Abstract
Previous studies have shown that the packaging RNA (pRNA) of bacteriophage phi29 DNA packaging motor folds into a compact structure, constituting a RNA nanoparticle that can be modularized with functional groups as a nanodelivery system. pRNA nanoparticles can also be self-assembled by the bipartite approach without altering folding property. The present study demonstrated that 2′-F-modified pRNA nanoparticles were readily manufactured through this scalable bipartite strategy, featuring total chemical synthesis and permitting diverse functional modularizations. The RNA nanoparticles were chemically and metabolically stable and demonstrated a favorable pharmacokinetic (PK) profile in mice (half-life (T1/2): 5–10 hours, clearance (Cl): <0.13 l/kg/hour, volume of distribution (Vd): 1.2 l/kg). It did not induce an interferon (IFN) response nor did it induce cytokine production in mice. Repeat intravenous administrations in mice up to 30 mg/kg did not result in any toxicity. Fluorescent folate-pRNA nanoparticles efficiently and specifically bound and internalized to folate receptor (FR)-bearing cancer cells in vitro. It also specifically and dose-dependently targeted to FR+ xenograft tumor in mice with minimal accumulation in normal tissues. This first comprehensive pharmacological study suggests that the pRNA nanoparticle had all the preferred pharmacological features to serve as an efficient nanodelivery platform for broad medical applications.
Introduction
Nanoparticles have been recognized as a preferred delivery approach for diagnostic reporters and pharmaceutics in recent decades. The concept of RNA nanotechnology has appeared for more than a decade1,2,3,4 (for review, see refs. 5,6). The first evidence was presented in 1998 showing that dimeric, trimeric, and hexameric RNA nanoparticles can be assembled through self-assembly of multiple reengineered natural RNA molecules in 1998.1 The field of RNA nanotechnology has emerging and become increasingly popular due to the recognition of its potential in the treatment of cancer, viral infection, and genetics diseases.5
Nanoparticles have several advantages over conventional delivery platforms including:7 (i) improve solubility and facilitate administration; (ii) have favorable pharmacokinetic (PK) profile [reduced renal clearance and increased half-life, T1/2, and exposure, area under the curve (AUC)], enhanced bioavailability (reduced extrasvasation—a smaller volume of distribution, Vd, and enhanced permeation and retention at disease tissues); (iii) can be readily engineered to be specifically self-delivered to the intended disease tissues/cells by decoration with ligands that target disease tissues/cells; and (iv) can readily provide multimodality of different mechanisms of actions in a single particle. All these features enhance its specificity by increasing activity and reducing side effects, and thus also reduce dose and costs.
Many factors influence the utility and effectiveness of a nanodelivery system. First, thermodynamic and chemical stability affect manufacturing, storage, and potency. Second, metabolic stability affects PK and in vivo activity. Third, the other physicochemical properties including size, rigidity, surface charge, and hydrophobicity play key roles in biocompatibility, including serum protein binding, tissue penetration, cellular uptake, clearance, and immunotoxicity. It is believed that the optimal particle size range for nanodelivery is 10–100 nm: large enough to avoid kidney filtration (>10 nm) but small enough to penetrate tissues, access cell surface receptors, and facilitate intracellular trafficking, while also minimizing reticuloendothelial system (RES)-mediated clearance.8 The commonly used lipid and polymer-based nanoparticles have large particle sizes and result in predominant accumulation in the liver, spleen, and lung distributions.8 Fourth, the engineered targeting mechanisms can specifically guide delivery to disease tissues/cells or pathogens. In certain therapeutic applications, cell internalization and intracellular trafficking of the nanoparticle to drug action sites (e.g., cytoplasm or nucleus) are also required. Fifth, the safety of nanoparticles is critical to all nanodelivery systems. For example, the induction of a strong innate immune response and certain organ toxicity (e.g., liver), causing dose-limiting toxicity, is a major obstacle for the majority of the liposomal nanotechnologies. Finally, manufacturability with the consistency of a defined particle size and functional modification is also a major challenge facing many of today's nanotechnology platforms.
