Abstract
A lentiviral vector encoding β-globin flanked by insulator elements has been used to treat β-thalassemia (β-Thal) successfully in one human subject. However, a clonal expansion was observed after integration in the HMGA2 locus, raising the question of how commonly lentiviral integration would be associated with possible insertional activation. Here, we report correcting β-Thal in a murine model using the same vector and a busulfan-conditioning regimen, allowing us to investigate efficacy and clonal evolution at 9.2 months after transplantation of bone marrow cells. The five gene-corrected recipient mice showed near normal levels of hemoglobin, reduced accumulation of reticulocytes, and normalization of spleen weights. Mapping of integration sites pretransplantation showed the expected favored integration in transcription units. The numbers of gene-corrected long-term repopulating cells deduced from the numbers of unique integrants indicated oligoclonal reconstitution. Clonal abundance was quantified using a Mu transposon-mediated method, indicating that clones with integration sites near growth-control genes were not enriched during growth. No integration sites involving HMGA2 were detected. Cells containing integration sites in genes became less common after prolonged growth, suggesting negative selection. Thus, β-Thal gene correction in mice can be achieved without expansion of cells harboring vectors integrated near genes involved in growth control.
Introduction
Retroviral vectors have been used successfully in human trials of gene transfer to treat a number of genetic diseases, including X-linked severe combined immunodeficiency disorder (X-SCID),1 adenosine deaminase deficiency (ADA-SCID),2 chronic granulomatous disease (CGD)3 X-linked adrenoleukodystrophy,4 and β-thalassemia major (β-Thal).5 However, several adverse events have occurred in which integration of the therapeutic vector resulted in insertional activation of proto-oncogenes, contributing to the development of leukemia.6,7,8 Thus, there has been intense interest in characterizing the integration profile of gene therapy vectors as a step toward improving their safety.
Although the majority of completed trials of retroviral gene transfer have employed γ-retroviral vectors, lentiviral vectors have several advantages. Lentiviral vectors, unlike γ-retroviral vectors, infect nondividing cells. Additionally, no convincing examples of insertional activation of oncogenes and consequent transformation have been reported associated with HIV infection, though HIV proviruses can affect the activity of nearby genes. Studies in tumor-prone mouse models9,10 and tissue culture systems11,12 have reported less genotoxicity resulting from transduction with lentiviral vectors than with γ-retroviral vectors. One possible contributor to the difference may be differences in their preferred sites of integration. Insertional activation in animal models and human patients is most commonly caused by integrated vector promoter or enhancer elements upregulating downstream cellular genes.7,13 γ-Retroviral vectors show a strong propensity to integrate at promoters and gene 5′ ends,14 and clustering near genes controlling cell growth and proliferation has been reported.15,16 Lentiviruses, on the other hand, favor integration in the bodies of transcription units, avoiding regulatory 5′ regions,17,18 with no bias for growth-associated genes. The lentiviral integration pattern may thus be less likely to result in insertional activation, though other variables such as cell type and cytopathicity of infection may also explain the lack of insertional mutagenesis during HIV infection. In the context of vectors, the engineered transcriptional elements controlling transgene expression likely influence vector genotoxicity.9,10
Three clinical trials have been conducted using lentiviral vectors in humans, and in each the genomic distribution of integration sites was monitored. The first tested an anti-HIV therapy by delivering an antisense HIV-env gene to mature T-cells.19 Integration events in these patients showed no evidence of enrichment of sites in proto-oncogenes following transduction.20 The second trial treated two patients with adrenoleukodystrophy by transduction of hematopoietic stem cells.4 Integration sites in these two patients also showed sustained polyclonality up to 24 months after transplantation without adverse events. Thirdly, one patient was treated for β-Thal by hematopoietic stem cell transduction with a lentiviral vector encoding β-globin with clinical success.5
The lentiviral vector used in the β-Thal trial and in mice studied here incorporated several features to improve efficacy and safety that were developed by work in multiple laboratories (see refs. 21,22,23 and references therein). The vector contained self-inactivating deletions in the long-terminal repeat (LTR) to block expression from LTR transcriptional signals and contained essential elements of the β-globin locus control region to allow erythroid-specific expression. Modification of hematopoietic cells with such vectors has resulted in high-level, erythroid-specific, and long-term expression of the human β- or γ-globin transgenes in several mouse models of hemoglobin disorders as well as in human hematopoietic cells transplanted in immune-compromised mice. As an additional safety feature, two copies of the cHS4 chromatin insulator were incorporated into the 3′ U3 of the vector used here and in the phase 1/2 clinical trial of β-Thal treatment.24 Additionally, while most of the preclinical transplant models to date have involved preconditioning with high-dose total body irradiation, busulfan treatment is favored with patients and was used in the mouse model studied here.
In the human β-Thal trial, the vector integration site distribution determined up to 3 years post-transplantation showed that an insertion site located within the proto-oncogene HMGA2 accounted for 30% of all integration sites in whole blood cells.5 The integration event was associated with increased transcription of HMGA2 in erythroid cells and expression of a transcript whose 3′ UTR was replaced by vector sequences. This 3′-subsituted transcript lacked the target of the repressive miRNA let-7, contributing to elevated HMGA2 expression together with an increase in the rate of transcriptional initiation. HMGA2 has been implicated in the persistence of stem cells25 and is disregulated in some cancers, including by disruption of the normal gene 3′ end (see ref. 26 and references therein). While the treated patient remains healthy 3 years after therapy, this finding has raised questions about the possibility of lentiviral integration in or near growth-control genes imparting a selective advantage and leading to preferential outgrowth of the gene-modified cell.
A batch of this vector [“Lentiglobin” (LG)], produced and processed under cGMP conditions for use in the clinical trial, was used to evaluate its therapeutic efficacy in β-thalassemic mice pretreated with busulfan. This allowed study of the mechanism of disease correction and possible genotoxicity, and also made possible a comparison of the distribution of integration sites in the mouse and human studies.
Results
Correction of hematological disease associated with β-Thal
The clinical-grade vector and the experimental design are depicted in Figure 1. Bone marrow cells from female β-Thal donor mice were transduced ex vivo with LG vector (or mock-transduced) for 24 hours, and then transplanted into male recipient β-Thal mice. Twenty-two mice were studied: five wild-type C57BL/6J mice, four untreated β-Thal mice, four receiving fresh donor cells, four mice receiving mock-transduced donor cells, and five receiving LG vector-transduced cells. The average vector copy number per cell in LG vector-transduced cells, measured by quantitative PCR was 0.68. Following initial recovery from enhanced anemia resulting from myeloablation (30–40 days), the recipients of LG-transduced bone marrow exhibited almost complete phenotype correction by 40–50 days, as assessed by hemoglobin level, reticulocyte, and red blood cell counts (Figure 2 and Supplementary Table S1). At 9.2 months post-transplant, recipients of gene-transduced bone marrow cells exhibited normalization of spleen weights (Figure 3a,b), decreased proportion of early erythroblasts (basophilic, Ter119high CD71high) as compared to late erythroblasts (orthochromatophilic, Ter119high CD71low) (Figure 3c), decreased iron accumulation in spleen and liver (Figure 3d) and decreased plasma Epo levels (Supplementary Table S1). No significant difference in the yield of bone marrow hematopoietic progenitors [(colony-forming unit in culture (CFU-Cs)] in the transplant inoculum was observed among the β-Thal groups (Supplementary Table S1). No apparent hematopoietic bias was detected in bone marrow according to a panel of cell surface markers on flow cytometry (Figure 3e).
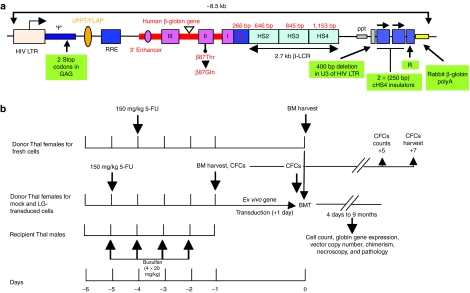
Figure 1.
