Abstract
Highly sensitive fluorescence microscopy techniques allow single nanoparticles to be tracked during their uptake into living cells with high temporal and spatial resolution. From analysis of the trajectories, random motion can be discriminated from active transport and the average transport velocity and/or diffusion coefficient determined. Such an analysis provides important information regarding the uptake pathway and location of viruses and nanoparticles. In this review, we give an introduction into single-particle tracking (SPT) and determination of the mean-squared displacement. We also give an overview of recent advances in SPT. These include millisecond alternating-laser excitation for removal of spectral crosstalk, alternating wide-field (WF), and total internal reflection fluorescence (TIRF) microscopy for sensitive experiments at the plasma membrane and three-dimensional tracking strategies. Throughout the review, we highlight recent advances regarding the entry (and egress) of natural and artificial viruses obtained via SPT.
Introduction
Nanoparticles intended for pharmaceutical use as well as viruses have to overcome several barriers in order to reach and enter their destined target cell. Whereas viruses have developed their mechanisms of cellular entry over millions of years, the entry of pharmaceutical nanoparticles is typically a limiting factor and still under development. In both cases, the entry pathways in living cells are partly unknown and understanding of the entry pathways will provide a better understanding of cell biology and virology and is essential for improving the design of pharmaceutical nanoparticles. Highly sensitive fluorescence microscopy techniques combined with live-cell imaging are powerful tools for elucidating the details of natural and artificial viral–cell interactions on the single-cell level with high temporal and spatial resolution in real time. They allow direct observation of the pathway a nanoparticle or virus particle takes on their way into and through the cell. Whereas bulk measurements like gene expression studies and/or flow cytometry only investigate the final outcome of cellular infection, high-resolution live-cell imaging provides detailed kinetic information over the entire pathway and directly spots bottlenecks in a chain of succeeding events. Although single-cell measurements are not yet capable of providing information about distribution of pharmaceuticals in a full body or full-tissue scenario after systemic application, they provide essential information with unprecedented detail on the interaction of the particle of interest with the ultimate target, the single cell. As such, they are an additional and very specific tool in the researcher's toolbox for unraveling the full pathway of viruses and nanoparticles from contact with a living body after systemic distribution down to the single cell.
In this review, we give an overview on the prospects of single-cell microscopy techniques and their impact on unraveling the interaction of nanoparticles and viruses with cells. We give a short introduction to single-particle tracking (SPT), the instrumental setups involved and trajectory analysis and discuss newly developed methodologies for improving and expanding the capabilities of SPT. Throughout the review, we highlight examples from the literature regarding recent advancements in understanding the entry (and egress) of natural and artificial viruses obtained via SPT.
Investigating the Internalization of Viruses and Nanoparticles into Cells
Besides their use as viral gene vectors, viruses are valuable tools for investigating the possible entry pathways into cells. As viruses are gene transfer particles per se, the discovery of virus entry pathways provides basic knowledge for potential entry portals into cells and contact points for improvements toward more efficient nonviral gene vectors. The main optical technique used to study the entry of viruses and nanoparticles into cells has been multicolor fluorescence microscopy on living or fixed cells.1,2,3,4,5 Compartments of the endocytic pathway in living cells can be visualized by fluorescently labeled markers (e.g., dye-coupled transferrin, cholera toxin B, or dextrans) or more specifically by cellular expression of marker proteins fused to fluorescent proteins (e.g., clathrin-green fluorescent protein, caveolin-green fluorescent protein).6 The entry of viruses or nonviral vectors labeled with a fluorescent dye of another color can then be followed in the living cell and colocalization analysis with cellular compartments used to analyze the entry route taken. This method can be extended by using chemical inhibitors or small interfering RNA to block-specific entry pathways and test for internalization of gene carriers. However, care should be taken as the use of markers and inhibitors on the single-cell level has some caveats and can give inconsistent results on the single-cell level.7 Based on studies of nanoparticle uptake, it was revealed that all tested gene carriers enter cells by endocytosis,8,9,10 even the ones connected to cell-penetrating peptides.11,12,13 Although viruses have the ability to penetrate the cellular membrane for productive infection—which can happen either at the plasma membrane or the limiting membrane of an intracellular organelle—the majority of viruses exploit the cellular endocytotic machinery and need it for productive infection. This may be due to the fact that endocytosis offers real advantages for infectivity such as directed movement toward the nucleus in vesicles by molecular motors,14 circumvention of barriers in the cell periphery,15 and the use of local cues such as low pH for timed escape into the cytosol at specific locations.16 However, there is no generalized internalization pathway for viruses as well as gene carriers. The exact endocytic pathway used is strongly dependent on the individual cell type and also the sort of virus or gene carrier.8,17,18 Several endocytosis pathways are often used simultaneously and inhibition of one pathway leads to an increase in internalization by alternative routes. In addition, for gene carriers, the pathway finally resulting in successful transgene expression varies with cell line and also gene carrier type.17 Therefore, knowledge of the exact internalization pathway of a specific gene carrier is required to improve its efficiency. One approach to increase the transfection efficiency of gene carriers is to target a pathway leading to successful gene expression by using a specific receptor. This can concomitantly be connected to cell-specific targeting.19,20,21,22 However, the exact successful entry pathway has to be defined for each target cell line and particle type individually. Such detailed understanding of the entry process of natural and artificial viruses on the single-cell level can be obtained using SPT. By following individual viruses and nanoparticles in real-time with high spatial and temporal resolution, the kinetics of transfection can be measured and the specific interactions can be visualized.
SPT Microscopy
Experimental setup
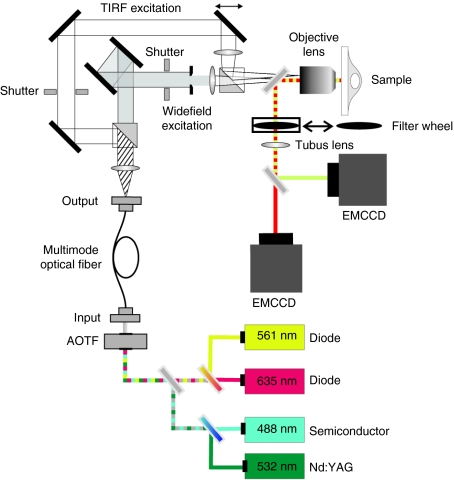
Single-particle techniques are typically based upon highly sensitive fluorescence wide-field (WF) microscopy. Internalization of single gene carriers can be continuously followed from their initial attachment to the cell membrane through the various steps of the uptake process and intracellular trafficking. The ability to image single gene carriers, which can eventually be labeled with only a few fluorophores, at high temporal resolution over several minutes requires ultrasensitive detection. The setup we currently use for SPT is shown in Figure 1. Excitation is performed by laser light which provides the necessary excitation intensity required for strong photon emission by the fluorophores and whose narrow spectrum makes it easy to suppress scattered light without blocking fluorescence signal. Typical laser lines for excitation of common fluorophores are 488, 532, 561, and 635 nm. The availability of several laser lines allows simultaneous imaging of two to three fluorophores, depending on emission spectra of the labels and filters available. An acousto-optical tunable filter is used to select the appropriate excitation wavelength or wavelengths without speed limitations or vibrational and mechanical constrains related to mechanical shutters or rotating wheels. The laser light is focused onto the back aperture of the objective for WF and total internal reflection fluorescence (TIRF) illumination. The cells are kept at 37 °C throughout the experiment by a temperature controlled stage (heating table). The objective is also heated to 39 °C (slightly higher than the temperature of the sample) as oil immersion objectives are in direct thermal contact with the sample and act as a strong heat sink for the cells. It is best to use objectives that have been designed for use near 37 °C. There are also microscope stage incubator chambers available that allow control of the CO2 level, but with the exception for long-time experiments, the use of CO2-independent medium is typically sufficient. Focal instabilities in the microscope mechanics due to temperature fluctuations in the surroundings after mounting the sample provide major difficulties for SPT. An auto-focus system keeps the sample at a constant z-position and is very beneficial for intermediate and long-term (1–2 hours) measurements. The emission light is typically collected by a ×60 or ×100 oil immersion objective with high numerical aperture (up to 1.49 NA) and separated from the excitation light with the appropriate dichroic mirror. The fluorescence emission is then directed to and detected by a highly sensitive (typically back illuminated) electron multiplied charge-coupled device camera. For multicolor imaging, either multiple regions can be imaged on different portions of a single camera or separate cameras can be used. Multiple cameras have the advantage of an increased field of view, but also increase the cost of the setup. The exact configuration of the setup, mainly the choice of lasers, dichroic mirrors and filters, depends on the combination of fluorophores used and may have to be adapted for the individual experiments by the user. Accessory optics for TIRF illumination (see Figure 1) or confocal detection can be added to increase the flexibility and variability of custom-built setups. TIRF illumination is especially interesting for fast tracking of cell surface associated events at the basal plasma membrane.
