced |
| 59 | 65 | M | Positive | 3 y | No | 0 | Low | Early |
| 60 | 75 | F | Positive | 12 mo | No | 0 | Low | Early |
| 61 | 70 | F | ND | Not reached | No | I | Intermediate | Early |
| 62 | 73 | M | Negative | Not reached | No | I | Intermediate | Early |
| 63 | 67 | M | Positive | 6 mo | No | I | Intermediate | Early |
| 64 | 82 | F | Negative | 3 mo | L/P | IV | High | Advanced |
| 65 | 76 | M | ND | 3 mo | COP | IV | High | Advanced |
| 66 | 60 | M | Negative | 8 mo | No | II | Intermediate | Early |
| 67 | 83 | F | Positive | 6 mo | L/P | I | Intermediate | Early |
| 68 | 80 | F | Negative | 6 mo | L/P | IV | High | Advanced |
| 69 | 65 | M | ND | NA | FCR, APSCT | IV | High | Advanced |
| 70 | 60 | F | Positive | 1 y | L/P | I | Intermediate | Early |
Patient's samples were drawn at least 6 wk after the last dose of chemotherapy.
APSCT, autologous peripheral stem cell transplantation; COP/CHOP, cyclophosphamide, oncovine, and prednisone, with or without an anthracycline; F, female; FC/R, fludarabine and cyclophosphamide with or without rituximab; L/P, leukeran/prednisone; M, male; NA, not applicable.
Cell purification
B lymphocytes were purified using a RosettSep Ab mixture (StemCell, Vancouver, British Columbia, Canada), as described previously (4, 17).
Mice
C57BL/6 female mice were used at 6 to 8 wk of age. All animal procedures were approved by the Animal Research Committee at the Weizmann Institute of Science, (Rehovot, Israel).
Reagents
The hLL1 mAb (IMMU-115; milatuzumab) was provided by Immunomedics (Morris Plains, NJ). Development and properties of hLL1 were described previously (11, 20). rMIF was purified from an expression system as previously described and contaminating endotoxin removed by C8 chromatography (21).
Stimulation and blocking
For MIF stimulation, assays were performed as described previously (4, 8). Briefly, 1 × 107 cells were incubated in RPMI medium containing 0.1% (v/v) FCS at 37°C for 3 h. CLL cells were then stimulated in the presence of 100 ng/ml of MIF at 37°C for 18 h. Blocking the CD74 pathway: CLL (1 × 107) cells were incubated in the presence of hLL1 (50 μg/ml) or (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1, 20 μM; Calbiochem, San Diego, CA) at 37°C for 18 h, as described previously (4).
Western blot analysis
Lysates were separated by SDS-PAGE. The proteins were transferred onto a nitrocellulose membrane and probed with anti-p63 (4A4; BD Biosciences, SanJose, CA), anti-p53 (BD Biosciences), anti-β1 (TS 2/16; Endogen, Woburn, MA), anti–VLA-4 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA), or anti–Bcl-2 (C-2; Santa Cruz Biotechnology) followed by HRP-conjugated anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA). The membrane then was stripped and reprobed with anti-tubulin Ab (Sigma-Aldrich, St. Louis, MO), followed by peroxidase-conjugated anti-mouse (Jackson ImmunoResearch Laboratories).
RNA isolation and reverse transcription
Total RNA was isolated from cells by using the TriReagent kit (Molecular Research Center, Cincinnati, OH), according to the manufacturer's instructions. Reverse transcription was carried out by using SuperScript II RT (Invitrogen, Carlsbad, CA) and oligo dT 15 (Promega, Madison, WI) primers. Primers that were used in PCR reactions included: Bcl-2: (sense) 5′-AGATCTCTGGTTGGGATTC-3′, (anti-sense) 5′-CACCGAACACTTGATTCTG-3′; TAp63: (sense) 5′-GCCCATTGACTTGAACTTTGTG-3′, (anti-sense) 5′-GCTGGGCTGTGCGTAGG-3′; ΔNp63: (sense) 5′-TTGTACCTGGAAAACAATGC-3′, (antisense) 5′-GCTGGGCTGTGCGTAGG-3′; β1: (sense) 5′-TGGAACAGATCTGATGAATG-3′, (anti-sense) 5′-AAGGTGAGCAATAGAAGGAT-3′; VLA-4: (sense) 5′-AGCCAGCATACTACCGAAGT-3′, (anti-sense) 5′-GCACGGCCATTGTAAATA-3′; CXCR4: (sense) 5′-CTG AGA AGC ATG ACG GAC AA-3′, (anti-sense) 5′-TGG AGT GTG ACA GCT TGG AG-3′; and actin: (sense) 5′-TGAAGTGTGACGTGGACATCCG-3′, (anti-sense) 5′-GCTGTCACCTTCACCGTTCCAG-3′.
The alternative promoters P1 and P2 of p63 drive the expression of trans-activating (TA) and the N-terminally truncated (ΔN) isoforms (22). Therefore, the 5′ primers were designed to recognize the specific exons unique to each isoform. The 3′ primers were identical and recognize both isoforms.