Lentiglobin vector and experimental design. (a) Diagram of the human β-globin (βA) lentiviral vector [Lentiglobin, (LG)]. The 3′ β-globin enhancer, the 372 base pairs (bp) IVS2 deletion, the βA-T87Q mutation (ACA[Thr] to CAG[Gln]) and DNase I hypersensitive sites (HS) 2, HS3, and HS4 of the human β-globin locus control region (LCR) are indicated. Safety modifications including the two stop codons in the ψ+ signal, the 400 bp deletion in the U3 of the right HIV LTR, the rabbit β-globin polyA signal and the 2 × 250 bp cHS4 chromatin insulators are indicated. βp, human β-globin promoter; cPPT/flap, central polypurine tract; HIV LTR, human immunodeficiency type-1 virus long-terminal repeat; ppt, polypurine tract; RRE, Rev-responsive element; ψ+, packaging signal. (b) Treatment and transplantation protocol. Experiments were conducted with busulfan conditioning of male β-thalassemia (β-Thal) mice followed by transplant of female β-Thal bone marrow cells (4 × 105 fresh, mock-transduced, or LG-transduced cells) and monitoring of peripheral blood and pathology.
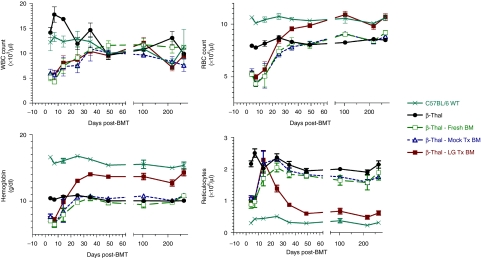
Figure 2.
Hematological recovery and improvement of erythroid parameters in β-thalassemia (β-Thal) recipients of Lentiglobin (LG)-transduced β-Thal bone marrow cells. Results shown are the mean ± SD for untransplanted, control β-Thal mice (n = 5), normal unmanipulated C57BL/6 mice (n = 4), and β-Thal mice transplanted with freshly harvested cells (n = 4), mock-transduced cells (n = 4), and LG-transduced cells (n = 5). Changes in red blood cell (RBC) number, hemoglobin concentration, and reticulocyte count seen at 30 days and beyond in β-Thal recipients of LG-transduced cells were highly significant in comparison with nontransplanted, or control-transplanted β-Thal mice (P < 0.001). BMT, bone marrow transplantation; RBC, red blood cells; WBC, white blood cells; WT, wild type.
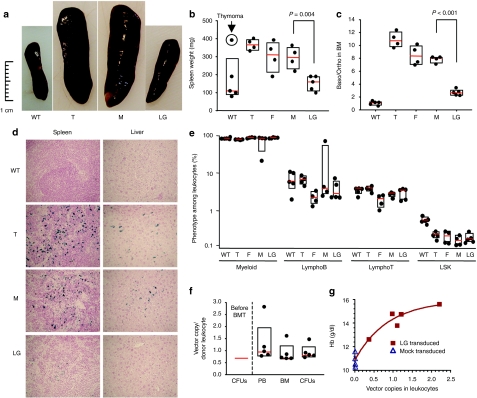
Figure 3.
Long-term correction of β-thalassemia (β-Thal) disease at 9.2 months post-bone marrow transplantation (BMT). (a) Spleen size and (b) spleen weight of normal C57BL/6J (WT) mice, unmanipulated β-Thal (T) and β-Thal recipients of fresh (F), mock (M), or Lentiglobin (LG)-transduced β-Thal bone marrow. The overall weight of each mouse was similar, so correction of spleen weight by body weight did not affect the conclusions (data not shown). (c) Proportion of early erythroblasts (basophilic, Ter119high CD71high) as compared to late erythroblasts (orthochromatophilic, Ter119high CD71low) from flow cytometric analysis of the bone marrow. (d) Photomicrographs of Mallory stained spleen and liver tissue sections from representative mice showing iron deposition related to β-Thal. (e) Phenotypic analysis of different cell populations in the bone marrow according to cell surface antibody staining and flow cytometry. For all histograms, results shown are the median values (red line), the 25th and 75th percentiles (bottom and top boundary of the boxes) and individual values (dots) for normal C57BL/6J (WT, n = 5), control β-Thal mice (T, n = 4), β-Thal mice transplanted with freshly harvested cells (F, n = 4), mock-transduced cells (M, n = 4), and LG-transduced cells (LG, n = 5). (f) Median copy number in the transplanted bone marrow (BM) progenitors [(colony-forming units (CFUs)] before BMT and in donor peripheral blood leukocytes (PB), total bone marrow cells (BM), and BM progenitors (CFUs) at 9.2 months post-BMT of β-Thal mice transplanted with LG-transduced cells (n = 5). Results shown are the median values (red line), the 25th and 75th percentiles (bottom and top boundary of the boxes) and individual values (dots). (g) Relationship between transgene copy number in the peripheral blood leukocytes and hemoglobin (Hb) concentration.
Analysis of female (donor) versus male (recipient) cells according to Y-chromosome real-time PCR at 9 months after bone marrow transplantation showed that an average of 72.7% (range 13.0–90.6) of the peripheral blood leukocytes were of donor origin in the group of mice receiving LG-transduced cells. The levels of donor-type chimerism were equivalent in mice receiving fresh (63.8%, range: 27.6–81.5) or mock-transduced (76.0%, range: 35.5–100.0) cells. Transgene copy numbers were determined in peripheral blood leukocytes, total bone marrow cells, and myeloid progenitors (CFUs). Median values of vector copy numbers normalized to female donor cells were not significantly different (P > 0.05) and were equivalent to that observed in transduced CFU-Cs before bone marrow transplantation (Figure 3f). Flow cytometric analysis revealed that the majority of red blood cells (86–97%) contained modified human β-globin contributing to 19.3–34.0% of all β-globin chains with a mean corpuscular human hemoglobin in modified red blood cells of 4.0 ± 0.7 pg (mean corpuscular hemoglobin in C57BL/6 and β-Thal of 14.5 ± 0.3 and 9.4 ± 0.4 pg, respectively). Analysis of transgene sequences in the overall peripheral leukocyte population showed that resolution of the disease can be attained at copy numbers of 0.36 and above including one recipient that exhibited only partial engraftment of donor (female) cells (13%). In this mouse, high-vector copy number per donor cell (2.8) but low donor chimerism (13%) allowed preferential expansion and/or prolonged survival of corrected erythroid cells (Figure 3 and Supplementary Table S1).
No gross abnormalities were observed on autopsy of the mice at 9.2 months with the exception of one control C57BL/6 mouse that presented with a thymic mass (170 mg) and enlarged spleen (390 mg). The bone marrow cellularity and body and tissue (liver, thymus kidney, and lung) weights were normal among the other mice. Histopathological evaluation of bone marrow, spleen, thymus, liver, lung, heart, kidney, stomach, and intestine were unremarkable with no evidence of neoplasia other than the malignant thymoma that appeared to spread to the spleen in the untreated C57BL/6 mouse. Mild inflammation of the kidneys was occasionally seen but not associated with treatment or transplant of LG-transduced bone marrow.
Isolation of integration sites from LG-transduced bone marrow before and after transplantation
In order to analyze vector integration sites for evidence of insertional activation, transduced bone marrow cells were harvested and analyzed pretransplantation and 9 months post-transplantation (at time of sacrifice). Before isolating genomic DNA, aliquots of pretransplant and post-transplant cells were subjected to methylcellulose culture to select committed hematopoietic progenitor cells. Culturing cells in this way allowed us to study the integration sites of progenitor cells, and compare them to sites from total bone marrow, the latter of which also contains terminally differentiated cells. A total of 500,000 pretransplantation cells were cultured in liquid media for 11 days. About 90,000 cells were cultured in methylcellulose-based media for 7 days, yielding about 2,000 colonies from myeloid progenitors. Post-transplantation cells were either cultured in methylcellulose as above, or DNA was extracted immediately. Three million bone marrow cells were used for immediate DNA extraction, and 90,000 bone marrow cells per mouse were cultured in methylcellulose, yielding about 50 colonies from myeloid progenitors per mouse.