Figure 1.
Schematic drawing of a wide-field/objective-type total internal reflection fluorescence (TIRF) microscope for single-particle tracking (SPT). For excitation, several excitation wavelengths are available and can be switched in microseconds with an acousto-optical tunable filter (AOTF). Laser light is coupled into the objective either in wide field (WF) or total Internal refection fluorescence microscopy (TIRFM) mode, which is selectable using mechanical shutters and can be alternated with each frame. A field aperture is used such that only the area detectable on the camera is illuminated. The emission light is separated from the excitation light by a dichroic mirror. For multicolor imaging, the emission light is spectrally separated by a second dichroic mirror and the two emission channels are detected on separate EMCCD cameras. DC, dichroic mirror; F, filter; L, lens; M, mirror; S, slit. Courtesy of Dr Sergey Ivanchenko and Jens Prescher.
Trajectory analysis and MSD plots
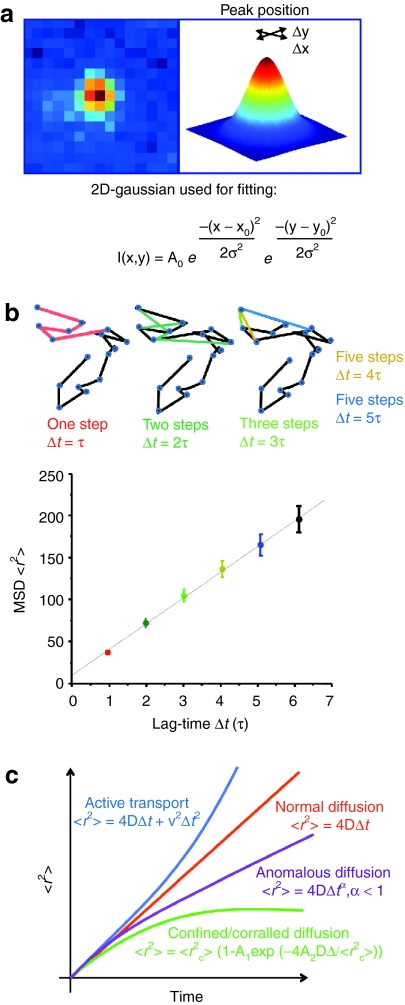
In the acquired image sequences, individual fluorescent particles can be identified as bright spots on a dark background. Each frame in the movie is a representation of the position of the particles at a certain time point. By extraction of the x- and y-coordinates of the particles from the centroids of their diffraction limited spots in all frames of the movie where the particle is detectible, the trajectories of the particles can be obtained. This is typically performed by first reducing background noise and then selecting particles by setting an intensity threshold on the filtered image. In a second step, particles for tracking are selected based on their intensity, size and shape. The x-and y-coordinates are obtained by fitting a 2D-Gaussian function to the particle's intensity profile (Figure 2a). In a final step, the particle coordinates are subsequently used for calculating the corresponding trajectories based on a nearest-neighbor algorithm.23,24 With this method, a positional accuracy well beyond the optical diffraction limit is achieved. The centroid position of a sufficiently bright fluorophore can be determined to within nanometer precision.25
Figure 2.
Tracking and mean-square displacement (MSD). (a) When a single particle is present in the psf, the x-, y-coordinates of the particle at a give time point are derived from the central position of its diffraction limited intensity profile by, for example, fitting it to a 2D-Gaussian function. Hence, a positional accuracy far below the optical resolution is obtainable. (b) MSD plots. Upper panel. A schematic trajectory showing the distance traveled between consecutive frames (blue circles in trajectory). The average of all steps within the trajectory for each time-lag Δt, with Δt = τ (red), Δt = 2τ (dark green), and so on (where τ = acquisition time interval from frame to frame) gives a point in the MSD plot. (c) The time dependence of the MSD allows the classification of the type of diffusional behavior. A linear plot indicates normal diffusion and can be described by <r2> = 4DΔt (D, diffusion coefficient) for a two-dimensional analysis and <r2> = 6DΔt in three-dimensions. A quadratic dependence of <r2> on Δt indicates directed motion and can be fitted by <r2> = v2Δt2 + 4DΔt (v, mean velocity). When the MSD asymptotically approaches a maximum value for larger Δt, the system is undergoing confined or corralled diffusion with <r2> = <r2c> [1 – A1 exp (−4A2DΔt/ <r2c>)]. Anomalous diffusion has the form <r2> = 4DΔtα and α < 1 (subdiffusive).
The resulting trajectories are usually analyzed for their type of motional behavior as the motion provides information on the surroundings and interactions of the particle. The most common analysis starts with calculation of the so called mean-square displacement (MSD). From the MSD, their type of motional behavior can be determined.26 A simplified way to calculate the MSD is depicted in Figure 2b. The MSD <r2> describes the average of the squared distances between a particle's start and end position for all time-lags of certain length Δt within one trajectory.
With increasing time-lag, however, less data points are available and the uncertainty of the MSD values increases.27 To account for this uncertainty, the MSD should always be calculated for time-lags corresponding to less than ~¼ of the total number of points in the trajectory.26 As a consequence, the time-axis of a MSD plot can only represent a fraction of the time scale of the trajectory. From evaluation of the MSD plot, information about the mode of motion can be obtained (Figure 2c). This mode of motion can then be interpreted in a biological context and conclusions on the location and environment of the tracked particle can be drawn.
Normal and Anomalous diffusion is described by
where D is the diffusion coefficient, Δt is the given time interval and the exponent, α, that distinguishes between normal or Brownian diffusion (α = 1) and anomalous diffusion (α < 1). The factor 4 is specific for diffusion in two dimensions and is replaced by 6 in all formulas for three-dimensional tracking. For Brownian motion, the MSD increases linearly with Δt. Anomalous diffusion is typically observed when the diffusive particle is hindered by obstacles.
Confined or corralled diffusion is indicated by an asymptotic behavior of <r2> for large Δt and implies a confinement for the diffusive particle. The relation between <r2> and Δt is given by:
where <r2c> is an approximation of the size of the confinement and the constants A1 and A2 are determined by the confinement geometry. The asymptotic value of <r2> for large Δt can be used to estimate of the size of the confinement <r2c>. We note that confinement within a certain region is only observable when the observation time is long compared to the time between successive contacts of the particle with the barrier. For short observation times, normal or anomalous diffusion within the confinement is observed.
Active transport is described by a quadratic dependence of the MSD <r2> on Δt:
where v is the velocity of the directed motion which is also called drift. Superimposed on this motion is normal diffusion with the diffusion coefficient D. The whole process can be described using the analogy of a conveyor belt, where particles are transported but also diffuse along the belt. By fitting formula (3) to an MSD curve, the mean velocity v of the directed motion and the diffusion coefficient D are obtained.
In the above equations, the impact of uncertainties and statistical errors has been ignored. Often the information extracted from the equations above is sufficient for the biological question of interest. However, it is still possible to perform a more quantitative analysis by taking the static and dynamic errors into account. Static errors, represented by σ, arise from the uncertainty in determining the position of the particle due to experimental noise. When static uncertainties are not taken into account, freely diffusing particles may be incorrectly categorized as undergoing anomalous diffusion.28 The static contribution can be determined by measuring immobilized particles at a signal-to-noise ratio similar to the actual experiments.29 Dynamic errors arise from the fact that the particle is diffusing within the integration time of each frame, leading to motional blurring of the fluorescence intensity on the camera. When the camera is constantly illuminated for the entire frame, the modified MSD for a freely diffusing particle including static and dynamic uncertainties is given by:29,30,31
where ΔtE is the frame integration time of the camera. When the illumination intensity is varied during the exposure time of a single frame, such as for stroboscopic illumination, the correction term for dynamic errors will vary.30 For tracking the entry of viruses and nanoparticles, the static uncertainty can be significant due to the limited signal-to-noise ratio of experiments performed in living cells and the lower illumination intensities used for measurements over extended time periods. Whether dynamic errors significantly contribute to the MSD depends on the mobility of the tracked particles and details of the experimental protocol.
Subtrajectory analysis
MSD curves of diffusive motion can be very heterogeneous and often change during the course of the trajectory. Therefore, it is important to perform a MSD over subregions of the trajectory. A careful description of the various modes of motion within one trajectory requires the separation of the trajectory in several parts e.g., manually according to morphological differences or by velocity thresholds32,33 as the shape of the MSD or effective diffusion coefficient curves are not sufficient. For example, particles showing hop diffusion may fulfill all analysis criteria for “diffusive” motion whereas the hop diffusion pattern is only visible in the trajectory.33
As early as 1995, Jacobson and co-workers applied a subtrajectory analysis to identify transient confinement zones of membrane proteins.34 They calculated the maximum displacement within multiple segments over the entire trajectory and calculated the probability that a freely diffusing particle remains within a confined region of the size of the maximum displacement for the entire segment. When the probability was reasonable, normal diffusion was assumed. However, when the log-likelihood that the diffusional motion was not random exceeded a critical threshold over a minimum critical duration, they would assign that region of the trajectory to a confinement zone. The optimal critical threshold and duration were determined from simulations.