Packaging RNA (pRNA), is a 117-nt RNA molecule that constitutes one of the six pRNA subunits (monomers) of the DNA packaging motor of bacteriophage phi29.1,9,10,11 The structures and functions of the pRNA molecule and its domains have been comprehensively characterized during the past decade.12 pRNA folds into a stable and unique secondary/tertiary structure that constitutes a nanoparticle ~11 nm in size.13 Atomic force microscopy studies have also revealed the uniform sizes and shapes of each type of pRNA nanoparticle depending on the specific configurations.14,15,16,17,18 pRNA contains three unique regions: the intermolecular interaction single-stranded right hand loop, left-hand loop, and a 3′/5′ double helix14,19,20,21 (for review, see ref. 12). The two intermolecular interaction regions mediate the interactions among pRNA subunits through interlocking loop interactions, thus facilitating the formation of dimer, trimer, and hexamer.1,2,4,14,16,17,22,23,24 Although the intermolecular domains tolerate little change in sequence, the double helix can accommodate any changes in sequences and lengths without affecting the pRNA structure, as long as double helix is maintained.20,22,25 This enables incorporations of diverse functional groups of targeting mechanisms, therapeutic payloads, and imaging/diagnostic agents (e.g., RNAi trigger, aptamer, ribozyme, and fluorescence/radioisotope reporters) (see refs. 6,8,16,24,26). Using in vitro transcription, several modifications and functional modules have been introduced into the pRNA molecules to demonstrate these utilities in vitro. RNA nanoparticles have several features that make them attractive nanodelivery platforms:4,16,17,23,24,26,27 (i) diverse and stable 2D, 3D, and 4D structures, and (ii) thus diverse biochemical/biological functions; (iii) metabolic stability achieved by modification; (iv) complete biocompatibility, biodegradability, and noninduction of antibodies; and (v) modular design and flexible fabrication. pRNA also tolerates 2′-F-modifications while maintaining its structures and functions.18 In addition, we have demonstrated a bipartite synthesis strategy followed by a self-assembly process without affecting the structure and functions known so far,28 suggesting a possibility of a total chemical process for pharmaceutical grade manufacturing. However, due to the extreme flexibility and variability of RNA structure, it is a major technical challenge to predict and design robust RNA nanoscaffolds that are thermodynamically stable and allow diverse modularizations so as to accommodate a variety of applications without affecting the core structure.29,30 As a nanodelivery system, pRNA has yet to be validated in vivo. Currently, one technical hurdle is the lack of a scalable process to generate a sufficient amount for evaluation in animal models. The current studies describe a total synthesis of functionalized pRNA nanoparticles. We demonstrated their favorable physicochemical properties and pharmacological profiles through a series of in vitro and in vivo evaluations. Our data confirm that the pRNA nanoparticle is indeed a robust RNA nanodelivery platform that can be applied broadly in a variety of medical applications.