Real-time RT-PCR analysis
Levels of mRNA of Rp2, TAp63, α4, aL, and Bcl-2 were analyzed by quantitative real-time RT-PCR using a Light-Cycler instrument (Roche Diagnostics, Mannheim, Germany). The reaction volume (10 ml) contained 3 mM MgCl2, LightCycler HotStart DNA SYBR Green I mix (Roche Diagnostics), specific primer pairs, and 2.5 ml cDNA. Conditions for PCR were as follows: 10 min at 95°C followed by 40–60 cycles of 15 s at 95°C, 15 s at 60°C, and 15 s at 72°C. PCR was performed in duplicate as previously described (23). Primer sequences were as follows: TAp63: (sense) 5′-GACTCGGACCTGAGTG-3′, (anti-sense) 5′-CTGGGTAGTCGGTGTT-3′; α4: (sense) 5′-ATCTGACTCTGCCTTCAT-3′, (anti-sense) 5′-CATTCCTCACCATCACTG-3′; Bcl-2: (sense) 5′-GGATCAGGGAGTTGGAAG-3′, (anti-sense) 5′-GCACTGCCAAACGGAG-3′; αL: (sense) 5′-GAGGGTGGATCACCTG-3′, (anti-sense) 5′-GCAATGGCGCAATCTT-3′; and RP2: (sense) 5′-GCACACGTCCAATGACAT-3′, (anti-sense) 5′-GTGCGGCTGCTTCCATAA-3′.
RP-2 levels were used as a housekeeping control to normalize samples for calculation of the relative expression levels of other genes.
Flow cytometry
Staining of CLL cells was performed as previously described (17). The following Abs were used: CD19 (Miltenyi Biotec, Auburn, CA), IgG1k isotype control (MOPC-21; BD Biosciences), β1 (TS 2/16; Endogen), CD49d (9F10; BD Biosciences), CXCR4 (12G5; BD Biosciences), LFA-1α (HI111; BD Biosciences). For Annexin/propidium iodide staining, cells were incubated with Annexin and propidium iodide (BD Biosciences) for 15 min at room temperature and analyzed by FACSCalibur (BD Biosciences).
Small interfering RNA
Down regulation of p63 expression was performed as described previously (12). The targeting oligomers used were small interfering-LacZ (5′-AAGTGACCAGCGAATACCTGT-3′) and small interfering p63 (5′-CAGATCAAGGTGATGACCCCA-3′).
Transwell migration
Chemotaxis was assayed as previously described (17, 24). Briefly, 5 × 106 CLL cells were added to the top chamber of a Transwell culture insert coated with fibronectin. Migration toward CXCL12 (100 ng/ml) was analyzed by FACSort (BD Biosciences) 3 h later. Percent migration was calculated as the number of migrating cells in the lower chamber as a fraction of the input cell number in the upper chamber.
Tracking of cells in vivo
After 24 h of stimulation, cells were labeled with 5 μM CFSE (Molecular Probes) or 5 μM benzoyl amino tetramethylrhodamine (CMTMR; Sigma-Aldrich) for 15 min at room temperature. Neither CFSE nor CMTMR had any effect on cell survival throughout the experiment. Then, 1 × 107 cells were i.v. injected into control C57BL/6 mice. Homing of CLL cells to the spleen, BM, and lymph nodes (LNs) was determined 3 h later by FACS analysis of isolated tissues.
In vitro shear flow assays
Laminar flow adhesion assays were performed as described (25).
Statistics
Statistical analysis was performed using SPSS (SPSS, Chicago, IL). The data were tested for normal distribution, and t tests were performed on normally distributed data sets, as described previously (4).
Results
TAp63 is a target gene of CD74 in CLL cells, inducing their survival
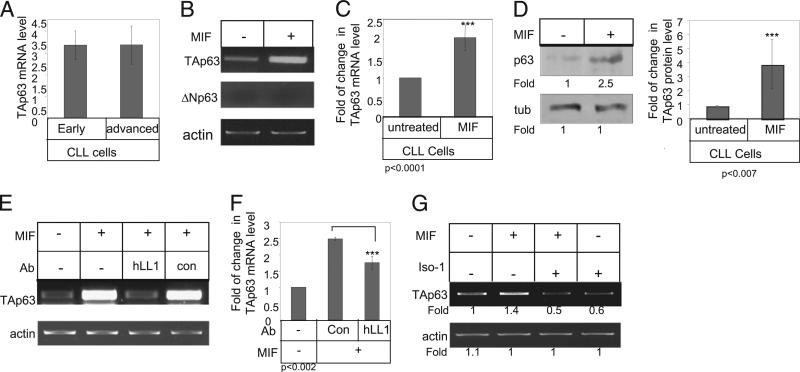
Our recent studies showed that murine CD74 induces TAp63 expression, resulting in increased cell survival (12). In this study, we first showed that hCD74 modulates TAp63 expression following its transfection into human HEK 293 cells (Supplemental Fig. 1). Based on these results in transfect cell lines, we next tested TAp63 expression in early and advanced CLL (Table I). TAp63 was detected at similar levels in both CLL cells derived from early- and advanced-stage patients (Fig. 1A). Next, CLL cells were incubated in the presence or absence of MIF for 18 h, and TAp63 and ΔNp63 mRNA levels were analyzed (Fig. 1B, 1C). CD74 stimulation had no effect on ΔNp63 mRNA levels (Fig. 1B). However, a significant elevation of TAp63 mRNA levels was detected in CLL cells from both early and advanced patients stimulated with MIF (Fig. 1B, 1C). This elevation resulted in augmented p63 protein expression (Fig. 1D), demonstrating that MIF specifically upregulates TAp63 expression in CLL cells.