Vector integration sites were initially amplified from the genomic DNA by restriction enzyme digestion of the genomic DNA and linker-mediated amplification as previously described.13 This method is sensitive, and allows reproducible characterization of genomic integration site distributions. However, one limitation of this method is its reliance on restriction digestion, which has been shown to introduce a significant recovery bias.13,27 Samples were processed with two different restriction enzymes in parallel, MseI and NlaIII, in an attempt to maximize site recovery and diversity. Additionally, in order to more carefully assess integration site abundance, a more quantitative method was used. This technique makes use of the phage Mu transposase to insert linker sequences at random sites in the genomic DNA to overcome the bias introduced by restriction enzymes. Extensive control studies show that the Mu-mediated method is less biased and so suitable for quantifying the most abundant clones in a population. Vector-host junctions were sequenced by 454/Roche pyrosequencing, using 8 bp barcodes in the primer to distinguish among samples sequenced in pools.
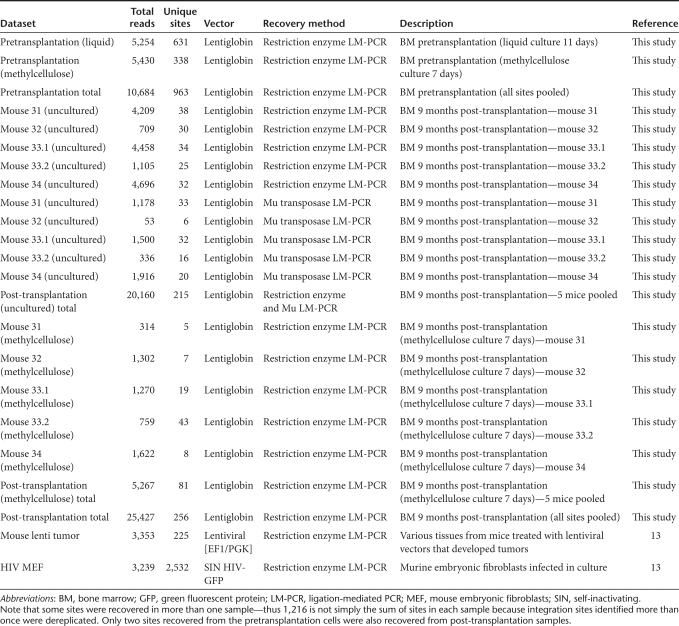
Table 1 summarizes the number of integration sites identified for samples from each mouse. Across all sets, we obtained a total of 36,111 integration site sequences passing quality control, representing 1,216 unique sites overall sets. There was a significant overlap between the sites recovered from uncultured and methylcellulose-cultured cells. Three of five methylcellulose sites in mouse 31 were found in uncultured cells, all 7 for mouse 32 were also found in uncultured cells, 13 of 19 for mouse 33.1, 9 of 43 for mouse 33.2, and 7 of 8 for mouse 34. Four sites were recovered in more than one mouse. Transduced cells were cultured for 24 hours before transplantation, so it is possible that these sites resulted from cell division during this time, but they may also be the result of PCR contamination during sample amplification.
Table 1. Integration site data sets used in this study.
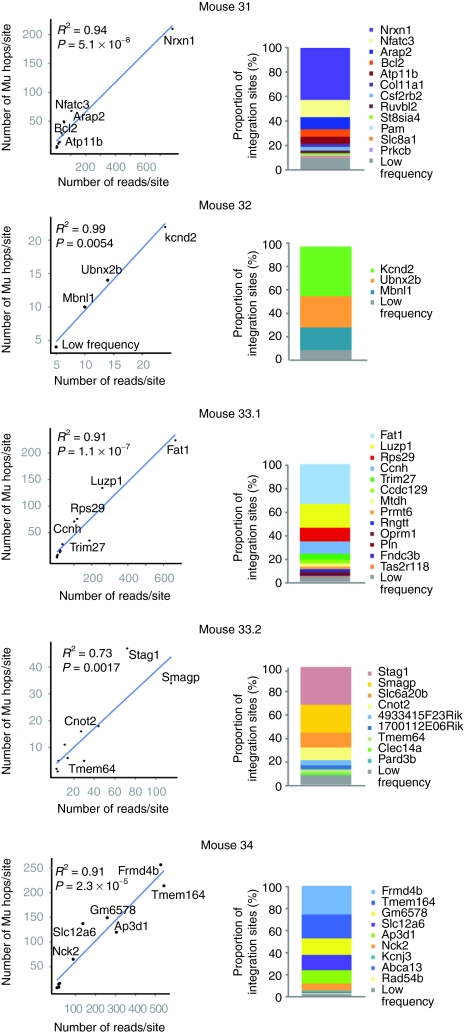
Use of Mu transposase-mediated linker insertion rather than restriction enzyme digestion allowed us to assess the relative abundances of different integration sites in post-transplantation bone marrow samples. Figure 4 shows analysis of integration site abundance from uncultured bone marrow from each treated mouse. Two approaches could be used to estimate site abundance. One method takes advantage of the fact that vector integration sites from expanded clones are more abundant targets for MuA transposition in vitro. Thus, one measure of abundance is to count how many times an integration site is isolated with an independent MuA transposition event (Mu hop). A second approach is to count the number of sequence reads obtained for each integration site. To assess the accuracy of our abundance estimation method, we compared results for the two. The scatter plots in Figure 4 show for each mouse the abundance of each site, as determined by the two methods. Abundant sites are labeled with the name of the RefSeq gene the site lies within or closest to. These plots show good agreement between the two estimation methods (correlation coefficients ranging from R2 = 0.73 to 0.99). The abundance of integration sites in each mouse, as estimated by counting Mu hops per site, is also displayed as a stacked bar graph, with each site labeled with the name of the RefSeq gene is it within or closest to. It can be seen that integration events post-transplantation were oligoclonal, with the apparent abundance of the most common clone varying from 30 to 40% in different mice.
Figure 4.
Relative proportions of integration sites in mouse bone marrow quantified using Mu-mediated recovery of integration sites. To illustrate robust recovery and quantification, the abundance of integration sites in each mouse are presented as scatter plots showing concordance between quantification methods (left column). The y axis shows the number of times an integration site was isolated with an independent transposition event catalyzed by MuA transposase in vitro. The x axis shows the number of sequence reads per integration site. The R2 values indicate the correlations between the two quantification methods for each mouse. Abundant integration sites are labeled with the name of the nearest RefSeq gene. Site abundance is also displayed for each mouse as a stacked bar graph (right column) showing the relative proportions of integration sites recovered (quantified by independent Mu-mediated integration events). Sites recovered fewer than 10 times were combined into the low frequency group.
Integration is not enriched near genes associated with growth control or oncogenesis
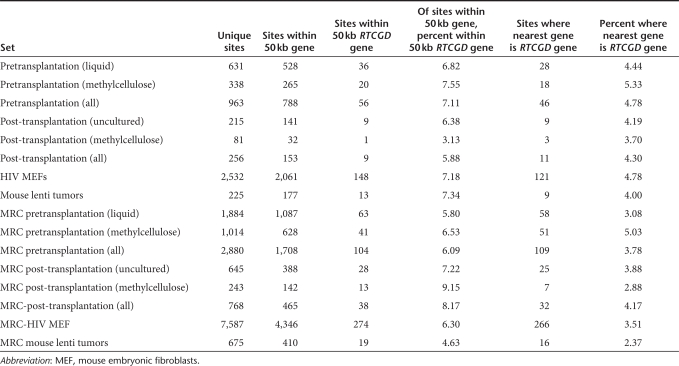
We next investigated the functional categories of the genes close to integration sites. Insertional oncogenesis by retroviruses most commonly results from activation of proto-oncogenes or, less commonly, inactivation of tumor-suppressors. Thus, cells harboring integration sites close to growth-control genes can potentially have a selective advantage and become enriched in vivo. We therefore asked whether integration sites post-transplantation showed evidence of such enrichment near genes associated with cell growth. Table 2 shows the proportion of unique integration sites that lie within 50 kb of a cancer-related gene, as defined by studies of insertional activation in mice (the RTCGD http://rtcgd.ncifcrf.gov/). Integration sites from all five mice were pooled in this analysis. The proportion of sites in each data set is compared to the proportion pretransplantation, and also to matched random control sites in the genome. No statistically significant differences between sets, experimental or control, were found by the Fisher's exact test for proximity to cancer-related genes. We also repeated these analyses with an extended list of cancer-related genes (the allOnco data set from http://microb230.med.upenn.edu/protocols/cancergenes.html). This list is a compilation of several gene lists of cancer-associated genes from diverse vertebrates, in which all genes in any organism were mapped to their murine homologs (see Materials and Methods section). Results with this expanded list also did not show any statistically significant differences between sets (data not shown).