Results
Chemical synthesis of bipartite 2′-fluoro- pyrimidine-modified pRNA molecules
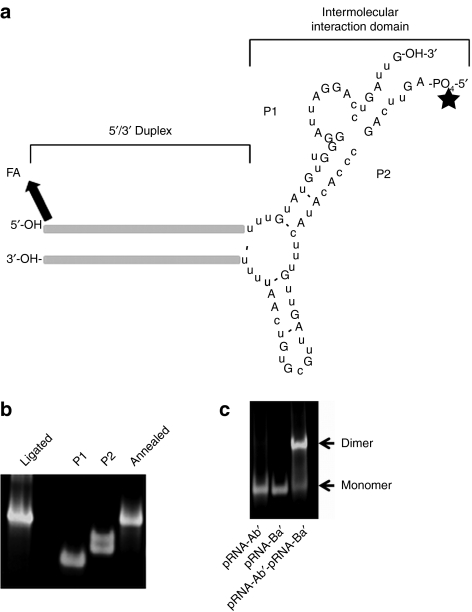
Since in vitro transcription cannot be scaled up for industrial production and cannot support the precise modifications, a scalable process of pRNA nanoparticles production was set out to develop by a total chemical synthesis to generate sufficient RNA for current animal studies and for industrial-scale manufacturing of pharmaceuticals in the future. However, the pRNA molecule (117 nt) is too long for current RNA chemical synthesis approach to produce reasonable yield and purity, particularly with the 2′-F-modification. Therefore, the bipartite strategy was adopted for the total synthesis of pRNA monomer, which was successfully shown using in vitro transcription.28 The full-length pRNA was divided into two pieces, P1 and P2, with a break point at 3′ of nucleotide 55 located within the 53–58 single-stranded loop region of the wild-type pRNA28 (Figure 1a). Each piece was approximately half the length (49–60 nt) of the full-length pRNA, rendering synthesis possible using conventional solid phase automated RNA synthetic chemistry, with a free hydroxyl on the 3′-end of P1 (P1-OH-3′) and 5′-phosphate group on P2 (5′-PO4-P2). This strategy creates two new additional termini to facilitate functionalization (see below).
Figure 1.
The structure and characterization of chemically synthesized bipartite packaging RNA (pRNA) (P1/P2). (a) The structure of a typical monomeric pRNA nanoparticle consisting of P1 and P2 RNAs. Folate and Alexa647 can be optionally conjugated to the 5′-ends of both P1 and P2. (b) Native PAGE analysis of P1, P2, unligated P1/P2, and ligated P1/P2. (c). Dimerization of pRNA-Ab′/pRNA-Ba′, as analyzed by TBM native PAGE as described previously.28
A chemical process to produce pRNA-P1- and -P2 was developed using TBDMS chemistry. 2′-F-pyrimidine phosphoramidites were used instead of the nonmodified monomers along the entire length of the pRNA, or at specified positions as desired. A process has been achieved that can yield P1 and P2 at subgram-scales with >80% purity as measured by high-performance liquid chromatography, which is sufficient for in vivo small animal trials. The chemically synthesized pRNA components (P1/P2), even with functional modules (see below), can self-assemble into a single noncovalently bound two-molecule complex by simple 1:1 molar ratio mixing28 as shown by native gel analysis (Figure 1b). MFold analysis (http://dinamelt.bioinfo.rpi.edu/) confirmed the identical folding pattern of the bipartite and one-piece (full-length) pRNA (data not shown). To confirm the assumed equivalence of the chemically synthesized bipartite molecule to the one-piece molecule synthesized by in vitro transcription, two additional experiments were also performed: ligation and dimerization.
2′-F-P1 and -P2 are efficiently ligated by T4-RNA ligase into the full-length pRNA. Our current synthetic strategy has produced two new termini (P1-OH-3′ and 5′-PO4-P2) that can be readily ligated to form a full-length molecule if desired. In principle, a correctly folded pRNA monomer would present the T4-RNA ligase with an intramolecular ligation step instead of the regular intermolecular ligation between two separate single RNA strands, which in turn should increase the rate of this chemical step during enzyme catalysis.31,32,33 Also the presence of a preformed unligated monomer would result in the lack of circular RNAs, a common side product during RNA ligation using T4-RNA ligase.33,34 This concept was tested by incubating the annealed mixture (P1/P2) with T4-RNA ligase at 37 °C, followed by denaturing PAGE analysis (Figure 1b). The results demonstrated that the two pieces were efficiently ligated almost to completion. The ligated full-length pRNA is indistinguishable from that generated by in vitro transcription as seen by gel electrophoresis. In addition, the absence of any circular side product that can be formed by ligation of the 3′-hydroxyl group and the 5′-phosphate group on P2 further confirms our theory.