A more sophisticated method for automated trajectory analysis and mode of motion detection utilizes a rolling-window algorithm. The algorithm described by Arcizet et al.35,36 reliably separates the active and passive states of particle motion and extracts the velocity during active states as well as the diffusion coefficients during passive states. It assumes that active transport, for example on microtubules, is characteristically directional over a certain time and measures the directional persistence of particle motion by the angular correlation of the following steps.35 The angle analysis provides an estimate of the probability of the particle undergoing active motion within the analyzed subregion. Of course, the higher the temporal resolution and the more data points acquired, the better and more detailed the analysis can be. As a consequence, for very fast transport events, the temporal resolution has to be high enough to acquire sufficient data points for analyzing the different modes of motion and for separating the break points between them.
Other elegant approaches have also been taken to extract heterogeneous diffusion characteristics within a single trajectory. Hang and co-workers developed a maximum likelihood approach to automatically recognize when a system undergoes a transition to a state with different diffusional properties and performs a subtrajectory analysis in the regions between transitions.37 Matsuoka and co-workers took a different approach and explicitly calculated the displacement probability density function assuming a single, freely diffusion species, two noninterconverting freely diffusing species and a species that interconverts between two freely diffusing states with different diffusion coefficients.38 Using their model, they could not only extract the diffusion coefficients of the different states but also the transition rate between the states. Although subtrajectory analyses are powerful for investigating the heterogeneous diffusional behavior of individual particles, other means such as the probability distribution of square displacement39 can also be used to detect diffusional heterogeneities. For example, Pinaud and co-workers have recently detected two diffusion regimes for the glycosylphosphatidylinositol-anchored protein and found that the slower mobility population correlated with glycosphingolipid-rich microdomains.40
Millisecond ALEX
Multicolor imaging is a powerful tool for extracting information regarding the uptake of gene carriers and viruses in living cells and can be used to visualize interactions or track multiple components. For example, the uptake pathway can be investigated in more detail by labeling different cellular components with fluorescent proteins (e.g., early or late endosomes41) or cargo delivery can be visualized by labeling the cargo and the carrier with different colors and visualizing when the cargo separates from the carrier. However, one difficulty of multicolor experiments is spectral overlap of the fluorophores, which can be especially troublesome for experiments restricted to the use of fluorescent proteins and/or where the labeling ratios of the different components vary dramatically. However, spectral overlap can be removed with the use of pulsed interleaved excitation42,43,44 in confocal setups equipped with such technologies or with millisecond alternating-laser excitation (ALEX)45 for camera-based imaging methods.
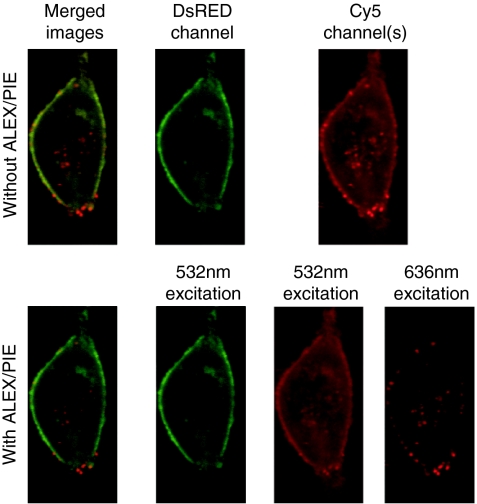
In millisecond ALEX, consecutive frames are collected using alternating excitation wavelengths. In a two-color experiment with DsRed and Cy5 (Figure 3), the odd frames will be recorded with 532 nm excitation for DsRed and the even frames collected with 636 nm excitation for Cy5. With the appropriate choice of the emission filter for the DsRed channel, no fluorescence from Cy5 will be visible in the DsRed channel. With the appropriate choice of excitation wavelengths, the higher energy fluorophore will not be excited with the lower energy wavelength. In the above example, 636 nm light did not excite DsRed. Thus, the DsRed detection channel with 532 nm excitation had no overlap with Cy5 and the Cy5 detection channel with 636 nm excitation has no contribution from DsRed. Even when the concentration of fluorophores are different by orders of magnitude, these combinations are free of spectral crosstalk.
Figure 3.
Removal of spectral crosstalk with alternating-laser excitation (ALEX)/pulsed interleaved excitation (PIE). An image of a live HeLa cell transfected with DsRed labeled actin and infected with Cy5-labeled polyplexes. The upper panels show the image as would be measured using normal dual-color excitation with two-channel detection. The lower panes show the image using ALEX/PIE where spectral crosstalk is separated from the Cy5 channel. A merged image (left pane), and images of the DsRed channel (left-middle pane), and Cy5 channels (right panes) are shown. The merged image with ALEX/PIE and the Cy5 channel with 636 nm excitation shows the improved contrast when spectral crosstalk is removed.
In addition to removal of spectral crosstalk, millisecond ALEX can be utilized to calculate stoichiometries or distinguish different fluorophores based on both their excitation and emission properties. For example, one characteristic typical of cellular autofluorescence is a broad excitation and emission spectrum. Autofluorescence will be detectible in multiple channels and hence, be distinguishable from the other fluorescence signals. This is particularly critical when exciting at wavelengths of 488 nm or shorter as shorter wavelengths generally result in a stronger autofluorescence signal.
Following the Dynamics of Single Nanoparticles in Living Cells
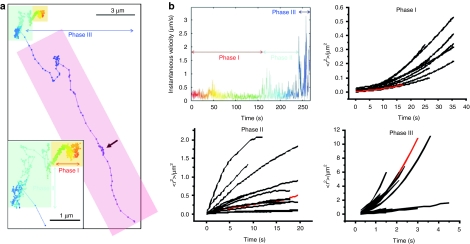
To follow the internalization process of single polymeric gene carriers (polyplexes) in living cells in real time, de Bruin and co-workers combined sensitive fluorescence WF microscopy and SPT with a temporal resolution of 300 ms.32 They obtained trajectories showing the full internalization process of epidermal growth factor receptor-targeted polyplexes starting with attachment to the cell membrane followed by uptake into the cell and finally active transport toward the nucleus. Typically, three phases of motion were found (Figure 4, the video corresponding to the trajectory can be found in the supplementary material of ref. 32). Phase I showed slow directed motion, phase II consisted either of normal, anomalous and confined diffusion or a combination of them, and phase III showed fast, active transport in the cytoplasm. Phase I occurred shortly after attachment to the cell membrane where the polyplexes showed slow directed transport with typical velocities of ~0.015 µm/s. This transport was attributed to the movement of the underlying actin cytoskeleton mediated by the EGF receptor and linker proteins as revealed by dual-color microscopy. The polyplexes were thus transported by the retrograde actin flow within the cell. A similar behavior was also observed for untargeted polyplexes by Bausinger et al.46 where the cell-surface attached polyplexes colocalized with actin fibers of the underlying cortical actin network. In these experiments, SPT of the membrane bound polyplexes revealed a variety of motion behavior including 2D-free diffusion, anomalous diffusion as well as complete immobilization. Also magnetic polyplexes display the three phases of internalization, independent of the applied magnetic field.47 From these studies, it can be concluded that application of a magnetic field only affected the extracellular distribution of the magnetic polyplexes whereas the cellular processes of uptake and trafficking were unaffected.
Figure 4.