Table 2. Integration site frequency in the vicinity of oncogenes.
A related question centered on whether the proportion of sites in each dataset where the closest gene was an RTCGD gene was increased after growth of cells in mice. These values also did not differ between groups, or from random (Table 2). Thus, we found no evidence of preferential integration near cancer-related genes compared with random, or accumulation of integration sites near cancer-related genes following transplantation and growth in mice. This held true both for uncultured cells, which likely contain repopulating and differentiated cells, and for cells subjected to methylcellulose culture, which selects for progenitor cells.
The above analysis did not take into account the frequency of recovery of different integration sites. An early indication of insertional activation could be the overrepresentation of integration sites close to genes involved in cancer or growth control, so we asked whether there was a correlation between the frequency of recovery of a particular site and its proximity to genes in the RTCGD. Integration sites from each set were grouped into three bins of increasing recovery frequency, then the proportion of sites in each bin that fell within 50 kb of an RTCGD gene end calculated. No significant correlations between frequency of recovery and proximity to cancer genes were found in any data set (data not shown). Performing this analysis on the 15 most abundant sites across all mice also did not show any significant increase in the frequency of sites within 50 kb of an RTCGD gene relative to less abundant sites. Thus, these data provide no evidence of clonal expansion of cells bearing integration sites near cancer-related or growth-control genes.
In the human β-Thal trial5 outgrowth was detected of an integration site in the HMGA2 gene. This was in the sense orientation in the third intron of the gene, upstream of miRNA-binding sites in the 3′UTR, leading to 3′ end substitution in the HMGA2 mRNA and potential release from post-transcriptional repression by miRNA binding. We thus examined the 15 most abundant sites across all mice, as determined by Mu transposition counts, looking for outgrowth of integration sites lying in the sense orientation in the intron of a gene with predicted miRNA binding in the 3′ UTR. Seven of the 15 integration sites (47%) met these criteria. By comparison, of all 256 unique post-transplantation sites, 47 (18%) were in the sense orientation in an intron and of 963 unique pretransplantation sites, 311 were (32%). Statistical analysis of the frequency of integration within an intron in the sense orientation showed that integration in such a way was more frequent in the 15 most abundant integration sites post-transplantation than in all post-transplantation or pretransplantation sites. However, none of the genes in which these seven sites lie has been associated with oncogenesis. We were thus unable to find any strong evidence associating clonal expansion with vector integration in the sense orientation within a growth-control gene subject to miRNA regulation.
Integration sites after growth of transduced cells in vivo or in methylcellulose culture are less frequently found within transcription units
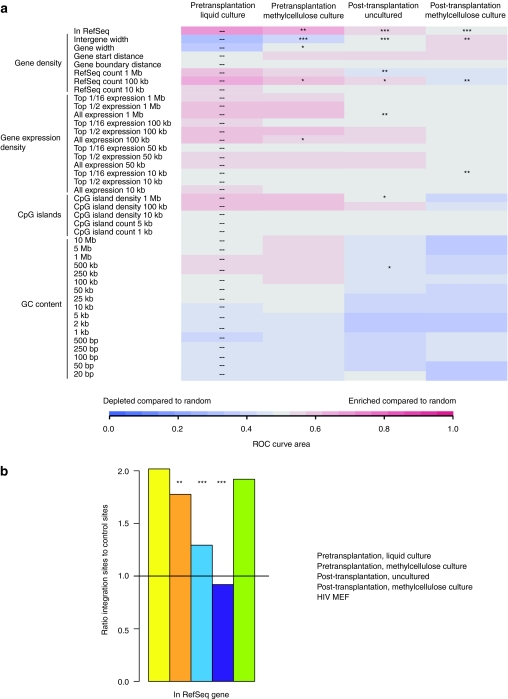
Lentiviral vectors show a preference for integration in particular genomic features, such as the bodies of transcription units and gene-rich regions of the genome.14,17,18,28 Figure 5a shows a heatmap representing the genomic distribution of unique integration sites in four types of samples—pretransplantation and post-transplantation, each with and without methylcellulose culture. For this analysis, unique integration sites from all mice were pooled within a sample type. For each genomic feature, favoring or disfavoring of integration in the feature compared to random is represented by red or blue coloring, respectively. The integration site distribution before transplantation was consistent with previously published studies, with favored targeting of transcription units and regions with high gene density, narrow genes, and high CpG island density.17,18,28 However, post-transplantation and particularly following methylcellulose culture these trends weakened and in some cases reversed (asterisks represent statistically significant deviations from the pretransplantation liquid culture dataset).
Figure 5.
Distribution of unique integration sites with respect to genomic features. (a) Heat map illustrating genomic distribution of unique integration sites. Favoring or disfavoring of a genomic feature within a window around integration sites in each data set is represented as a colored tile. The color is determined by receiver operating characteristic (ROC) curve area comparing the density of the feature near experimental sites and matched random control sites.28 See Materials and Methods section for explanation of genomic features. The P value for the comparison with pretransplantation liquid culture, determined by a logistic regression method that respects the pairing in the data (c-logit), is overlaid on the heatmap tile (*P < 0.05; **P < 0.01; ***P < 0.001). (b) Integration in transcription units. The proportion of unique integration sites in each data set within transcription units are shown, normalized to matched random control sites (indicated by the horizontal line). Significant differences from pretransplantation (liquid culture) sites is denoted by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001).
Figure 5b shows a graphical representation of the frequency of integration in transcription units in each set. It can be seen that while integration pretransplantation is significantly enriched in RefSeq transcription units, cell growth both in mice and in methylcellulose decreased the preference for provirus accumulation in transcription units. 67.7% of integration sites pretransplantation were found in transcription units. In pretransplantation cells cultured in methylcellulose, 64.8% of sites were in transcription units (P = 0.0097 for the difference between the two). Integration in post-transplantation cells was 47.9% in genes (P < 0.0001 for the difference with pretransplantation cells) and in post-transplantation cells grown in methylcellulose culture 27.7% (P < 0.0001 for the difference with pretransplantation cells). Integration in transcription units is thus below the level expected by chance after growth in mice and culture in methylcellulose, suggestive of selection against cells with integrated proviruses disrupting transcription units. We therefore asked whether any bias in the orientation of the provirus relative to transcription or integration in introns versus exons could be seen, since proviruses in different orientations may have different effects on host gene activity. No statistically significant effects were seen (Supplementary Table S2). We thus conclude that provirus accumulation within transcription units was lower after prolonged growth, but that this was not associated with a bias in targeting introns versus exons or provirus orientation relative to the host transcription unit.
Frequent loss of vector insulator elements
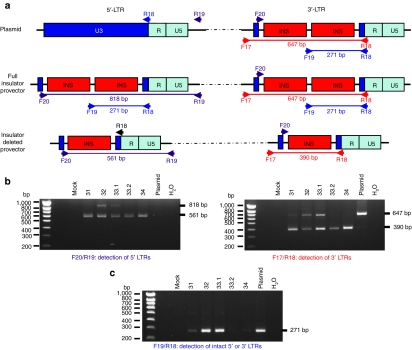
The LG vector used contained two copies of the 250 bp cHS4 chromatin insulator in U3, included to reduce the chances of insertional activation (see Figure 1). In the human patient treated with this vector, analysis of the vector integrated in the dominant clone showed that only one 250-bp insulator core was retained.5 We performed PCR across the insulator regions to determine whether in transduced mice, as in the treated patient, recombination of the insulator sequences had occurred, leading to loss of one of the copies. Figure 6 shows that in all mice a single copy of the insulator is the major form detected at both the 5′ and 3′ LTRs, and in mice 31, 33.2, and 34, the double copy form is barely detectable. Thus, the insulator was mostly lost from the vector.
Figure 6.