The chemically synthesized P1/P2 complex with or without ligation can form a dimer with other pRNA. A correctly folded two-piece pRNA monomer (Ba′) or the 2-F modified pRNA monomer have been shown to form a dimer with a complementary (Ab′) monomer.18,28 To test whether the pRNA monomer generated from the 2-F two-piece folded correctly, a bipartite 2′-F-Sur-pRNAi Ba′ monomer was mixed with a 2′F-pRNA Ab′ monomer at a 1:1 molar ratio, followed by native PAGE analysis. The results demonstrated that the two monomers, whether ligated or nonligated, formed a dimer (Figure 1d). Fluorescent-based capillary electrophoresis, a technique milder and more sensitive than regular slab gel, indicated that the efficiency of 2′-F-pRNA dimer formation was as high as 99% (Supplementary Figure S1).
The 2′-F-pRNA nanoparticle (bipartite) is stable. The stability of a pharmaceutical product is particularly important for manufacturing, storage, transportation, and potency maintenance (expiration date), etc. Thermodynamically, stability is also the foundation for diverse and robust modularizations. Furthermore, thermostability can also affect the behavior of nanoparticles inside the body upon administration. For a functional nucleic acid agent such as pRNA, the most important aspect of its stability lies in the stability of its folding, particularly the two pieces folding together. Melting temperature (Tm) studies reveal that the nonligated 2′-F bipartite pRNA is stable (S. Abdelmawla and Q.-X. Li, manuscript in preparation).
The 2′-F-pRNA is metabolically stable in the presence of serum or RNase A. The purpose of chemical modification is to make the pRNA nanoparticle metabolically stable in biological fluids (e.g., plasma). Our previous data have shown that the 2′-F-modified pRNA generated by in vitro transcription is indeed significantly stabilized in the biological milieus.18 To test the chemically synthesized and self-assembled bipartite 2′-F-pRNA, this particle was incubated with culture media containing 10% and 50% human serum for various periods, followed by denaturing urea-PAGE analysis. We found that a large proportion of the 2′-F-pRNAi remained intact for up to 24 hours (Supplementary Figure S2), in contrast to the nonmodified RNA, which degraded rapidly. The enhanced stability was further confirmed by incubating the 2′-F-pRNA with RNaseA, where the majority of full-length pRNAi remained intact (Supplementary Figure S2). These results demonstrated that the chemically synthesized 2′-F-pRNA is metabolically stable.
The pRNA nanoparticles did not induce IFN response in cultured cells in vitro
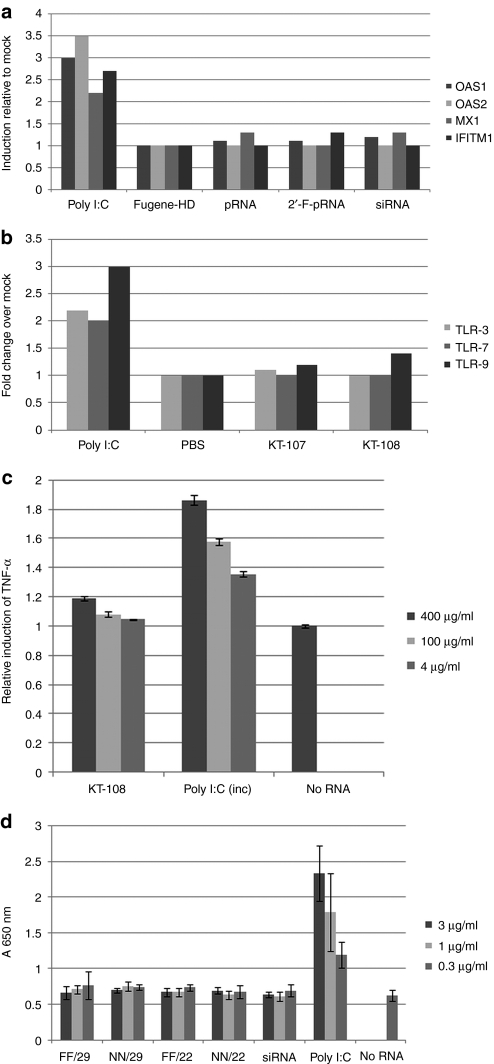
A safety profile is a key aspect of any nanodelivery system for systemic applications. The main concern with a nucleic acid based system is the nonspecific activation of interferon response (IFN) pathways. To this end, the potential induction of IFN responses was set out to be assessed in vitro using several cell-based systems. Because polyinosinic:polycytidylic acid (poly I:C), a synthetic double-stranded RNA, is a ligand to Toll-like receptor (TLR)-3 and a potential inducer of cytokines [IFN, interleukin-6, and tumor necrosis factor-α (TNF-α), etc.],35 it was used as a positive control in all tests. First, the gene expression of several key IFN response genes were examined including OAS1, 2 (2′, 5′-oligoadenylate synthetase 1, 2), MX1, and IFITM136 in KB cells upon transfection with pRNA. Our data showed that the expression of these genes did not increase in KB cells transfected with