FIGURE 1.
p63 is a target gene of CD74 in CLL cells. A, Early- and advanced-stage B cells derived from CLL patients were purified. Quantitative real-time PCR was performed using primers for TAp63 and RP-2, as described in Materials and Methods. B–D, CLL cells were incubated in the presence or absence of MIF. B and C, After 18 h, RNA was purified. B, TAp63, ΔNp63, and actin mRNA levels were analyzed by RT-PCR. n = 23 patients. C, Quantitative RT-PCR: results are expressed as a fold of change in TAp63 expression by stimulated cells compared with nonstimulated cells, which was defined as 1. n = 5 patients. D, Cells were lysed after 24 h exposure to MIF, and p63 and tubulin expression were analyzed by Western blot analysis. Graph summarizes the results of four different experiments. E and F, CLL cells were incubated in the presence or absence of MIF, hLL1, or a control Ab for 18 h. E, RNA was purified, and levels of TAp63 and actin mRNA were analyzed. The results presented are representative of six CLL patients. F, Quantitative RT-PCR: results are expressed as a fold of change in TAp63 expression in stimulated cells compared with nonstimulated cells, which was defined as 1. Results shown are a summary of three separate experiments. G, CLL cells were incubated in the presence or absence of MIF or ISO-1 for 18 h. RNA was purified, and levels of TAp63 and actin mRNA were analyzed. The results presented are representative of five CLL patients.
In addition, blocking CD74 by the humanized anti-CD74 blocking Ab (hLL1) specifically downregulated TAp63 mRNA levels, demonstrating that blocking CD74 activity inhibits TAp63 expression (Fig. 1E, 1F). To further demonstrate that MIF secreted from CLL regulates TAp63 expression, CLL cells were incubated in the presence or absence of ISO-1 (a nontoxic inhibitor of MIF that binds to the bioactive MIF at its catalytically active N-terminal tautomeraze site) (26), which inhibits MIF binding to CD74 (27). As shown in Fig. 1G, lower levels of TAp63 mRNA were detected in cells treated with ISO-1, showing that MIF secreted by CLL cells regulates their TAp63 expression through an autocrine loop involving CD74.
Because CD74 regulates CLL survival, we wished to confirm that it supports survival through its target gene, TAp63. Knockdown of p63 downregulated Bcl-2 mRNA and protein levels, resulting in an increased apoptotic subpopulation (Supplemental Fig. 2). In addition, downregulation of p63 specifically inhibited the MIF-induced elevation of Bcl-2 mRNA levels, showing that the MIF/CD74-induced survival cascade is mediated through TAp63 (Supplemental Fig. 2).
Migration of advanced CLL cells to the BM is enhanced due to elevation in VLA-4 integrin
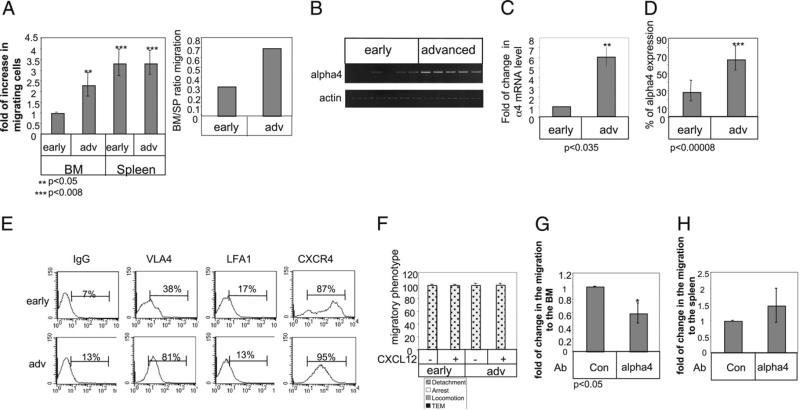
It was shown recently that in addition to its role in cell survival, p63 regulates the expression of an array of proteins that mediate cell adhesion (16). We therefore speculated that in CLL cells, TAp63 might be similarly involved in expression of genes regulating cell adhesion and migration. Thus, we wished to determine whether TAp63 might regulate CLL migration to lymphoid organs in an MIF-dependent manner. Our previous results demonstrated that early- versus advanced-stage CLL exhibits different migratory patterns (17). We first compared in vivo homing of early and advanced stage human CLL cells to the mouse BM, LN, and spleen 3 h after their i.v. injection. Early CLL cells stained with CFSE and advanced CLL cells stained with CMTMR were injected i.v. into C57BL/6 mice. After 3 h, recovered BM, LN, and spleen populations were analyzed by FACS. CLL cells from early- and advanced-stage patients could not be detected in the LN, in agreement with recent results (28). In contrast, similar levels of early- and advanced-stage CLL cells homed to the spleen. Strikingly, advanced CLL exhibited significantly enhanced homing to the BM (Fig. 2A).
FIGURE 2.
Migration of advanced CLL cells to the BM is enhanced due to elevation of VLA-4 integrin. A, Early CLL cells stained with CFSE and advanced CLL cells stained with CMTMR were injected i.v. into C57BL/6 mice. BM and spleen populations were analyzed by FACS after 3 h. The graph presents the averages of four early and three advanced CLL patients. B and C, Early- and advanced-stage B cells derived from CLL patients were purified, and RNA was purified. B, VLA-4 and actin (10 early and 15 advanced CLL patients) mRNA were analyzed. C, Quantitative RT-PCR was performed using primers for α4 integrin (11 early and 9 advanced) and RP-2 (as described in Materials and Methods). D, Cells were stained with anti-α4 and analyzed by FACS. Graph shows α4 integrin (17 advanced and 21 early) expression on CLL cells. E, Cells were stained with anti–LFA-1, anti-α4, CXCR4, and CD19 and analyzed by FACS. Histograms show the expression of various receptors on CD19-positive cells. The results presented are representative of two early and two advanced CLL patients. F, CLL cells accumulated at 0.75 dyn/cm2 for 60 s on HUVECs alone, or HUVECs overlaid with CXCL12 were subjected to physiological shear stress (5 dyn/cm2) for 15 min. Results show relative numbers of each migratory phenotype. The graph is representative of two early (VLA-4 and LFA-1 low) and two advanced (VLA-4 high and LFA-1 low) patients. G and H, CLL cells were incubated in the presence of anti–VLA-4 or a control Ab for 30 min. Cells were stained with CFSE and were injected i.v. into C57BL/6 mice for 3 h. Bone marrow (G) and spleen (H) populations were analyzed by FACS. n = 3. TEM, transendothelial migration.