Internalization of epidermal growth factor receptor (EGFR)-targeted polyplexes. (a) Trajectory of a polyplex during uptake into a HuH7 cell. The particle was tracked for 4 minutes and 30 seconds at a temporal resolution of 300 ms/frame. Three phases are typically observed during internalization. The trajectory starts immediately after the polyplex attaches to the cell membrane. Time information is given by the color of the trajectory changing from red to blue-violet during the movie. The arrow indicates a spot of back-and-forward movement. The corresponding video is available as supplementary information in ref. 28. The overlaying color boxes mark the phases: Yellow: phase I—slow active transport; blue: phase II—anomalous and confined diffusion; purple: phase III—active transport. (b) A plot of the instantaneous velocity of the trajectory shown in a and mean-square displacement (MSD) plots of the three phases analyzed from several trajectories. The plots corresponding to the trajectory presented in panel (a) are drawn in red. Based on the MSD analysis, the particle is undergoing active transport with a mean drift velocity for all plots in phase I of vI = 0.015 µm/s and a corresponding diffusion coefficient of DI = 4 × 10−4 µm2/s. For phase II, the plots could not be averaged due to heterogeneous appearance showing anomalous, normal and confined diffusion. Phase III plots show again active transport by this time with a significantly higher mean velocity of vIII = 0.7 µm2/s with a corresponding mean diffusion coefficient of DIII = 0.1 µm2/s. Adapted with permission from the American Society of Gene Therapy.32
For untargeted cationic gene carriers, cell-surface proteoglycans such as heparan sulfate proteoglycans have been suggested as a kind of “unspecific receptor” with binding characteristics based on electrostatic interactions.10,46,48 The results of the SPT from Bausinger et al. support the model of receptor binding by the polyplexes and receptor clustering before endocytosis occurs. Recent experiments using colocalization with fluorescent markers, inhibitor, and small interfering RNA treatment by Payne et al.49 provides strong evidence for proteoglycans as the unspecific receptors for polyplexes. Targeting gene carriers to specific cell surface receptors can influence the internalization behavior. de Bruin32 and co-workers demonstrated in their study that targeting of polyplexes to the EGF receptor lead to increased and accelerated endocytosis of the polyplexes. Instead of the unspecific proteoglycan mediated endocytosis, the polyplexes were quickly internalized by the specific EGF receptor mediated endocytosis. The fast endocytosis was represented in the trajectories by a significantly shortened duration of phase I.
The internalization event itself—namely the pinching off of an endocytic vesicle from the plasma membrane—cannot be resolved by light microscopy. However, by combining SPT with fluorescence quenching of extracellular polyplexes at different time points, de Bruin et al.32 showed that the internalization into an endocytic vesicle occurs during phase I. More interestingly, some polyplexes defined as intracellular continued the slow directed movement characteristic for phase I for a certain time before switching to the movement characteristic for intracellular particles. This suggested that the newly formed endocytic vesicles close to the membrane may be trapped in the cortical actin cytoskeleton and therefore show motion similar to the polyplexes bound to the membrane and connected to the actin cytoskeleton by transmembrane proteins.
Typically, most internalized particles show anomalous or confined diffusion followed by active transport at later time points. The anomalous diffusion and confinement displayed by the MSD analysis during phase II represent the local microenvironment of the particles in the cytoplasm where the cytoskeleton, organelles and large macromolecules are local obstacles for free diffusion. Long-range active transport (phase III) occurs with motor proteins (kinesins or dyneins) along microtubules with velocities up to 4 µm/s and is a signature of internalized particles. Typically, this long-range active transport displays the so called stop-and-go motion as a result from binding and unbinding of motor proteins to the microtubules.32,33,50 Reversal of the transport direction can frequently be observed (see arrow in Figure 4a). Motor protein driven transport of nanoparticles usually occurs as vesicular transport. This is in contrast to many viruses, where the virus particles show active transport in the cytosol after fusion with the plasma membrane or endosomal membrane.51,52,53 This is due to the ability of specific proteins from the virus particle to bind motor proteins.
For murine polyoma virus-like particles (VLP), motional behavior on the plasma membrane similar to the phase I observed in polyplexes was found during entry.3 After binding to the cell surface, the VLP showed free lateral diffusion followed by confinement through an actin cytoskeleton-dependent mechanism as well as confined movement in zones with a slow drift. The results suggested that the multivalent VLPs bound to cell-surface gangliosides, which clustered and induced transmembrane coupling to the underlying actin cytoskeleton leading to confinement of the VLP–ganglioside complex.
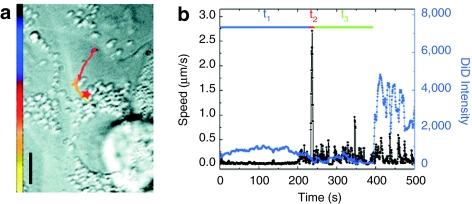
For the entry of influenza viruses, a three-stage entry process was found, which is slightly different than that for the polyplexes.5 Shortly after addition of the viruses to the cells, virus particles showed a slow actin-dependent movement in the cell periphery in stage I. Stage II consists of a short unidirectional movement toward the perinuclear region and is followed by intermittent and bidirectional movement in the nuclear periphery in stage III (Figure 5). Interestingly, the rapid stage II movement was followed by the initial acidification of the endosomes. As a consequence, virus fusion occurred at the end of stage III. To visualize the fusion event by fluorescence microscopy, Lakadamyali et al.5 took advantage of the self-quenching effect of the lipophilic dye DiD. The viral membrane of influenza virus can be loaded with DiD at sufficiently high concentrations for self-quenching and viral fusion is then detected by a significant increase in dye fluorescence intensity by dequenching due to lateral diffusion of dye molecules in the cell membrane. By using multicolor fluorescence microscopy, also mechanistic details of the internalization process of influenza virus were resolved. Influenza virus can enter cells by a clathrin-mediated as well as clathrin- and caveolin-independent endocytic pathway. Using fluorescently labeled influenza virus particles and EYFP-clathrin expressing cells, Rust et al.4 visualized the clathrin-mediated entry in great detail. Binding of virus particles to the cell membrane induced de novo formation of clathrin-coated pits at the virus binding site. After internalization, clathrin uncoating of the vesicles was characteristically followed by the rapid movement associated with stage II.
Figure 5.
Entry of influenza virus. (a) The trajectory of a DiD-labeled virus inside a cell. The color of the trajectory codes time with the colored bar indicating a uniform time axis from 0 s (black) to 500 s (yellow). The red star indicates the fusion site. Bar = 10 µm. (b) The instantaneous velocity (black) and DiD fluorescence intensity (blue) during the trajectory of the virus. t1, t2, and t3 are the durations of stages I, II, and III, respectively. Stage I starts with contact of the virus with the cell, stage II consists of a rapid unidirectional translocation from the cell periphery to the perinuclear region, stage III is the period between stage II and fusion and shows intermittent and bidirectional movement in the nuclear periphery. Fusion of the virus with the acidified endosome always occurs after the rapid movement in stage II. Copyright (2003) National Academy of Science, USA.5
Using approaches similar to those outlined above, the exact pathways of nanoparticle endocytosis and transfection can be elucidated. Currently, how gene carries enter and transfect cells is a black box. By tracking individual viruses and nanoparticles throughout the entire transfection pathway, it is possible to define different stages in the uptake and infection process. For example, rapid directed transport visualized from a subtrajectory analysis of individual particles is a clear indicator that the particles have been internalized by the cell. By combining SPT with multichannel detection, it becomes possible to investigate additional steps in the internalization process such as separation of DNA from polymer, to study the interaction of nanoparticles with the cellular organelles and proteins, and determine where in the kinetic scheme these additional processes and interactions occur. These types of investigations promise to improve the functionality of gene carriers by visualizing which steps occur efficiently and recognizing where adjustments need to be made in the design of the nanoparticles.
Advanced Microscopy Methods
Alternating WF and TIRF microscopy
Many interactions during the uptake of gene carriers occur at the plasma membrane of cells. TIRF microscopy (TIRFM) is an excellent method for live-cell measurements at the plasma membrane, particularly in the presence of a high cytosolic background. In TIRFM, the sample is illuminated through the evanescent wave generated at the glass–medium interface when illuminated at angles higher than the critical angle and penetrates ~100–250 nm into the sample dependent on the wavelength and angle of excitation:
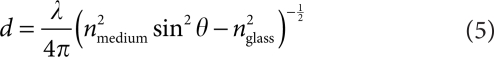 |
where θ and λ are the incident angle with respect to the normal and wavelength of illumination, respectively and ni the index of refraction of the corresponding substance. Due to the exponential decay of the evanescent field in TIRFM, it is very sensitive to the z-position of the fluorophore. To avoid misinterpretation of the fluorescence intensity in the TIRF, it is beneficial to simultaneously measure the fluorescence intensity using WF microscopy. A comparison of the fluorescence intensity in both modes allows one to distinguish between real changes in fluorescence intensity and axial moment of the tracked object.