either the nonmodified or the 2′-F modified pRNAi (Figure 2a). Second, double-stranded RNA has been reported to stimulate the expression of TLRs.37,38 The effect of pRNAi on the expression of TLR-3, -7, and -9 genes was examined then in human peripheral blood mononuclear cell. Again, the results indicated that 2′-F-pRNA did not induce these genes, as compared to the mock treatment, and in contrast to poly I:C (Figure 2b). Third, we tested whether the exposure to the pRNA nanoparticles would cause cytokine induction in mouse macrophages. The RAW-264 mouse macrophage cell line was incubated with pRNA, at the concentrations indicated, for 3 hours at 37 °C. The TNF-α production in the culture media of the treated cells were then analyzed. The results demonstrated that the exposures to pRNA levels upto 100 ng/ml resulted in little induction, in contrast to that of poly I:C that caused dose-dependent induction of TNF-α (Figure 2c). Fourth, HEK-Blue-hTLR3 is a reporter 293-cell line that overexpresses the human TLR3 gene (hTLR3) and secretes reporter embryonic alkaline phosphatase (SEAP), whose gene is under the control of an nuclear factor-κB and activating protein-1 inducible promoter. Upon stimulation of the TLR3 receptor by double-stranded RNA, the nuclear factor-κB and the activating protein-1 pathways are activated, which in turn activates the production and subsequent secretion of SEAP. The SEAP in the culture media provides a colorimetric readout of the activation of TLR3-induced pathways. pRNA was incubated with HEK-Blue-hTLR3 overnight, and then quantified the SEAP production. We found that pRNA did not induce the TLR-3 pathway, in contrast to the poly I:C-induced dose responsive induction of the TLR-3 pathway (Figure 2d). In summary, pRNA nanoparticles do not induce IFN response in multiple cell types at all tested dose levels in vitro. Interestingly, pRNA with a longer double-strand region (29 bp long 5′/3′ duplex) did not cause any IFN response. The reason that pRNA nanoparticles do not induce IFN response is unclear.
Figure 2.
The packaging RNA (pRNA) nanoparticle did not induce an interferon response in vitro. (a) KB cells were transfected with pRNA (50 nmol/l) (2′-F modified versus nonmodified), small interfering RNA (siRNA) (50 nmol/l), and polyinosinic:polycytidylic acid (poly I:C) 1 (µg/ml) using Fugene-HD. The RNAs were harvested 24 hours later for semiquantitative reverse transcriptase (RT)-PCR analysis for the interferon (IFN) responsive genes expressions. (b) Human peripheral blood mononuclear cell (PBMC) were incubated with 50 nmol/l pRNA or 1 µg/ml poly I:C. TLR-3, 7, and 9 gene expression were analyzed as in a. KT-107 and KT-108 are pRNA monomers that differ in the sequence of the 5′/3′ helical region. (c) Production of tumor necrosis factor-α (TNF-α) in the mouse macrophage cell line RAW-647 after incubation with different concentrations of pRNA and poly I:C. Three hours postincubation, the culture media were analyzed for TNF-α by enzyme-linked immunosorbent assay (ELISA). (d) Activation of Toll-like receptor (TLR)-3 pathway by pRNAs were measured using HEK-Blue-hTLR3 reporter cell line. pRNA differing in the length (29 nucleotides or 22 nucleotides) and the extent of modifications (FF, 2′-F modified helical region; NN, nonmodified helical region; all pRNA constructs used had a modified intermolecular interaction domain) in the 5′/3′ region were tested. After 24 hours incubation, the media were analyzed for TLR-3 activation. Poly I:C was used in all these assays as positive control.