Homing of normal and malignant B cells to the marrow is mediated by the chemokine receptor CXCR4 (29). We therefore assessed the expression of CXCR4 and integrins expressed on early- and advanced-stage cells. Similar levels of LFA-1 (αL integrin), β1 integrin, and the chemokine receptor CXCR4 (Supplemental Fig. 3) were detected in bothearly- andadvanced-stage CLL cells at the mRNA and cell-surface protein level. However, differences in α4 integrin (VLA-4) expression levels were observed. Early-stage CLL expressed low levels of the α4 mRNA (Fig. 2B, 2C, Table II) and cell surface protein (Fig. 2D, 2E, Table II) compared with their elevated levels in advanced-stage CLL cells. Thus, VLA-4 might play a role in the enhanced homing of advanced-stage CLL cells to the BM.
Table II.
α4 expression in CLL patients according to stage
| Total | Low Expression of α4 | High Expression of α4 | |
|---|---|---|---|
| Total | 52 | 30 | 22 |
| Early | 29 | 28 | 1 |
| Advanced | 23 | 2 | 21 |
Patients 1–3, 5–8, 10–15, 17, 20–34, 40–61, 68.
We next asked whether the increased expression and function of VLA-4 in BMhoming of advanced-stage CLL cellsare also reflected in the generally enhanced adhesive and migratory capacities of these cells in an in vitro model of TNF-α–activated HUVECs, a prototype for cytokine-activated vascular endothelium. HUVECs express high levels of both ICAM-1 and VCAM-1, ligands of LFA-1 and VLA-4 integrins, respectively (30). Early-stage CLL cells, which expressed low levels of VLA-4 and LFA-1 (Fig. 2E), as well as cells from advanced disease, which expressed high levels of VLA-4 and low levels of LFA-1 (Fig. 2, Supplemental Fig. 3), readily detached from the endothelial surface and were unable to transmigrate across the monolayer, even in the presence of the stimulatory chemokine CXCL12 (Fig. 2F). These results suggest that the increased levels of VLA-4 on advanced-stage CLL are not sufficient to promote their adhesion and transendothelial migration across an inflamed ICAM-1–and VCAM-1–expressing endothelial barrier. However, blocking of VLA-4 resulted in inhibition of homing of advanced-stage CLL to the BM (Fig. 2G), whereas homing to the spleen, which is considered an integrin-independent process, was not affected (Fig. 2H).
Activation of cell-surface CD74 upregulates CLL migration to the BM in a TAp63- and VLA-4–dependent manner
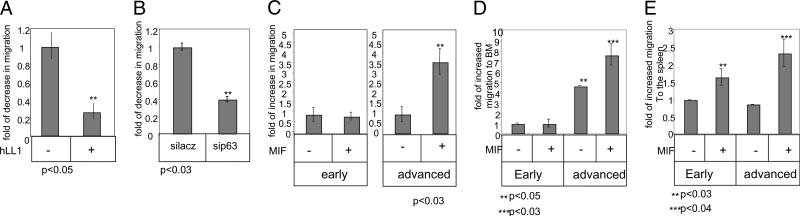
We next determined whether CD74 regulates CLL migration. To this end, we analyzed the effect of CD74 blockade on migration of advanced CLL using the blocking Ab hLL1. As seen in Fig. 3A, incubation with hLL1 for 24 h significantly downregulated the ability of advanced-stage CLL to migrate through a Transwell plate coated with fibronectin, suggesting that CD74 is involved in the BM homing of advanced cells. Furthermore, the migratory response toward CXCL12 of cells lacking p63 was lower compared with control cells (Fig. 3B). Thus, CD74 regulates TAp63 expression, which, in addition to its role in survival, regulates migration of advanced-stage CLL cells.
FIGURE 3.
Activation of cell surface CD74 upregulates CLL migration to the BM in a TAp63-dependent manner. A, CLL cells were incubated in the presence or absence of hLL1 for 24 h. Cell migration toward CXCL12 was analyzed for 3 h through a Transwell plate coated with fibronectin. The graph shows one experiment out of three advanced CLL patients tested. B, CLL cells were treated with siRNA for p63 or for a control gene, lacz, for 26 h. Cell migration toward CXCL12 was analyzed for 3 h in a Transwell plate coated with fibronectin. The graph shows one experiment, representative of three. C–E, Early- and advanced-stage CLL cells were incubated in the presence or absence of MIF for 24 h. C, Cell migration toward CXCL12 was analyzed for 3 h in a Transwell plate coated with fibronectin. The graph shows one experiment representative offive early and four advanced patients tested. D and E, Cells were stained with CFSE and injected i.v. into C57BL/6 mice for 3 h. BM (D) and spleen (E) populations were analyzed by FACS. The graph shows the results of six early and seven advanced CLL samples.