Analysis of the number of insulator sequences in vector long-terminal repeats (LTRs) after transduction. (a) Diagram of the 5′ and 3′-LTRs, indicating the sizes expected for amplification products with one or two insulators. Diagrams show (i) the plasmid used to produce the vector, (ii) the vector after reverse transcription and integration into the chromatin, and (iii) the vector structure after recombination between core insulator sequences. Recombination leads to the deletion of 257 bp. Primers used for PCR are indicated; R19 and F17 specifically amplify 5′ and 3′-LTR regions, respectively. F19 recognizes sequences located between the two core insulators and amplify LTRs only if two insulators are present. (b) Amplification of 5′- (left) and 3′-LTRs (right) from mouse bone marrow DNA with specific primers. In mice 31, 33.2, and 34 most of the PCR product corresponds to amplification of 5′- and 3′-LTRs with only one insulator. The smallest DNA fragments were purified an sequenced, confirming that recombination deleted 257 bp (data not shown). (c) Amplification of DNA with primers specifically amplifying intact double core insulator.
Discussion
Trials of γ-retroviral vectors in humans have led to adverse clinical events associated with clonal enrichment of integration sites near cancer-associated genes, but the three human trials of lentiviral gene therapy have not led to clinical adverse events to date. In a recent trial involving transduction of hematopoietic stem cells for the treatment of β-Thal major, relative clonal dominance of cells containing a site in the proto-oncogene HMGA2 was observed,5 though the patient remains healthy. Here, we present a study of treatment of a mouse model of β-Thal with the lentiviral vector used in the β-Thal clinical trial. The lentiviral vector used here differs from vectors previously studied in small animal models by addition of transcriptional insulator elements, and was produced under stringent cGMP conditions. Another distinction of the present study is the use of busulfan in place of irradiation to deplete endogenous bone marrow stem cells, also as used in our clinical trial. Although the methods used in the murine study paralleled those used in the clinical trial, no evidence for vector driving of cellular proliferation associated with vector integration in growth-control genes was detected.
The level of transduction achieved in our study resulted in the expression of the modified human hemoglobin in the majority of the circulating red blood cells at a level sufficient to ameliorate the erythroid abnormalities associated with β-Thal. The transduction efficiency was around 0.7 LG vector copies per cell (according to real-time PCR analysis of the CFU-Cs), which was predictive of the average vector copy number in the donor cells of blood and bone marrow up to 9.2 months after bone marrow transplantation. The β-Thal recipient mice varied in their extent of donor cell engraftment, with levels of chimerism as low as 13% in one mouse. Previous studies have shown that the level of donor-type chimerism is dependent on both the dose of transplanted bone marrow and the degree of ablation of competing endogenous stem cells by the conditioning regimen in the recipient.29 Thus a dose of 4 × 105 transplanted cells per mouse appears to represent a critical threshold at which significant competitive repopulation may arise from endogenous stem cells that survived busulfan treatment.
In spite of the low level of engraftment seen in one β-Thal mouse, the percent of red blood cells expressing human hemoglobin remained high, at 85%, with significant resolution of the disease. The ability of a relatively small proportion of gene-corrected stem cells to produce a therapeutic effect is in keeping with mixed chimerism experiments in other murine β-Thal studies30,31 as well as in models of SCA.32 In these models, preferential enrichment of erythrocyte over leukocyte chimerism is attributed to both the survival advantage of normal red blood cells and more effective late erythropoiesis. Mixed stable chimerism is also found in β-Thal patients receiving allogeneic stem cell transplants, where relatively low levels of donor leukocyte engraftment (10–30%) can be associated with amplification of the donor erythroid compartment, rendering patients transfusion independent.33,34
Two insulator elements were included in the vector LTRs with the goal of minimizing effects of vector transcriptional signals on adjoining cellular genes. However, characterization of integrated vectors showed that most contained only single copies of the insulators. Reverse transcription is known to result in efficient recombination between directly repeated sequences, providing a likely mechanism for insulator loss. It will be useful going forward to redesign the insulator system to minimize the potential for recombinational loss.
Integration sites in the bone marrow were oligoclonal, but the genes closest to integration sites did not show evidence of enrichment in growth-related genes relative to sites in pretransplantation samples. No enrichment of integration sites within 50 kb of proto-oncogenes was found post-transplantation, and no relationship was seen between the proximity of integration sites to oncogenes and site abundance. Thus the oligoclonal nature of reconstitution seen is likely a consequence of the biology of hematopoietic reconstitution in mice,35 and not a consequence of specific integrated vectors driving cellular proliferation. A rough calculation of the expected numbers of transduced stem cells is consistent with this idea. A total of 400,000 bone marrow cells were transplanted into each mouse. Following treatment with 5-fluorouracil, mouse bone marrow is estimated to contain at least 1 in 10,000 stem cells (see ref. 36 and references therein). In the transduction conducted in this study, an average of 70% of cells were transduced, so recovery of ~30 clones would be expected on average per mouse. The numbers of clones per mouse detected as unique integration sites was in the same range (31–69 unique clones per mouse), though this number is a lower bound due to probable incomplete recovery of the integration site population. Thus, the number of clones predicted and the number counted by vector marking were of the same order of magnitude, emphasizing that vector-induced clonal skewing is not needed to explain the observed oligoclonal reconstitution.
When initiating the integration site analysis, one question was whether we would see in mice the dominance of an integration site similar to that found in HMGA2 in the human trial, where integration of the LG vector was associated with 3′ end substitution to generate chimeric HMGA2 mRNAs and also an increase in the rate of HMGA2 transcription initiation.5 The 3′ end substitution removed a miRNA-binding site in the HMGA2 3′ UTR that promotes degradation of the message. In the mice studied, there were no examples of HMGA2 integration, though only five mice were surveyed.
However, there are other examples of insertional activation by 3′ end substitution in the literature, raising the question of whether such genes might be targeted here. Pim1 and Gfi1 are oncogenes activated in murine T-cell lymphomas by substitution of their normal 3′UTRs, which contain miRNA-binding sites.37,38 Retroviral integration downstream of the oncogenes Fgf3 (int-2)39 and c-myc40 has also been associated with oncogenic transformation. No enrichment of integration sites near these genes was seen here. The closest site to Pim1 was 80,427 bp upstream of the 5′ end in an intergenic region, the closest to Fgf3 was over 3 Mb upstream and lying within another gene, the closest to c-myc was over 10 Mb upstream in an intergenic region. Of the 15 most abundant sites recovered from the five mice, none were strong candidates for activation and clonal skewing by 3′ end substitution of known growth-control genes. Overall, the results in the five mice did not detectably recapitulate the clonal skewing associated with insertion in the HMGA2 third intron seen in the human β-Thal trial. Indeed, previous studies have argued that cell-intrinsic factors can contribute to the development of clonal dominance following lentiviral vector insertion, without any bias for integration near proto-oncogenes.35 The difference between the human and mouse studies of the same LG vector also illustrate potential limitations of mouse models in predicting the consequences of lentiviral gene therapy in preclinical tests.41
We found that growth of transduced cells in mice or, to a greater extent, in methylcellulose, was associated with a decrease in the favoring of integration in transcription units. We carried out a meta-analysis of previously published integration site datasets that have compared the proportion of lentiviral integration sites in transcription units pretransplantation and post-transplantation into mice or humans. Shown in Supplementary Table S3 are data from three published studies where cells were transduced by a lentiviral vector and transplanted into mice or humans, and vector integration sites were analyzed before and after transplantation. We found that two of the three studies (one to treat Wiskott–Aldrich syndrome in mice,42 and one to treat HIV in humans20) showed a decrease in the frequency of integration in transcription units after transplantation, though only one difference was statistically significant (Supplementary Table S3). Evidence of selection against other retroviral elements integrated in genes can also be found. Reduced preference for integration in transcription units has been observed in a clinical trial to treat ADA-SCID with a γ-retroviral vector.43 Similarly, HTLV integration sites from chronically infected individuals are less frequently found in genes compared with ex vivo infections.44 It has also been reported that evolutionarily older endogenous retroviruses are found less frequently in transcription units, in both mouse and human,45,46 likely reflecting selection against cells bearing integrated proviruses in transcription units. A potential explanation for our results would be that integration of lentiviral vectors into transcription units disrupted expression of the gene, and that this more often leads to a fitness cost for that cell than a fitness advantage. The effect could be at the transcriptional level, involving a change in mRNA levels, or via production of abnormal proteins. Data from previous studies of the effect of lentiviral integration on the transcription level of nearby genes support this explanation. Maruggi et al.47 showed that lentiviral vectors with several different promoter elements caused transcriptional deregulation of genes within 200 kb of integration events. The frequency of this effect for lentiviral vectors was lower than for γ-retroviral, and the nature of the internal promoter appeared to be important. Hargrove et al. studied the effect of integration by a lentiviral vector similar to that used in the present study, expressing γ-globin and enhancer elements from the β-globin locus control region.48 They reported altered gene expression, both up- and downregulation, in 28% of clones, with 11% being deregulated in a gene within 600 kb of an integration site. Taken together, our data and that of other groups are consistent with the idea that integration of lentiviral vectors may perturb gene expression and cause a growth disadvantage that becomes evident after long-term proliferation.