Such an alternating TIRFM/WF setup was constructed to investigate the assembly of HIV-1 at the plasma membrane.54 Although this review focuses on the entry of viral and nanoparticles, egress has many similarities to entry and profits from the same technical approaches. During HIV egress, the individual assembly sites showed motion at the plasma membrane, making it necessary to track the individual particles during assembly and release. For these experiments, HeLa cells were transfected with the complete HIV-1 plasmid expressing all viral proteins with the exception of NEF and the long-terminal repeat regions were removed to render the virus noninfectious.55 The structural protein gag was fused with enhanced green fluorescent protein inserted between the matrix and capsid moieties and mixed with an equimolar mixture of nonlabeled constructs to guarantee wild-type like properties.56
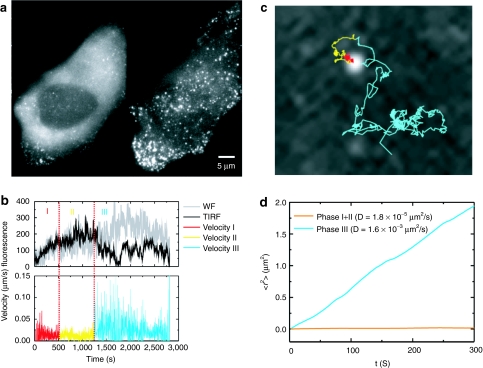
The excitation modality was alternated frame-by-frame such that the odd frames were collected with TIRFM excitation and the even frames with WF excitation. The evanescent field was sensitive to distances up to ~150 nm from the coverslip whereas the depth-of-field for WF is ~1–2 µm. Figure 6a shows a WF image along with the corresponding TIRFM image collected during the assembly of HIV-1-budding sites. HIV assembly was observed to occur in three-phases as exemplified in the trace shown in Figure 6b. Phase I involves the recruitment of gag proteins to the assembly site. Typically, the increase in fluorescence intensity follows the form of a saturating exponential. The duration of phase I from the start of assembly to 90% saturation was 8–9 minutes. In phase II, gag delivery to the assembly site is complete as shown in photobleaching experiments of Jouvenet et al.57 and interactions with some of the cellular ESCRT machinery takes place.58,59 The average duration of phase II was ~12 minutes. In phase III, a decrease in fluorescence signal is observed which, for at least some cases, can be attributed to particle release. The average timescale from assembly to release was measured to be on the order of 30 minutes.54
Figure 6.
Budding of HIV. (a) A wide-field (left) and total Internal refection fluorescence microscopy (TIRFM) (right) image of HeLa cells during HIV-1 assembly. The individual fluorescent punctea are individual assembly sites. (b) The fluorescence intensity and instantaneous velocity for an individual assembly site highlighting the three phases observed during assembly. The color code is red for phase I, yellow for phase II, and cyan for phase III. (c) The trajectory of the assembly site shown in panel (b) superimposed upon an image of the assembly site. The trajectory is color-coded as in b. (d) Mean-squared-displacements of phase I and II (orange) and phase III (cyan) from the trajectory shown in panel (c).
For the above experiments, the fluorescence intensity of the individual budding sites and the surrounding background were monitored in TIRFM and WF modes. When large fluctuations of the plasma membrane were seen or the particle was moving axially during the assembly process, the traces were not further analyzed. However, when axial motion was observed after assembly, it can be used as one possible indication of release. For example, Figure 6c shows a decrease in TIRFM signal during phase III while the WF signal remains constant, suggesting that the particle is moving in and out of the evanescent field in a small pocket between the coverslip and the plasma membrane of the cell. Release was verified by analyzing the mean-squared displacement of the particle during the different phases (Figure 6d). In phase III, the diffusion coefficient increases ~100 fold over phase I and II to 1.6 × 10−3 µm2/s and shows random, nondirected motion. Such a large diffusion coefficient is not possible in the cytosol for HIV-1 particles, which have a diameter of ~170 nm.60 Therefore, it is clear that the particle has been released.
The advantage of TIRFM is that only a thin sheet above the interface is illuminated by the evanescent field providing high contrast image of interactions at and near the plasma membrane. This can also be a limitation for experiments that investigate the entry process of artificial viruses or gene carriers where very few particles are able to reach the extracellular region illuminated by the evanescent field between the coverslip and plasma membrane. However, TIRFM/WF microscopy can still play an important role in investigating the uptake of particles by using innovative approaches to get the nanoparticles between the coverslip and the cell plasma membrane. One alterative is to use reverse transfection where the particles are first attached to the coverslip and the cells are then seeded on top of the particles.61 A second possible alternative is to use nano- and/or micropatterning to functionalize the surface of the coverslip such that there are pockets between the coverslip and cellular membrane that are easily accessible for nanoparticles.
Three-dimensional tracking
A comparison of the fluorescence intensity in TIRFM and WF excitation makes it possible to distinguish axial motion. Particularly during virus and artificial virus uptake, tracking in three-dimensions is important. A multitude of methods for three-dimensional tracking are emerging, but can be classified into two general approaches. In the first approach, data from a particular region is collected in three-dimensions and the particles are tracked manually or with automatic tracking programs afterwards. The second approach is to track single particles in real time using a feedback loop.
z-Stacks. The most straight-forward approach for three-dimensional tracking is to collect data in three dimensions using one type of scanning confocal microscopy. Often, a spinning disk confocal microscope is used because of its speed, however, laser-scanning confocal microscopes and two-photon microscopy can also be used. Images in x-y are collected at different z-positions. Hence, this technique is often referred to as z-stacks. Several z-stacks are collected and the particle tracked ex post facto. In our research group, we typically make a z-projection of each stack, taking the maximum intensity for each x-y position in the z-stack and use the software we have developed for two-dimensional imaging to extract the x-y coordinates from the projected image. To determine the z-position, a z-slice is taken at the x-y coordinates of the particle and fit with a one-dimensional Gaussian function. The z-resolution depends on the signal-to-noise ratio, but resolutions on the order of 60 nm can be easily achieved.
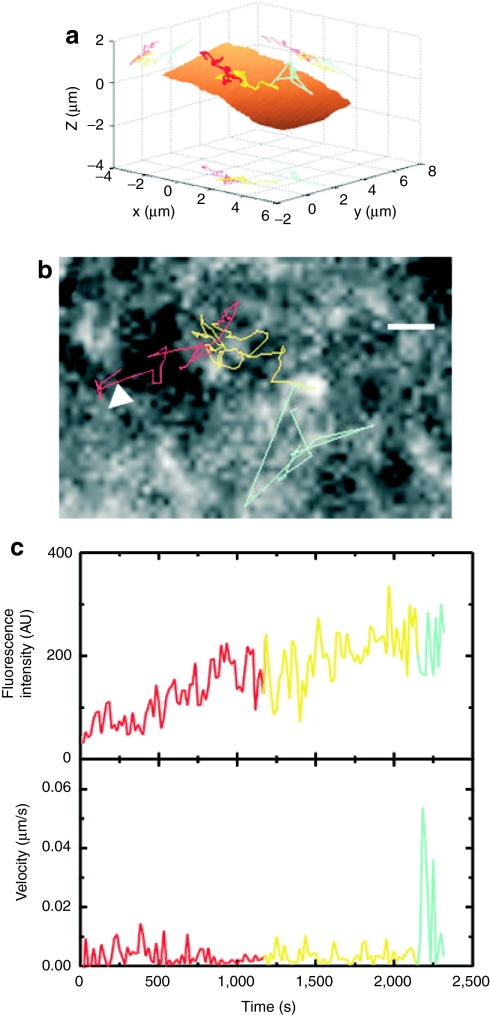
Spinning disk confocal microscopy was used to track the assembly of individual HIV-1 particles on the dorsal cell surface. Figure 7a shows a three-dimensional trajectory of a nascent assembly site from the beginning of assembly through to release. Below is a two-dimensional projection of the trajectory, the fluorescence intensity during the assembly process and the instantaneous velocity (Figure 7b,c). The phases are color-coded with phase I shown in red, phase II in yellow, and phase III in cyan. The residual fluorescence of gag molecules in the plasma membrane made it possible to reconstruct the cell membrane, shown as the brown surface in Figure 7a. From the measurements using the spinning disk confocal microscope, we could verify that the total assembly time of HIV-1, ~ 30 minutes, is not influenced by the presence of the coverslip in TIRFM measurements.
Figure 7.
Thee-dimensional tracking of HIV-1 assembly and release. (a) The trajectory of an HIV-1 particle observed using a spinning disk confocal microscope shown in three-dimensions. The plasma membrane is shown in brown. The trajectory is color-coded as in Figure 6 (phase I, red; phase II, yellow; phase III cyan). (b) A two-dimensional projection of the trajectory superimposed on a single confocal image of the assembly site. Bar = 1 µm. (c) The fluorescence intensity (upper panel) and instantaneous velocity (low panel) during assembly and release of the HIV-1 particle tracked in panel (a).