To determine whether stimulation of CD74 by its ligand MIF regulates advanced-stage CLL migration, we first analyzed the in vitro migration of CLL cells treated with MIF versus untreated cells in a Transwell plate coated with fibronectin. As seen in Fig. 3C, incubation with MIF enhanced the ability of advanced cells to migrate toward CXCL12, whereas it had no effect on migration of early-stage cells. Next, to determine whether CD74 stimulation regulates CLL migration in vivo, early- and advanced-stage CLL cells were incubated in the presence or absence of MIF for 24 h. Cells were then stained with CFSE and injected i.v into C57BL/6 mice. Spleen and BM were collected 3 h following injections, and cells migrating to these compartments were analyzed by FACS. As shown in Fig. 3D, MIF upregulated migration of advanced cells to the BM, whereas it had almost no effect on the migration of early-stage cells (Table III). However, MIF elevated migration to the spleen of both early- and advanced-stage cells (Fig. 3E), suggesting that the differential accumulation of early- versus late-stage CLL cells in the BM results from elevation of migration to this compartment and not survival alone (Table III).
Table III.
Migration to the BM according to α4 expression
| α4 Expression | Migration to the BM | Increased Migration to the BM Following MIF | |
|---|---|---|---|
| Low | 8 | 1 | 1 |
| High | 6 | 6 | 6 |
Patients 10, 25, 40–42, 45–47, 49, 53, 54, 58, 61, 68.
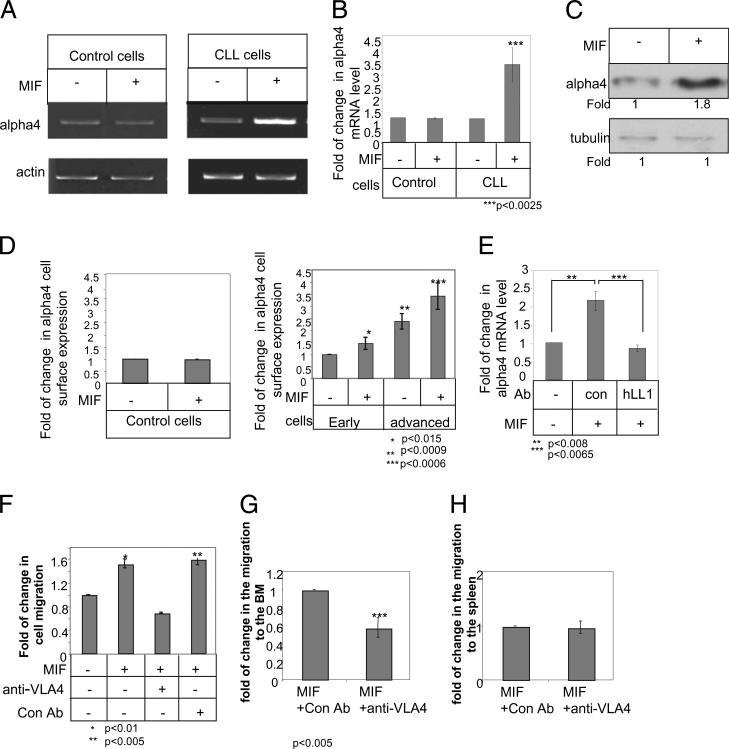
We then determined whether the upregulation of a4 (VLA-4) integrin expression is induced by CD74. Surface CD74 on control and early- and advanced-stage CLL cells was stimulated with MIF for 18 h, and VLA-4 expression was followed. Although no change was observed in VLA-4 mRNA levels in control B cells (Fig. 4A, 4B), MIF upregulated α4 mRNA (Fig. 4A, 4B), protein (Fig.4C), and cell-surface protein (Fig. 4D) expression in both early- and advanced-stage CLL cells. However, despite MIF upregulation of α4 expression in both early- and advanced-stage cells, the expression levels of this chain in MIF-stimulated early -stage CLL cells was still significantly lower compared with untreated advanced-stage cells (Fig. 4D). To directly determine whether CD74 regulates MIF-induced VLA-4 expression, MIF-stimulated CLL cells were incubatedin the presence or absence of hLL1, and VLA-4 mRNA levels were analyzed by real-time PCR. As shown in Fig. 4E, hLL1 downregulated the MIF-induced elevation of α4 mRNA levels. Thus, MIF binding to CD74 induces a cascade that elevates α4 expression.
FIGURE 4.
MIF upregulates VLA-4 expression that is essential for homing of CLL cells to the BM. A–D, B cells derived from healthy individuals (control) or CLL patients were purified. Cells were incubated in the presence or absence of MIF. A and B, After 18 h, RNA was purified. A, α4 and actin mRNA were analyzed by PCR. n = 8 patients. B, Quantitative RT-PCR: results are expressed as a fold change in α4 expression in stimulated cells compared with nonstimulated cells, which was defined as 1. Results shown are a summary of two controls and three CLL patients. C and D, Cells were lysed after 24 h. C, α4 integrin and tubulin were analyzed by Western blot analysis. n = 4 patients. D, Cells were stained with anti–VLA-4 and analyzed by FACS. The graph shows the average VLA-4 expression on 6 advanced and 10 early CLL patients. E, CLL cells were incubated in the presence or absence of MIF, hLL1, or a control Ab. After 18 h, RNA was purified. Quantitative RT-PCR: results are expressed as a fold change in α4 expression by stimulated cells compared with nonstimulated cells, which was defined as 1. Results shown are a summary of three separate experiments. F–H, Advanced cells were incubated with MIF for 24 h and then with anti–VLA-4 for 30 min. F, Cell migration toward CXCL12 was analyzed on a Transwell plate coated with fibronectin for 3 h. The graph shows a single experiment, representative of four. G and H, Cells were stained with CFSE and were injected i.v. into C57BL/6 mice for 3 h. BM (G) and spleen (H) populations were analyzed by FACS. The graph shows an average of three advanced CLL patients.