In summary, our study of reconstitution in mice showed that gene transfer with the LG vector was effective and not associated with detectable vector driven clonal skewing. Oligoclonal reconstitution was observed, but integration site analysis indicated that clonal dominance was probably not a result of insertional activation. Unexpectedly, our observations are also consistent with the idea that integration of the lentiviral vector caused a slight growth disadvantage in some cells containing integration sites in transcription units.
Materials and Methods
Lentiviral vector design and production. The version of the lentiviral vector containing a mutated human β-globin gene used in this study (Figure 1a) is based on the design previously detailed and proven successful for correcting both β-Thal and sickle cell disease in lethally irradiated mice49,50 but with the incorporation of the cHS4 chromatin insulator in the right self-inactivating LTR. Clinical-grade vesicular stomatitis virus glycoprotein pseudotyped lentiviral supernatant was produced in a cGMP facility (Indiana University Vector Production Facility, Indianapolis, IN) by transient transfection of HEK293T cells with the 5—plasmid system (LG, HPV 275—gag-pol, ψN 15—vsvG env, p633—rev, HPV601—tat) by calcium phosphate coprecipitation in Dulbecco's modified Eagle's media supplemented with 5% fetal bovine serum (Invitrogen, Carlsbad, CA) followed by harvest in serum-free media (OptiPRO; Invitrogen). Concentrated virus was then obtained after ultrafiltration and diafiltration into X-Vivo 20 medium (Cambrex, East Rutherford, NJ). An infectious titer of 11 × 107 transducing units/ml was determined by transducing NIH 3T3 mouse fibroblasts with serial dilutions of the viral supernatant in the presence of polybrene (8 µg/ml) and, after 7 days in culture, the total DNA was prepared (Qiagen, Valencia, CA) and integrated copies were quantified by TaqMan assay using GAG primers (Supplementary Table S4) and normalized with mouse β-actin primers (Applied Biosystems, Courtaboeuf, France).
Animals. All animal experiments were approved by the ethical committee of Ile-de-France and were in accordance with the French decree 2001-464. We employed a mouse model for β-Thal intermedia (Hbbth-1/th-1), referred to as β-Thal mice, which bear a homozygous deletion of the mouse βmaj-globin gene and manifest clinical and biological features similar to those observed in human β-Thal. β-Thal mice, originally provided by F. Constantini (Columbia University, New York, NY), were backcrossed in C57BL/6J mice and maintained in our animal facility. Normal C57BL/6J mice were purchased from Charles River Laboratory (L'Abresle, France).
Bone marrow transduction and transplantation. At 3 months of age, bone marrow cells from female β-Thal mice injected intravenously 4 days previously with 5-fluorouracil (150 mg/kg; Sigma-Aldrich, Lyon, France) were harvested and nucleated cells isolated on a discontinuous density gradient (Lympholyte M; Cedarlane, Burlington, Ontario, Canada) and resuspended at a cell concentration of 3 × 106 cell/ml in X-Vivo 20 medium alone (mock) or in 100% LG supernatant (transduced at final multiplicity of infection of 30) supplemented with 10 ng/ml recombinant mouse stem cell factor (PeproTech, Neuilly sur Seine, France), 100 ng/ml recombinant mouse thrombopoietin (rmTPO; PeproTech) and 8 µg/ml protamine sulfate (Sigma-Aldrich, Lyon, France). Transductions were performed in nontissue culture plates that had been treated with 100 µg/ml Retronectin (Takara Shuzo, Otsu, Japan) 16 hours at 4 °C and rinsed according to the manufacturer's recommendations. After 24 hours incubation at 37 °C with 5% CO2, cells were washed and resuspended in phosphate-buffered saline and injected intravenously at a dose of 4 × 105 cells via the retro-orbital sinus into anesthetized 6-month-old recipient male β-Thal mice that had been pretreated with four daily intraperitoneal doses of 20 mg/kg Busulfan (Busilvex; Pierre Fabre, Castres, France). To assess the effect of cell culture on hematological engraftment and recovery, a third group of busulfan pretreated male β-Thal mice was transplanted with 4 × 105 bone marrow cells harvested directly from 5-FU pretreated female β-Thal donors (fresh). Figure 1 describes the treatment and transplantation protocol.
Post-transplant monitoring. Blood samples (50 µl obtained by retro-orbital puncture with heparinized capillary tubes) were analyzed for hemoglobin and blood cell counts using an automated cell counter (Cell Dyn 3700; Abbott Diagnostic, Rungis, France) and hematocrit values were obtained by standard manual centrifugation method. Reticulocytes were determined after Thiazole orange staining and flow cytometric analysis. Erythropoietin (Epo) concentration in blood plasma was determined using the Epo monoclonal enzyme immuno-assay kit (Medac, Wedel, Germany) according to manufacturer instructions.
High-performance liquid chromatography. Percentages of human hemoglobin were determined by high-performance liquid chromatography with a Prominence chromatograph (Shimadzu, Champs sur Marne, France): DGU-20A3 online degasser connected with two LC-20AD solvent delivery units, in series with a CBM-20A system controller, a SIL-20AC HT autosampler, a CTO-20AC column oven and a SPD-20AD UV-VIS detector. Automated sample injections were performed with SIL-20AC HT autosampler: blood cells from 10 µl blood were washed with 1 ml 0.9% NaCl and sedimented by centrifugation at 1,000g for 10 minutes at room temperature. Packed red cells were lysed by addition of 200 µl of distilled water and centrifuged at 15,000g for 10 minutes at 4 °C. Supernatants were transferred in new tubes and frozen in liquid nitrogen. Hemoglobin concentrations were determined by converting total hemoglobin to cyano derivatives with the Drabkin's solution (Sigma-Aldrich, Lyon, France) and reading the absorbance of cyano derivatives at 546 nm. Mouse and human hemoglobins were separated by cation-exchange high-performance liquid chromatography. Twenty microliter of hemolysate (0.01 g Hb/dl) were injected onto a 100 × 2.1 mm, 5 µm-diameter particle size, PolyCAT A column (PolyLC, Columbia, MD). Elution was achieved with a linear gradient of two Tris buffers (buffer A: Tris 40 mmol/l, KCN 3 mmol/l. Adjust pH at 6.5 with acetic acid—buffer B: Tris 40 mmol/l, KCN 3 mmol/l, NaCl 200 mmol/l. Adjust pH at 6.5 with acetic acid) of different ionic strength at a flow rate of 0.4 ml/minute at room temperature. The gradient was 0–1 minute, 7% B; 1–5 minutes, 20% B; 8–12 minutes, 70% B; 13 minutes, 7% B. The detection wavelength was 420 nm. Data acquisition was performed with LC Solution software from Shimadzu.
CFU-C assay. Bone marrow cells in 0.5 ml α-MEM (Cambrex, Emerainville, France) were plated in triplicate in 2.8 ml methyl cellulose medium (MethoCult M3234; Stem Cell Technologies, Grenoble, France) supplemented with 50 ng/ml recombinant mouse stem cell factor, 10 ng/ml rmIL-3, 10 ng/ml rmIL-6 (Peprotech, Neuilly sur Seine, France), and 3 U/ml erythropoietin (Epoietin beta; Roche Pharmaceuticals, Neuilly sur Seine, France). Colonies were counted on day 5 on an inverted microscope. Following enumeration, all colonies were harvested at day 7 for evaluation of transgene copy number from individual plates by scraping the methylcellulose medium into at least four volumes of phosphate-buffered saline.