One disadvantage of collecting three-dimensional data via z-stacks is the slower time resolution. This can be seen by comparing the trajectories in Figures 6c and 7b. Although the individual images were collected with similar rates, the overall time resolution in three-dimensions is decreased by at least the number of x-y planes collected in the z-stack (when other hardware difficulties with z-scanning do not arise). In the last years, several methods have evolved to allow three-dimensional SPT on faster time scales. One method collects two image planes, one above and one below the depth of interest, and uses the shape of the psf in both images to extract the z-position of the particle.62,63 A second possibility uses a weak cylindrical lens into the emission pathway and determines the three-dimensional position of the particle from the axial astigmatism.64,65,66 Recently, Pavani and Piestun have developed an efficient double-helix point-spread function using a phase mask generated by a spatial light modulator67 and used it for three-dimensional SPT.68 This approach has been further championed by the group of Moerner.69,70 One drawback of all the above methods is that they are limited to z movement over a couple of micrometers.
Orbital tracking. An elegant alternative for three-dimensional SPT is orbital tracking. Orbital tracking is an extension of scanning-FCS and was proposed by Enderlein71 in two-dimensions and applied in two- and three-dimensions by Gratton72,73,74 and further adapted in our group.75
In orbital tracking, the laser is orbited around the object of interest with a radius roughly equivalent to the width of point-spread-function of the laser beam using a specially equipped (homebuilt) laser-scanning confocal microscope. When the particle is located at the center of the orbit, the fluorescence intensity will not change during the orbit. The measured intensity will be less than when the particle was centered in the laser beam, but the system is more sensitive to movement of the particle. Deviation of the particle from the center of the orbit is detectable as a modulation in the fluorescence signal. One or a few orbits are then averaged together and a fast Fourier transformation routine is used to determine the phase and amplitude of the modulation in microseconds. The phase of the modulation determines in what direction the molecule has moved and the amplitude of the modulation provides information on how far the particle has moved. Using a feedback loop, the orbit is then moved to the new location of the particle and the coordinates written to disk. With such a routine, it is possible to track particles with a time resolution of a few milliseconds and the trajectory is written to disk in real-time with no post processing necessary.
To track particles in three-dimensions, the detection pathway is split between two confocal channels. The confocal pinholes (or multimode fiber entrances, which act effectively as the confocal pinholes) are adjusted to be in focus ~500 nm above and ~500 nm below the tracked particle. The signals from the two channels are added together for determination of the x- and y-coordinates of the particle and the z-position determined from the difference of the two channels. The objective is then moved using a piezo stage to keep the particle focused between the two channels. With an appropriate signal-to-noise ratio, an accuracy of better than 10 nm is obtainable in all three dimensions at a time resolution of 32 ms. The z-position of the particle is determined from a single orbit and the position of the objective only changed when the tracked particle moves in the axial dimension. Thus, the objective is always focused in the plane of the tracked particle. Hence, a WF setup was built to allow the simultaneous observation of the environment in the focal plane about the tracked particle.
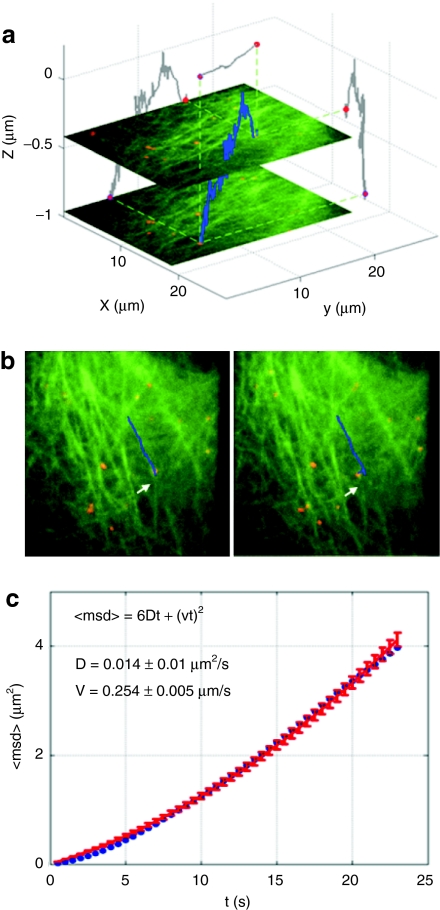
Using orbital tracking, we followed the transport of DNA/polyethylenimine polyplexes on microtubules. The polyplexes were prepared as previously described in ref. 76. The DNA plasmids were labeled with Cy3 and Cy5 so that they could be simultaneously observed in the WF setup (using Cy3) and tracked with orbital tracking (using Cy5). HuH7 cells were used where the microtubules were labeled with enhanced green fluorescent protein. After the polyplex has been taken up by the cell via endocytosis, the endosome is transported along the microtubules. Figure 8a displays a three-dimensional trajectory of a polyplex moving along a microtubule. Figure 8b shows two frames from the WF movie depicting the sideways movement of the polyplex through rearrangement of the microtubules network before being transported along a new microtubule filament. The MSD of the overall movement is plotted in Figure 8c clearly indicating active transport of the polyplex. The average transport velocity, in this case, was ~ 0.25 µm/s.
Figure 8.
Three-dimensional orbital tracking of polyplex transport along microtubules. (a) The three-dimensional trajectory (blue) of a polyplex was tracked in HuH7 cells with enhanced green fluorescent protein (EGFP) labeled tubulin (green structures in the image). Overlayed onto the 3D trajectory are two wide-field images taken at different z-positions during the measurement. Two-dimensional projections of the three-dimensional trajectory are shown in grey on the respective axes. (b) Two frames from the wide-field movie that was concomitantly recorded during orbital tracking of the polyplex (red dots). The corresponding video is available as supplementary information in ref. 75. The wide-field information allows correlation of the changes in the environment with motion of the tracked particle. (c) Mean-square displacement (MSD) plot (blue curve) of the trajectory presented in a. Fitting the MSD to directed motion (<r2> = v2Δt2 + 6DΔt for three-dimensions) yielded an average transport velocity of 0.25 µm/s. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with the permission.75
Another feedback approach is tetrahedral tracking by Berg77 and Werner78 where four detectors are used to measure different volumes arranged in a tetrahedral geometry about the focus of a confocal microscope. When a particle moves away from the focus, this motion is detected and the sample stage is then moved to place the tracked particle back into the center of the confocal volume.
Photomanipulation of Cells
In addition to the development of advanced tracking methods that allow multiple modalities to be measured quasi simultaneously or three-dimensional tracking in real-time, tracking methods can be combined with other optical methods for increased functionality. One of the major bottlenecks in nonviral gene transfer is the low efficiency of endosomal release.9,79 One approach to increase endosomal release is the use of membrane destabilizing polymers and peptides.80,81,82 Here, we discuss two optical strategies for getting genetic material into the cytosol where it can eventually reach the nucleus. Although these methods would only be applicable for optically accessible tissues, they also provide an example of how release can be followed using SPT.
Photoinduced release
One strategy to directly manipulate endosomal release is photochemically.83 In the work of de Bruin et al., endosomal membranes were loaded with a photosensitizer that was activated by a 405 nm laser pulse to produce singlet oxygen resulting in the rupture of endosomal membranes and release of the endosomal content. Polyplexes with fluorescently labeled polymer (Alexa488) and fluorescently labeled DNA (Cy5) were incubated for 12 hours with live cells to allow uptake into endosomes before the cells were illuminated with 405 nm light. By imaging the release process of single endosomes at high temporal resolution, de Bruin et al. showed that the release of the DNA is dependent on the polymer used for complexation. In the case of polyplexes based on PEI and DNA, the fluorescence signal of the labeled PEI vanished in less than a second due to diffusion into the cytosol. The fluorescence signal of the labeled DNA, in contrast, remained at the location of the ruptured endosome. As DNA molecules larger than ~1,000 bp cannot diffuse within the crowded cytoplasm,84 released plasmid DNA is basically immobile in contrast to the smaller sized polymer. However, the DNA was protected from degradation until endosomal rupture occurred and successfully released from the PEI. For polyplexes composed of PLL and DNA, the endosomal escape was different. The DNA remained either tightly bound to the polymer or vanished quickly by diffusion simultaneously with the polymer due to degradation within the endosome before its rupture.
Photothermally induced cell entry
A possibility to avoid entrapment in endosomes altogether is to fuse with the plasma membrane. Viruses have devised various means of getting into the cytosol. Single gold nanoparticles provide a possibility for getting pharmaceutical nanoparticles into cells. Feldmann and co-workers have used 80 nm large gold particles as local heaters to induce phase transitions in membranes.85 Nanometer sized gold particles have plasmon resonances at visible wavelengths. The plasmon resonance efficiently absorbs light and converts a significant fraction of light into heat. Thus, nanogold particles can be used to effectively heat the local surroundings. Using artificial membranes, phase transitions from gel phase to fluid phase were induced locally about the nanoparticle.85 Hence, one future strategy for getting pharmaceutical nanoparticles into cells is building them about metal particles. Absorption of light from the metal nanoparticles will cause local heating, transiently melting the plasma membrane and allowing controlled entry of the nanoparticles. This is just one example of optical functionality and addressability that can be added to the next generation of gene carriers.