We next wished to confirm that the upregulation of CLL migration following CD74 activation is a VLA-4–dependent process. Advanced-stage CLL cells were treated with MIF for 24 h, or left untreated, followed by incubation with an α4-blocking Ab for 30 min. Cell migration toward CXCL12 was analyzed in a Transwell plate coated with the VLA-4 ligand fibronectin for 3 h. As seen in Fig. 4F, VLA-4 blocking with an α4-blocking Ab specifically inhibited the elevated migration of MIF-treated cells toward CXCL12 to levels that were even lower than the levels of untreated cells. Thus, inhibition of α4 integrin resulted in a significant reduction in the MIF-stimulated and nonstimulated in vitro migration of advanced-stage CLL cells. This suggests that activation of cell-surface CD74 expressed on advanced-stage CLL cells augments their VLA-4–dependent migration over a VLA-4 ligand and toward a chemokine gradient. Next, to determine the role of VLA-4 in vivo, CLL cells treated for 24 h with MIF followed by incubation with an α4-blocking Ab for 30 min were injected i.v. into mice, and their numbers in the BM and spleen were analyzed. As shown in Fig. 4G, α4-blocking Ab inhibited the elevated migration of MIF-treated cells toward the BM, whereas it had no affect on the migration of MIF-treated cells toward the spleen (Fig. 4H), suggesting that VLA-4 indeed controls homing of CLL cells to the BM.
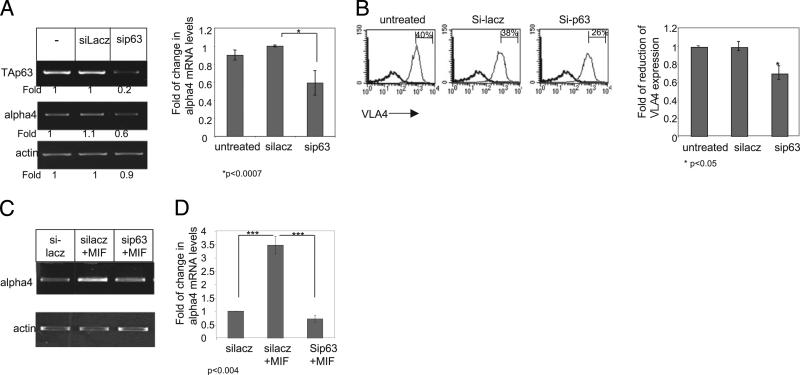
Finally, to directly show that CD74 induces CLL migration by elevating VLA-4 in a TAp63-dependent manner, p63 expression was knocked down by small interfering p63, and α4 expression on CLL cells was analyzed. Downregulation of p63 mRNA levels resulted in reduced levels of α4 mRNA (Fig. 5A) and cell-surface expression (Fig. 5B) compared with untreated cells or cells treated with control small interfering RNA (siRNA). In addition, reduced levels of p63 expression specifically abrogated the MIF-elevated expression of VLA-4 mRNA (Fig. 5C, 5D). Thus, our data demonstrate that VLA-4 is an additional target gene of the MIF/CD74 complex. Induction of the CD74 cascade results in elevation of TAp63 expression, enabling CLL cell homing to the BM.
FIGURE 5.
Downregulation of p63 expression inhibits the MIF-induced VLA-4 expression. CLL cells were incubated in the presence or absence of siRNA for p63 and for a control gene, lacz, for 6 h. A, Following an additional 18 h, RNA was purified, and levels of p63, α4, and actin mRNA were analyzed by PCR. The graph summarizes the results of three separate experiments. B, After 24 h, cells were stained with anti–VLA-4 (gray) or a control Ab (black) and analyzed by FACS. The results presented are representative of five CLL patients. The graph summarizes the results of five separate experiments. C and D, The cells were then stimulated with MIF, and RNA was purified after an additional 18 h. C, VLA-4 and actin mRNA were analyzed. The results presented are representative of two CLL patients. D, Quantitative RT-PCR: results are expressed as a fold-change in α4 expression in stimulated cells compared with nonstimulated cells, which was defined as 1. Results shown represent a summary of two separate experiments.
Discussion
The increased accumulation of CLL cells in the BM, with the resultant cytopenia during disease progression, suggests a change in the migratory and homing pattern of the cells, gradual occupation of normal hematopoietic cell niches, and displacement of normal cells. The results of our current study shed new light on those processes as well as their correlation with the survival of the malignant cells. In this study, we followed the role of TAp63 in the MIF-induced survival of CLL cells. We show that TAp63 has a dual role in CLL. It not only directly affects cell survival, but also has an effect on the migratory and invasive properties of the cells to the BM, indirectly regulating their survival as well (Supplemental Fig. 4).