Determination of copy number and donor chimerism. Genomic DNA was extracted with the Nucleospin Blood kit (Macherey Nagel, Hoerdt, France). The fraction of donor male cells among leukocytes and the vector copy number in bone marrow, peripheral blood, and methylcellulose progenitors were determined by quantitative PCR. Results were compared with those for serial dilutions of genomic DNA from male and female cells and of genomic DNA from a mouse cell line containing one copy of an integrated vector per haploid genome. Real-time PCR were performed for 40 cycles with denaturation at 94 °C for 15 seconds and annealing at 60 °C for 1 minute following an activation step of 10 minutes at 95 °C using the 7300 ABI Prism Detection system (Applied Biosystems, Courtaboeuf, France) and a 2× quantitative PCR MasterMix containing ROX (Eurogentec, Liege, Belgium). Primers and probes are described in Supplementary Table S4.
Flow cytometry. The relative percentages of leukocyte subsets in blood, spleen, thymus, and bone marrow cells were determined by antibody staining and flow cytometry. Erythrocytes in blood samples were removed by treating with ACK lysis buffer at room temperature for 10 minutes. Washing and dilutions were performed in labeling buffer containing phosphate-buffered saline with 0.1% bovine serum albumin (Roche Diagnostics, Meylan, France) and 2 mmol/l EDTA. Samples were preincubated 10 minutes with rat immunoglobulin G (20 µg/ml; Sigma-Aldrich) in labeling buffer. All antibodies were obtained from eBioscience (Paris, France) and added at 0.2–2 µg/ml in 50 µl labeling buffer. Samples were incubated 30 minutes at 4 °C and then washed twice. The following antibodies were used: phycoerythrin anti-mouse CD3e, GR-1, CD71 and Sca-1, fluorescein isothiocyanate anti-mouse B220 and Mac-1, allophycocyanin (APC) anti-mouse CD45.2, Ter119 and APC-Alexa Fluor 750 (APC-AF750) c-kit (clone ACK2). Biotinylated lineage (Lin) antibodies consisting of B220, Mac-1, Gr-1, Ter119, CD8a, CD4, and CD5 was used followed by streptavidin–Alexa Fluor 488 (Invitrogen) staining. Corresponding phycoerythrin-, fluorescein isothiocyanate-, APC- and APC-AF750-conjugated and biotinylated isotype control antibodies were used to assess negatively stained cells. Acquisition and analysis were performed on a Becton–Dickinson LSR machine with CellQuest software. Only viable cells were included in the data analysis as determined by 7-amino actinomycin D exclusion.
Pathology. On termination of the experiment at 9.2 months after bone marrow transplantation, gross autopsy was performed and the spleen, thymus, lung and liver weighed and along with stomach and ribs, tissue samples were placed in neutral-buffered formalin for fixation, processed and embedded in paraffin blocks for subsequent histopathological evaluation of hematoxylin and eosin stained slides by light microscopy (Dr Lawrence Fisher, Regulatory & Toxicology Solutions, Clearwater, FL). Bones were decalcified before embedding and Mallory's iron staining was additionally applied to liver and spleen sections.
Isolation of integration site sequences. Integration site isolation was performed by ligation-mediated PCR essentially as described previously.3,17,18 Each DNA sample (420–1,000 ng) was digested with MseI and NlaIII separately. Linkers were ligated to the digested samples and samples treated with ApoI to limit amplification of the internal vector fragment downstream of the 5′ LTR. Samples were then amplified by nested PCR and sequenced by 454/Roche pyrosequencing at the University of Pennsylvania DNA sequencing center. In order to sequence all amplicons in one sequencing run, PCR primers contained 8 bp barcodes between the 454 sequencing primer and the region complementary to the LTR. Primer sequences are shown in Supplementary Table S5.
Mu-mediated recovery of integration sites. Mu-mediated recovery of integration sites was performed by substituting MuA transposase for restriction enzymes and ligase at the step of PCR linker addition.51 Briefly, BanI digested genomic DNA was incubated with annealed Mu adaptor [Mu(+) and Mu(−); see Supplementary Table S5] and purified MuA transposase (Finnzymes, Espoo, Finland) in reaction buffer (25 mmol/l Tris–HCl pH 8.0; 10 mmol/l MgCl2; 110 mmol/l NaCl; 0.05% Triton X-100; 10% glycerol) at 30 °C for ~4 hours. PCR conditions were the same as with standard ligation-mediated PCR.
Amplification of insulator elements. PCRs were performed on 100 ng genomic DNA or 15 pg plasmid DNA for 30 cycles at 94 °C (15 minutes), 59 °C (30 minutes), 72 °C (60 minutes) with 3% dimethyl sulfoxide for amplification with F20/R19 or 3% formamide for amplification with F17/R18 and F19/R18. Primer sequences are shown in Supplementary Table S6.
Bioinformatic analysis. Integration sites were determined to be authentic if the sequences began within 3 bp of vector LTR ends, had a >98% match to the mouse genome (mm8 draft), and had a unique best hit when aligned to the mouse genome by BLAT. Integration site sequences are available at Genbank (http://www.ncbi.nlm.nih.gov/genbank/) under accession number LIBGSS_012199. Two integration sites were found in sequences barcoded as more than one mouse and deduced to be likely due to crossover between samples during PCR (though transduced cells were pooled before transplantation, providing another candidate explanation). The probable origin of each site was assigned based on sequence abundance and recovery in multiple samples from the same mouse, and excluded from analysis of the other mouse from which it was recovered. For each experimental integration site, three matched random control sites were computationally generated. These sites were matched to the experimental sites in their distance to the nearest MseI or NlaIII site as appropriate.
The RTCGD cancer gene database is available at http://rtcgd.ncifcrf.gov/. The expanded all onco cancer genes list is described at http://microb230.med.upenn.edu/protocols/cancergenes.html. Distance to the nearest gene in Table 2 was calculated by measuring the distance of the integration site to the closest the 5′ end or the 3′ end of a gene and analyzing the single closest gene. In the rare event that a site was equidistant from two gene ends, the upstream gene was chosen.
Enrichment of integration events relative to various genomic features was compared between datasets by Fisher's exact (where stated) or by multiple regression models for integration intensity and a c-logit test for significance, as described in Berry et al.28 Receiver operating characteristic analysis was used to represent the relationship between integration and various genomic features by a single numerical value,28 which was used to generate a colored heat map. Statistical methods for analysis of receiver operating characteristic curves are described in detail in Supplemental Material 3 of ref. 46. Genomic features analyzed in Figure 5 are as follows. “In RefSeq” shows the preference for integration within genes called by the RefSeq gene catalog. “Gene width” shows the relationship with the length of the gene harboring the integration site and integration frequency. “Intergene width” shows the relationship with the length of the interval between genes. Short genes and short intergene lengths are associated with gene-rich regions. “Gene start distance” shows the relationship to the distance to the nearest gene 5′ end; “Gene boundary distance” to the nearest gene 5′ or 3′ end. “RefSeq count” shows the relationship to the number of RefSeq genes within a given window around each integration site (windows shown as 1 Mb, 100 kb etc.). “Expression” quantifies the relationship with gene activity, measured in mouse embryonic fibroblasts using Affymetrix microarrays. Genes were ranked for relative expression and the relationship is shown between integration and the expression level over given windows around each site. “Top 1/2” and “Top 1/16” means only genes in the upper half or sixteenth were scored in each interval, respectively. “CpG count” is calculated analogously to “RefSeq count,” counting the number of CpG islands in the window specified. “CpG density” takes the number of CpG islands in the given window and divides by the number of base pairs for a density measure. “GC content” denotes the percentage G/C residues in the sequence surrounding each integration site, in the window shown. Analysis was carried out in R (http://www.r-project.org).
For the analysis in Figure 4, the closest gene was identified by starting at the integration site and traversing along the genome until a transcription start site was encountered. The nearest start sites in both directions were scored, and the single closest selected.
miRNA predictions were annotated based on three online tools: miRbase (http://microrna.sanger.ac.uk/sequences/), TargetScan (http://www.targetscan.org/mmu_50/) and miRDB (http://mirdb.org/miRDB/index.html).