Conclusions
Other methods for investigating particle motility
SPT provides a maximum amount of information when individual particles are easily discernable and move slow enough that they can be tracked. However, SPT is limited when the particles diffuse too quickly (for example, small proteins and peptides) or the particle density is so high that individual particles often cross paths, making it difficult to follow an individual particle. Many other methods are available to gain information over the mobility of particles when the particles are too quick or the concentrations too high for SPT. Fluorescence Fluctuation Spectroscopy is one class of methods which analyzes the fluctuations in fluorescence intensity to extract information regarding the number, mobility and brightness of the fluorescence particles. These methods include Fluorescence Correlation Spectroscopy,86,87,88 Image Correlation Spectroscopy,89,90 and Raster Image Correlation Spectroscopy.91,92 Another possibility is quantitative fluorescence speckle microscopy.93,94,95 Rather than tracking individual particles, speckles from a small number of fluorescent molecules are tracked and thereby the dynamics of macromolecular structures elucidated. These alternative methods that can be used at particle concentrations well beyond what is possible with SPT are being further developed and have great potential in the future of biophysical live-cell investigations.
Outlook
Highly sensitive single-cell fluorescence microscopy techniques are powerful tools for investigating the uptake of pharmaceutical nanoparticles as well as viruses into cells with high resolution. They can help elucidate the entry pathway of viruses or be used to unravel the bottlenecks of specific nanoparticle applications on the cellular level and provide feedback for improvements of nanoparticles. This is especially relevant for smart nanosystems with programmed functionalities that are designed to act on defined spots in the cell at the right time. By addition of functionalities like specific targeting, redox- or pH-sensitivity, nanoparticles can be rendered more “virus-like” and be made more effective. However, to evaluate the functionality of those gained complexities, also more complex techniques to observe them are required and novel microscopy techniques as described here are valuable tools.
Acknowledgments
We gratefully acknowledge the financial support of the Nanosystems Initiative Munich (NIM), the Deutsche Forschungsgemeinschaft through the Schwerpunktprogramm 1175 and 1313, the SFB 646 and the SFB 749, and the Ludwig-Maximilians-University Munich (LMUInnovativ BioImaging Network).
REFERENCES
- Payne CK. Imaging gene delivery with fluorescence microscopy. Nanomedicine (Lond) 2007;2:847–860. doi: 10.2217/17435889.2.6.847. [DOI] [PubMed] [Google Scholar]
- Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X., and, Hogle JM. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P., and, Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc Natl Acad Sci USA. 2005;102:15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Lakadamyali M, Zhang F., and, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP., and, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Jones AT., and, Stephens DJ. Intracellular trafficking pathways and drug delivery: fluorescence imaging of living and fixed cells. Adv Drug Deliv Rev. 2005;57:43–61. doi: 10.1016/j.addr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC.et al. (2010The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls Mol Ther 18561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J, Bragonzi A., and, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Rémy-Kristensen A, Clamme JP, Vuilleumier C, Kuhry JG., and, Mély Y. Role of endocytosis in the transfection of L929 fibroblasts by polyethylenimine/DNA complexes. Biochim Biophys Acta. 2001;1514:21–32. doi: 10.1016/s0005-2736(01)00359-5. [DOI] [PubMed] [Google Scholar]
- Kopatz I, Remy JS., and, Behr JP. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- Lundin P, Johansson H, Guterstam P, Holm T, Hansen M, Langel U.et al. (2008Distinct uptake routes of cell-penetrating peptide conjugates Bioconjug Chem 192535–2542. [DOI] [PubMed] [Google Scholar]
- Mäe M, Andaloussi SE, Lehto T., and, Langel U. Chemically modified cell-penetrating peptides for the delivery of nucleic acids. Expert Opin Drug Deliv. 2009;6:1195–1205. doi: 10.1517/17425240903213688. [DOI] [PubMed] [Google Scholar]
- Rinne J, Albarran B, Jylhävä J, Ihalainen TO, Kankaanpää P, Hytönen VP.et al. (2007Internalization of novel non-viral vector TAT-streptavidin into human cells BMC Biotechnol 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner K., and, Sodeik B. The role of the cytoskeleton during viral infection. Curr Top Microbiol Immunol. 2005;285:67–108. doi: 10.1007/3-540-26764-6_3. [DOI] [PubMed] [Google Scholar]
- Marsh M., and, Bron R. SFV infection in CHO cells: cell-type specific restrictions to productive virus entry at the cell surface. J Cell Sci. 1997;110 (Pt 1):95–103. doi: 10.1242/jcs.110.1.95. [DOI] [PubMed] [Google Scholar]
- Helenius A, Kartenbeck J, Simons K., and, Fries E. On the entry of Semliki forest virus into BHK-21 cells. J Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E., and, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Marsh M., and, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benns JM., and, Kim SW. Tailoring new gene delivery designs for specific targets. J Drug Target. 2000;8:1–12. doi: 10.3109/10611860009009205. [DOI] [PubMed] [Google Scholar]
- Cheng H, Zhu JL, Zeng X, Jing Y, Zhang XZ., and, Zhuo RX. Targeted gene delivery mediated by folate-polyethylenimine-block-poly(ethylene glycol) with receptor selectivity. Bioconjug Chem. 2009;20:481–487. doi: 10.1021/bc8004057. [DOI] [PubMed] [Google Scholar]
- Frederiksen KS, Abrahamsen N, Cristiano RJ, Damstrup L., and, Poulsen HS. Gene delivery by an epidermal growth factor/DNA polyplex to small cell lung cancer cell lines expressing low levels of epidermal growth factor receptor. Cancer Gene Ther. 2000;7:262–268. doi: 10.1038/sj.cgt.7700098. [DOI] [PubMed] [Google Scholar]
- Lu T, Sun J, Chen X, Zhang P., and, Jing X. Folate-conjugated micelles and their folate-receptor-mediated endocytosis. Macromol Biosci. 2009;9:1059–1068. doi: 10.1002/mabi.200900134. [DOI] [PubMed] [Google Scholar]
- Godinez WJ, Lampe M, Wörz S, Müller B, Eils R., and, Rohr K. Deterministic and probabilistic approaches for tracking virus particles in time-lapse fluorescence microscopy image sequences. Med Image Anal. 2009;13:325–342. doi: 10.1016/j.media.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Sbalzarini IF., and, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE., and, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Saxton MJ., and, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Qian H, Sheetz MP., and, Elson EL. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys J. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DS, Forstner MB., and, Käs JA. Apparent subdiffusion inherent to single particle tracking. Biophys J. 2002;83:2109–2117. doi: 10.1016/S0006-3495(02)73971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savin T., and, Doyle PS. Static and dynamic errors in particle tracking microrheology. Biophys J. 2005;88:623–638. doi: 10.1529/biophysj.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund AJ. Statistics of camera-based single-particle tracking. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82 1 Pt 1:011917. doi: 10.1103/PhysRevE.82.011917. [DOI] [PubMed] [Google Scholar]
- Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82 4 Pt 1:041914. doi: 10.1103/PhysRevE.82.041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris M.et al. (2007Cellular dynamics of EGF receptor-targeted synthetic viruses Mol Ther 151297–1305. [DOI] [PubMed] [Google Scholar]
- Suh J, Wirtz D., and, Hanes J. Real-time intracellular transport of gene nanocarriers studied by multiple particle tracking. Biotechnol Prog. 2004;20:598–602. doi: 10.1021/bp034251y. [DOI] [PubMed] [Google Scholar]
- Simson R, Sheets ED., and, Jacobson K. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys J. 1995;69:989–993. doi: 10.1016/S0006-3495(95)79972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcizet D, Meier B, Sackmann E, Rädler JO., and, Heinrich D. Temporal analysis of active and passive transport in living cells. Phys Rev Lett. 2008;101:248103. doi: 10.1103/PhysRevLett.101.248103. [DOI] [PubMed] [Google Scholar]
- Mahowald J, Arcizet D., and, Heinrich D. Impact of external stimuli and cell micro-architecture on intracellular transport states. Chemphyschem. 2009;10:1559–1566. doi: 10.1002/cphc.200900226. [DOI] [PubMed] [Google Scholar]