Our studies show that TAp63 directly regulates CLL cell survival from the early disease stage. We further demonstrate that TAp63 indirectly regulates advanced CLL cell survival by inducing their migration and homing to the BM. The BM stroma plays an essential role in B lymphopoiesis and can provide survival niches for both normal and leukemic mature B cells. The adhesion of CLL cells to BM stromal cells or to the BM vasculature has been shown to rescue these lymphocytes from apoptosis and to extend their life span (31–33). The increased accumulation of CLL cells in the BM during disease progression suggests a change in the migratory and homing pattern of the cells. We show that advanced-stage CLL cells express higher levels of a4 integrin compared with early-stage cells. Because CLL cells express only the α4β1 (VLA-4) integrin and not α4β7 (28), our results indicate an elevation of VLA-4 expression levels. Higher VLA-4 expression levels correlate with augmented homing to the BM, which might additionally lead to retention of CLL in the BM environment, leading to enhanced survival. Recently, it was demonstrated that VLA-4 is an independent prognostic indicator for overall survival, along with IGHV mutational status (34–36). In complete accordance with these recent reports, we observed significantly higher VLA-4 expression in advanced-stage cells compared with cells from patients with early disease. The low expression of VLA-4 might explain the inability of circulating early stage CLL cells to re-enter the BM.
Our results show that MIF and CD74 play a significant role in the regulation of VLA-4 expression and therefore affect homing and survival of CLL cells. MIF is secreted from all types of cells; therefore, CLL cells are stimulated by this chemokine in all compartments. MIF stimulation elevates VLA-4 cell-surface expression levels during advanced-stage disease. The mechanism that regulates high VLA-4 expression at the advance stage is still not known. It is likely that only after their progress to the advanced disease stage do the cells increase their VLA-4 expression to levels that support their homing to the BM. These results suggest that homing to the BM requires threshold levels of VLA-4 expression that enable retention and survival of CLL in BM, an environment that is enriched with the VLA-4ligands VCAM-1and fibronectinand supportstheirretention and survival. It is possible that CLL exposure to systemic MIF redirects circulating CLL back to the BM, where they may encounter more MIF and additionally elevate their VLA-4 expression and retention on stromal VLA-4 ligands. This may create a cycle that can further promote disease-associated BM failure.
Our results suggest that blocking MIF expression or its receptor, such as with an antagonistic anti-CD74 Ab (20), might inhibit homing of CLL cells to the BM and decrease their survival in this compartment. Thus, our results suggest that novel therapeutic strategies aimed at blocking the MIF/CD74 pathway could lead to better and more targeted eradication of the disease due to decreased cell survival and/ or alteration of disease progression by decreasing BM homing.
Supplementary Material
Acknowledgments
We thank Prof. Moshe Oren for helpful discussions, reviewing of this manuscript, and providing reagents for the study.
This work was supported by the Minerva Foundation, the Israel Cancer Association, the Moross Institute, the National Institutes of Health, the Alliance for Lupus Research (to R.B and L.L.), and in part by U.S. Public Health Service Grant PO1-CA103985 from the National Cancer Institute, National Institutes of Health (to D.M.G.), and by the United Israel Appeal of Canada. I.S. is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair.
Abbreviations used in this paper
- APSCT
autologous peripheral stem cell transplantation
- BM
bone marrow
- CLL
chronic lymphocytic leukemia
- CMTMR
benzoyl amino tetramethylrhodamine
- COP/CHOP
cyclophosphamide, oncovine, and prednisone, with or without an anthracycline
- F
female
- FC/R
fludarabine and cyclophosphamide with or without rituximab
- ISO-1
(S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester
- LN
lymph node
- L/P
leukeran/prednisone
- M
male
- MIF
macrophage migration inhibitory factor
- NA
not applicable
- siRNA
small interfering RNA
- TEM
transendothelial migration
Footnotes
Disclosures
D.M.G. is a director and stockholder of Immunomedics, Inc., which is developing the hLL1 Ab.
References
- 1.Caligaris-Cappio F. B-chronic lymphocytic leukemia: a malignancy of anti-self B cells. Blood. 1996;87:2615–2620. [PubMed] [Google Scholar]
- 2.Binet JL, Lepoprier M, Dighiero G, Charron D, D'Athis P, Vaugier G, Beral HM, Natali JC, Raphael M, Nizet B, Follezou JY. A clinical staging systemfor chronic lymphocytic leukemia: prognostic significance. Cancer. 1977;40:855–864. doi: 10.1002/1097-0142(197708)40:2<855::aid-cncr2820400239>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Gray JL, Jacobs A, Block M. Bone marrow and peripheral blood lymphocytosis in the prognosis of chronic lymphocytic leukemia. Cancer. 1974;33:1169–1178. doi: 10.1002/1097-0142(197404)33:4<1169::aid-cncr2820330441>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Binsky I, Haran M, Starlets D, Gore Y, Lantner F, Harpaz N, Leng L, Goldenberg DM, Shvidel L, Berrebi A, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim. Biophys. Acta. 2002;1542:1–13. doi: 10.1016/s0167-4889(01)00166-5. [DOI] [PubMed] [Google Scholar]