SUPPLEMENTARY MATERIAL Table S1. Clinical measures of hematological disease correction. Table S2. Integration with respect to transcriptional orientation and exons. Table S3. Frequency of integration in transcription units in published studies. Table S4. Primers and probes used for real-time PCR. Table S5. Primers used for integration site analysis. Table S6. Primers used for insulator PCR.
Acknowledgments
This work was supported by NIH grants AI052845 and AI082020, the French Association against Myopathies (AFM), the French Institute for Health and Medical Research (INSERM) and Bluebird bio. K.R. was supported by NIH training grant AI-07324-17. O.N., M.D., B.G.-L., K.H. and J.D.D. are employees of Bluebird bio. Y.B., P.L. and E.P. have a financial interest in Bluebird bio, although they are not employees thereof. This work was supported by NIH grant HL090921 to P.L.
Supplementary Material
Clinical measures of hematological disease correction.
Integration with respect to transcriptional orientation and exons.
Frequency of integration in transcription units in published studies.
Primers and probes used for real-time PCR.
Primers used for integration site analysis.
Primers used for insulator PCR.
REFERENCES
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P.et al. (2000Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease Science 288669–672. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A.et al. (2002Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning Science 2962410–2413. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U.et al. (2006Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1 Nat Med 12401–409. [DOI] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I.et al. (2009Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy Science 326818–823. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F.et al. (2010Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia Nature 467318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E.et al. (2008Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1 J Clin Invest 1183132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A.et al. (2010Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease Nat Med 16198–204. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C.et al. (2006Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration Nat Biotechnol 24687–696. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M.et al. (2009The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy J Clin Invest 119964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH.et al. (2009Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors Mol Ther 171919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam PI, Higashimoto T, Urbinati F, Modlich U, Nestheide S, Xia P.et al. (2009Genotoxic potential of lineage-specific lentivirus vectors carrying the β-globin locus control region Mol Ther 171929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C.et al. (2008DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer Nucleic Acids Res 36e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B.et al. (2007Hot spots of retroviral integration in human CD34+ hematopoietic cells Blood 1101770–1778. [DOI] [PubMed] [Google Scholar]
- Wang GP, Berry CC, Malani N, Leboulch P, Fischer A, Hacein-Bey-Abina S.et al. (2010Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial Blood 1154356–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC., and, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine BL, Humeau LM, Boyer J, MacGregor RR, Rebello T, Lu X.et al. (2006Gene transfer in humans using a conditionally replicating lentiviral vector Proc Natl Acad Sci USA 10317372–17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G.et al. (2009Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells Mol Ther 17844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imren S, Fabry ME, Westerman KA, Pawliuk R, Tang P, Rosten PM.et al. (2004High-level β-globin expression and preferred intragenic integration after lentiviral transduction of human cord blood stem cells J Clin Invest 114953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P, Arumugam PI, Yee JK., and, Puthenveetil G. Successful correction of the human Cooley's anemia β-thalassemia major phenotype using a lentiviral vector flanked by the chicken hypersensitive site 4 chromatin insulator. Ann N Y Acad Sci. 2005;1054:238–249. doi: 10.1196/annals.1345.030. [DOI] [PubMed] [Google Scholar]
- Puthenveetil G, Scholes J, Carbonell D, Qureshi N, Xia P, Zeng L.et al. (2004Successful correction of the human β-thalassemia major phenotype using a lentiviral vector Blood 1043445–3453. [DOI] [PubMed] [Google Scholar]
- Bank A, Dorazio R., and, Leboulch P. A phase I/II clinical trial of β-globin gene therapy for β-thalassemia. Ann N Y Acad Sci. 2005;1054:308–316. doi: 10.1196/annals.1345.007. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K., and, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I., and, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A.et al. (2009Comprehensive genomic access to vector integration in clinical gene therapy Nat Med 151431–1436. [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down JD., and, Ploemacher RE. Transient and permanent engraftment potential of murine hematopoietic stem cell subsets: differential effects of host conditioning with γ radiation and cytotoxic drugs. Exp Hematol. 1993;21:913–921. [PubMed] [Google Scholar]
- Persons DA, Allay ER, Sabatino DE, Kelly P, Bodine DM., and, Nienhuis AW. Functional requirements for phenotypic correction of murine β-thalassemia: implications for human gene therapy. Blood. 2001;97:3275–3282. doi: 10.1182/blood.v97.10.3275. [DOI] [PubMed] [Google Scholar]
- Roberts C, Kean L, Archer D, Balkan C., and, Hsu LL. Murine and math models for the level of stable mixed chimerism to cure β-thalassemia by nonmyeloablative bone marrow transplantation. Ann N Y Acad Sci. 2005;1054:423–428. doi: 10.1196/annals.1345.061. [DOI] [PubMed] [Google Scholar]
- Kean LS, Manci EA, Perry J, Balkan C, Coley S, Holtzclaw D.et al. (2003Chimerism and cure: hematologic and pathologic correction of murine sickle cell disease Blood 1024582–4593. [DOI] [PubMed] [Google Scholar]
- Li CK, Chik KW, Tsang KS, Pong H, Shing MM., and, Yuen PM. Mixed chimerism after bone marrow transplantation for thalassemia major. Haematologica. 2002;87:781–782. [PubMed] [Google Scholar]
- Lisini D, Zecca M, Giorgiani G, Montagna D, Cristantielli R, Labirio M.et al. (2008Donor/recipient mixed chimerism does not predict graft failure in children with β-thalassemia given an allogeneic cord blood transplant from an HLA-identical sibling Haematologica 931859–1867. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z.et al. (2009Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis Mol Ther 171537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Petek LM., and, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet. 2004;36:767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- Selten G, Cuypers HT., and, Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985;4:1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowska MJ, Dybkaer K, Johnsen HE, Wang B, Wabl M., and, Pedersen FS. Loss of MicroRNA targets in the 3' untranslated region as a mechanism of retroviral insertional activation of growth factor independence 1. J Virol. 2009;83:8051–8061. doi: 10.1128/JVI.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C, Smith R, Brookes S., and, Peters G. Proviral insertions within the int-2 gene can generate multiple anomalous transcripts but leave the protein-coding domain intact. J Virol. 1990;64:784–793. doi: 10.1128/jvi.64.2.784-793.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran LM, Adams JM, Dunn AR., and, Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984;37:113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Sorrentino B. Assessing the risk of T-cell malignancies in mouse models of SCID-X1. Mol Ther. 2010;18:868–870. doi: 10.1038/mt.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani J, Charrier S, Eckenberg R, Saurin W, Danos O, Perea J.et al. (2009Diverse genomic integration of a lentiviral vector developed for the treatment of Wiskott-Aldrich syndrome J Gene Med 11645–654. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A.et al. (2007Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy J Clin Invest 1172233–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekings KN, Leipzig J, Bushman FD, Taylor GP., and, Bangham CR. HTLV-1 integration into transcriptionally active genomic regions is associated with proviral expression and with HAM/TSP. PLoS Pathog. 2008;4:e1000027. doi: 10.1371/journal.ppat.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other momentos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Brady T, Lee YN, Ronen K, Malani N, Berry CC, Bieniasz PD.et al. (2009Integration target site selection by a resurrected human endogenous retrovirus Genes Dev 23633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, Sartori D.et al. (2009Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design Mol Ther 17851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C.et al. (2008Globin lentiviral vector insertions can perturb the expression of endogenous genes in β-thalassemic hematopoietic cells Mol Ther 16525–533. [DOI] [PubMed] [Google Scholar]
- Imren S, Payen E, Westerman KA, Pawliuk R, Fabry ME, Eaves CJ.et al. (2002Permanent and panerythroid correction of murine β thalassemia by multiple lentiviral integration in hematopoietic stem cells Proc Natl Acad Sci USA 9914380–14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawliuk R, Westerman KA, Fabry ME, Payen E, Tighe R, Bouhassira EE.et al. (2001Correction of sickle cell disease in transgenic mouse models by gene therapy Science 2942368–2371. [DOI] [PubMed] [Google Scholar]
- Brady T, Roth SL, Malani N, Wang GP, Berry CC, Leboulch P.et al. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy Nucleic Acids Resin press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical measures of hematological disease correction.
Integration with respect to transcriptional orientation and exons.
Frequency of integration in transcription units in published studies.
Primers and probes used for real-time PCR.
Primers used for integration site analysis.
Primers used for insulator PCR.