- Montiel D, Cang H., and, Yang H. Quantitative characterization of changes in dynamical behavior for single-particle tracking studies. J Phys Chem B. 2006;110:19763–19770. doi: 10.1021/jp062024j. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Shibata T., and, Ueda M. Statistical analysis of lateral diffusion and multistate kinetics in single-molecule imaging. Biophys J. 2009;97:1115–1124. doi: 10.1016/j.bpj.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz GJ, Schindler H., and, Schmidt T. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys J. 1997;73:1073–1080. doi: 10.1016/S0006-3495(97)78139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud F, Michalet X, Iyer G, Margeat E, Moore HP., and, Weiss S. Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot tracking. Traffic. 2009;10:691–712. doi: 10.1111/j.1600-0854.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ., and, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E., and, Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller BK, Zaychikov E, Bräuchle C., and, Lamb DC. Pulsed interleaved excitation. Biophys J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright RH.et al. (2005Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation Biophys J 882939–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong X., and, Weiss S. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- Bausinger R, von Gersdorff K, Braeckmans K, Ogris M, Wagner E, Bräuchle C.et al. (2006The transport of nanosized gene carriers unraveled by live-cell imaging Angew Chem Int Ed Engl 451568–1572. [DOI] [PubMed] [Google Scholar]
- Sauer AM, de Bruin KG, Ruthardt N, Mykhaylyk O, Plank C., and, Bräuchle C. Dynamics of magnetic lipoplexes studied by single particle tracking in living cells. J Control Release. 2009;137:136–145. doi: 10.1016/j.jconrel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Mislick KA., and, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CK, Jones SA, Chen C., and, Zhuang X. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP.et al. (2007One at a time, live tracking of NGF axonal transport using quantum dots Proc Natl Acad Sci USA 10413666–13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner K, Radtke K, Schmidt S., and, Sodeik B. Eclipse phase of herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J Virol. 2006;80:8211–8224. doi: 10.1128/JVI.02528-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R.et al. (2002Function of dynein and dynactin in herpes simplex virus capsid transport Mol Biol Cell 132795–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E.et al. (2000Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis Hum Gene Ther 11151–165. [DOI] [PubMed] [Google Scholar]
- Ivanchenko S, Godinez WJ, Lampe M, Kräusslich HG, Eils R, Rohr K.et al. (2009Dynamics of HIV-1 assembly and release PLoS Pathog 5e1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Daecke J, Fackler OT, Dittmar MT, Zentgraf H., and, Kräusslich HG. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol. 2004;78:10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe M, Briggs JA, Endress T, Glass B, Riegelsberger S, Kräusslich HG.et al. (2007Double-labelled HIV-1 particles for study of virus-cell interaction Virology 36092–104. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD., and, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD., and, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Kräusslich HG.et al. (2011Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component Nat Cell Biol 13469–474. [DOI] [PubMed] [Google Scholar]
- Endress T, Lampe M, Briggs JA, Kräusslich HG, Bräuchle C, Müller B.et al. (2008HIV-1-cellular interactions analyzed by single virus tracing Eur Biophys J 371291–1301. [DOI] [PubMed] [Google Scholar]
- Koch P, Lampe M, Godinez WJ, Müller B, Rohr K, Kräusslich HG.et al. (2009Visualizing fusion of pseudotyped HIV-1 particles in real time by live cell microscopy Retrovirology 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toprak E, Balci H, Blehm BH., and, Selvin PR. Three-dimensional particle tracking via bifocal imaging. Nano Lett. 2007;7:2043–2045. doi: 10.1021/nl0709120. [DOI] [PubMed] [Google Scholar]
- Prabhat P, Gan Z, Chao J, Ram S, Vaccaro C, Gibbons S.et al. (2007Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy Proc Natl Acad Sci USA 1045889–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HP., and, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzer L, Meckel T., and, Schmidt, T. Nanometric three-dimensional tracking of individual quantum dots in cells. Appl Phys Lett. 2007;90:1–3. [Google Scholar]
- Huang B, Jones SA, Brandenburg B., and, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavani SR., and, Piestun R. High-efficiency rotating point spread functions. Opt Express. 2008;16:3484–3489. doi: 10.1364/oe.16.003484. [DOI] [PubMed] [Google Scholar]
- Pavani SR., and, Piestun R. Three dimensional tracking of fluorescent microparticles using a photon-limited double-helix response system. Opt Express. 2008;16:22048–22057. doi: 10.1364/oe.16.022048. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Casolari JM, Badieirostami M, Brown PO., and, Moerner WE. Three-dimensional tracking of single mRNA particles in Saccharomyces cerevisiae using a double-helix point spread function. Proc Natl Acad Sci USA. 2010;107:17864–17871. doi: 10.1073/pnas.1012868107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MA, Lew MD, Badieirostami M., and, Moerner WE. Localizing and tracking single nanoscale emitters in three dimensions with high spatiotemporal resolution using a double-helix point spread function. Nano Lett. 2010;10:211–218. doi: 10.1021/nl903295p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderlein J. Tracking of fluorescent molecules diffusing within membranes. Appl Phys B. 2000;71:773–777. [Google Scholar]
- Kis-Petikova K., and, Gratton E. Distance measurement by circular scanning of the excitation beam in the two-photon microscope. Microsc Res Tech. 2004;63:34–49. doi: 10.1002/jemt.10417. [DOI] [PubMed] [Google Scholar]
- Levi V, Ruan Q, Kis-Petikova K., and, Gratton E. Scanning FCS, a novel method for three-dimensional particle tracking. Biochem Soc Trans. 2003;31 Pt 5:997–1000. doi: 10.1042/bst0310997. [DOI] [PubMed] [Google Scholar]
- Levi V, Ruan Q., and, Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Burkacky O, Meyer M, Bräuchle C, Gratton E., and, Lamb DC. Real-time nanomicroscopy via three-dimensional single-particle tracking. Chemphyschem. 2009;10:2458–2464. doi: 10.1002/cphc.200900436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gersdorff K, Ogris M., and, Wagner E. Cryoconserved shielded and EGF receptor targeted DNA polyplexes: cellular mechanisms. Eur J Pharm Biopharm. 2005;60:279–285. doi: 10.1016/j.ejpb.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Berg HC. How to track bacteria. Rev Sci Instrum. 1971;42:868–871. doi: 10.1063/1.1685246. [DOI] [PubMed] [Google Scholar]
- Lessard GA, Goodwin PM., and, Werner JH. Three-dimensional tracking of individual quantum dots. Appl Phys Lett. 2007;91:224106. [Google Scholar]
- Lechardeur D, Verkman AS., and, Lukacs GL. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv Drug Deliv Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Boeckle S, Fahrmeir J, Roedl W, Ogris M., and, Wagner E. Melittin analogs with high lytic activity at endosomal pH enhance transfection with purified targeted PEI polyplexes. J Control Release. 2006;112:240–248. doi: 10.1016/j.jconrel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Ogris M, Carlisle RC, Bettinger T., and, Seymour LW. Melittin enables efficient vesicular escape and enhanced nuclear access of nonviral gene delivery vectors. J Biol Chem. 2001;276:47550–47555. doi: 10.1074/jbc.M108331200. [DOI] [PubMed] [Google Scholar]
- Stayton PS, El-Sayed ME, Murthy N, Bulmus V, Lackey C, Cheung C.et al. (2005‘Smart' delivery systems for biomolecular therapeutics Orthod Craniofac Res 8219–225. [DOI] [PubMed] [Google Scholar]
- de Bruin KG, Fella C, Ogris M, Wagner E, Ruthardt N., and, Bräuchle C. Dynamics of photoinduced endosomal release of polyplexes. J Control Release. 2008;130:175–182. doi: 10.1016/j.jconrel.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N., and, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- Urban AS, Fedoruk M, Horton MR, Rädler JO, Stefani FD., and, Feldmann J. Controlled nanometric phase transitions of phospholipid membranes by plasmonic heating of single gold nanoparticles. Nano Lett. 2009;9:2903–2908. doi: 10.1021/nl901201h. [DOI] [PubMed] [Google Scholar]
- Magde D, Elson EL., and, Webb WW. Thermodynamic fluctuations in a reacting system - measurement by fluorescence correlation spectroscopy. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- Elson EL., and, Magde D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Magde D, Elson EL., and, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Petersen NO, Höddelius PL, Wiseman PW, Seger O., and, Magnusson KE. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman PW., and, Petersen NO. Image correlation spectroscopy. II. Optimization for ultrasensitive detection of preexisting platelet-derived growth factor-beta receptor oligomers on intact cells. Biophys J. 1999;76:963–977. doi: 10.1016/S0006-3495(99)77260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR., and, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Sengupta P, Wiseman PW, Brown CM, Horwitz AR., and, Gratton E. Fluctuation correlation spectroscopy with a laser-scanning microscope: exploiting the hidden time structure. Biophys J. 2005;88:L33–L36. doi: 10.1529/biophysj.105.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser G., and, Waterman-Storer CM. Quantitative fluorescent speckle microscopy: where it came from and where it is going. J Microsc. 2003;211 Pt 3:191–207. doi: 10.1046/j.1365-2818.2003.01222.x. [DOI] [PubMed] [Google Scholar]
- Danuser G., and, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC., and, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]