- 6.Matza D, Kerem A, Shachar I. Invariant chain, a chain of command. Trends Immunol. 2003;24:264–268. doi: 10.1016/s1471-4906(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 7.Starlets D, Gore Y, Binsky I, Haran M, Harpaz N, Shvidel L, Becker-Herman S, Berrebi A, Shachar I. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107:4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- 8.Gore Y, Starlets D, Maharshak N, Becker-Herman S, Kaneyuki U, Leng L, Bucala R, Shachar I. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J. Biol. Chem. 2008;283:2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- 9.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein R, Qu Z, Cardillo TM, Chen S, Rosario A, Horak ID, Hansen HJ, Goldenberg DM. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood. 2004;104:3705–3711. doi: 10.1182/blood-2004-03-0890. [DOI] [PubMed] [Google Scholar]
- 12.Lantner F, Starlets D, Gore Y, Flaishon L, Yamit-Hezi A, Dikstein R, Leng L, Bucala R, Machluf Y, Oren M, Shachar I. CD74 induces TAp63 expression leading to B-cell survival. Blood. 2007;110:4303–4311. doi: 10.1182/blood-2007-04-087486. [DOI] [PubMed] [Google Scholar]
- 13.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 14.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 15.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 16.Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 17.Haran M, Chebatco S, Flaishon L, Lantner F, Harpaz N, Valinsky L, Berrebi A, Shachar I. Grb7 expression and cellular migration in chronic lymphocytic leukemia: a comparative study of early and advanced stage disease. Leukemia. 2004;18:1948–1950. doi: 10.1038/sj.leu.2403512. [DOI] [PubMed] [Google Scholar]
- 18.Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O'Brien S, Rai KR. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–4997. [PubMed] [Google Scholar]
- 19.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein R, Mattes MJ, Cardillo TM, Hansen HJ, Chang CH, Burton J, Govindan S, Goldenberg DM. CD74: a new candidate target for the immunotherapy of B-cell neoplasms. Clin. Cancer Res. 2007;13:5556s–5563s. doi: 10.1158/1078-0432.CCR-07-1167. [DOI] [PubMed] [Google Scholar]
- 21.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF). Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 22.Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R. p63 in epithelial development. Cell. Mol. Life Sci. 2008;65:3126–3133. doi: 10.1007/s00018-008-8119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger D, Dayan M, Zinger H, Liu JP, Mozes E. A peptide based on the complementarity determining region 1 of a human monoclonal autoantibody ameliorates spontaneous and induced lupus manifestations in correlation with cytokine immunomodulation. J. Clin. Immunol. 2004;24:579–590. doi: 10.1007/s10875-004-6245-2. [DOI] [PubMed] [Google Scholar]
- 24.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. [PubMed] [Google Scholar]
- 25.Warnock RA, Askari S, Butcher EC, von Andrian UH. Molecular mechanisms of lymphocyte homing to peripheral lymph nodes. J. Exp. Med. 1998;187:205–216. doi: 10.1084/jem.187.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dios A, Mitchell RA, Aljabari B, Lubetsky J, O'Connor K, Liao H, Senter PD, Manogue KR, Lolis E, Metz C, et al. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J. Med. Chem. 2002;45:2410–2416. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- 27.Cournia Z, Leng L, Gandavadi S, Du X, Bucala R, Jorgensen WL. Discovery of human macrophage migration inhibitory factor (MIF)-CD74 antagonists via virtual screening. J. Med. Chem. 2009;52:416–424. doi: 10.1021/jm801100v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann TN, Grabovsky V, Wang W, Desch P, Rubenzer G, Wollner S, Binsky I, Vallon-Eberhard A, Sapoznikov AB, Burger M, et al. Circulating B-cell chronic lymphocytic leukemia cells display impaired migration to lymph nodes and bone marrow. Cancer Res. 2009;69:3121–3130. doi: 10.1158/0008-5472.CAN-08-4136. [DOI] [PubMed] [Google Scholar]
- 29.Sipkins DA, Wei X, Wu JW, Runnels JM, Côté D, Means TK, Luster AD, Scadden DT, Lin CP. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cinamon G, Shinder V, Shamri R, Alon R. Chemoattractant signals and beta 2 integrin occupancy at apical endothelial contacts combine with shear stress signals to promote transendothelial neutrophil migration. J. Immunol. 2004;173:7282–7291. doi: 10.4049/jimmunol.173.12.7282. [DOI] [PubMed] [Google Scholar]
- 31.Ghia P, Granziero L, Chilosi M, Caligaris-Cappio F. Chronic B cell malignancies and bone marrow microenvironment. Semin. Cancer Biol. 2002;12:149–155. doi: 10.1006/scbi.2001.0423. [DOI] [PubMed] [Google Scholar]
- 32.Chappell CP, Clark EA. Survival niches: B cells get MIFed as well as BAFFled by dendritic cells. Immunol. Cell Biol. 2008;86:487–488. doi: 10.1038/icb.2008.40. [DOI] [PubMed] [Google Scholar]
- 33.Sapoznikov A, Pewzner-Jung Y, Kalchenko V, Krauthgamer R, Shachar I, Jung S. Perivascular clusters of dendritic cells provide critical survival signals to B cells in bone marrow niches. Nat. Immunol. 2008;9:388–395. doi: 10.1038/ni1571. [DOI] [PubMed] [Google Scholar]
- 34.Shanafelt TD, Geyer SM, Bone ND, Tschumper RC, Witzig TE, Nowakowski GS, Zent CS, Call TG, Laplant B, Dewald GW, et al. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: a prognostic parameter with therapeutic potential. Br. J. Haematol. 2008;140:537–546. doi: 10.1111/j.1365-2141.2007.06965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattei V, Bulian P, Del Principe MI, Zucchetto A, Maurillo L, Buccisano F, Bomben R, Dal-Bo M, Luciano F, Rossi FM, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111:865–873. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- 36.Rossi D, Zucchetto A, Rossi FM, Capello D, Cerri M, Deambrogi C, Cresta S, Rasi S, De Paoli L, Bodoni CL, et al. CD49d expression is an independent risk factor of progressive disease in early stage chronic lymphocytic leukemia. Haematologica. 2008;93:1575–1579. doi: 10.3324/haematol.13103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.