Functionalization of pRNA nanoparticles
One important utility of nanoparticles for therapeutic or diagnostic applications is that different functional modules can be incorporated into a single nanoparticle. A robust nanodelivery system requires: (i) the incorporation should not change the structures and functions of either the nanoparticle core or the modules; (ii) the modularization can still be achieved by a simple bottom-up self-assembly process using the same basic building blocks but with different preincorporated functional modules; (iii) a chemical synthesis process can facilitates precisely controlled modulation of the building blocks. We believe that pRNA nanoparticles have these three features, exemplified by the following functional modularizations.
Incorporation of fluorescent molecules at the 5′-end of pRNA-P1 or P2. Fluorescence-labeled nanoparticles can be used as imaging reporters for diagnostic application as well as for other imaging applications in research. RNA can be readily labeled by fluorescence using a commercially available kit, such as the one that uses a fluorescent platinum reagent that can nonspecifically label any G base in a nucleic acid sequence (e.g., the Silencer Labeling Kit from Ambion, Austin, TX). However, this process is hardly stoichiometric, and the number of fluorescent tags per pRNA monomer is difficult to control, which in turn may affect folding, cause physical hindrance in intermolecular interaction, and/or disturb interaction with cell surface receptors. These difficulties can be more readily overcome during automated RNA synthesis by coupling the phosphoramidite of the desired fluorescent tag at a specific position within the RNA molecule. Fluorescent tags were able to be introduced (such as Cy5, Cy3, FITC, and AlexaFluor647) at the 5′-end as well as at the 3′-end or in the middle of the sequence of both P1 and P2. The fluorescence-labeled pRNA nanoparticle can be used in investigating the cellular uptake and intracellular trafficking (see below) and can be used for in vivo targeting for diagnostic applications. Figure 1a shows examples of these fluorescent tags incorporated at the 5′-end of P1 and P2 using this method. These fluorescently labeled P1 or P2 were self-assembled with 5′-folate-P1 or P2 to form a fluorescently labeled folate-pRNA monomeric pRNA nanoparticle to be used for in vitro and in vivo imaging experiments (see below).
Incorporation of folate at the 5′-end of P1 or P2 through direct coupling at the last step of chemical synthesis. Folate receptors (FR) are widely overexpressed in many cancer cell surfaces. Folate ligand binds to FR with high affinity (Kd of 0.1–1 nmol/l).39 Recently, folate has been broadly tested as a targeting mechanism for delivering chemotherapeutic drugs, liposome, small interfering RNA, or imaging reporters to tumors.40,41,42,43,44 Therefore, it is interesting to test the folate-pRNA nanoparticle for specific targeting to FR+ tumors. Previously, we have shown that 5′-folate-AMP can be incorporated into pRNA during in vitro transcription to generate 5′-folate-pRNA. However, this process is not quantitative (usually 10–30%), and ultimately, it is not a scalable procedure. Therefore, the folate was decided to be incorporated during the more reliable, scalable, and less labor-intensive automated chemical synthesis. To introduce a folate moiety at the 5′-end, folate phosphoramidite was first synthesized in collaboration with Berry & Associates (Dexter, MI). The folate phosphoramidite was then coupled into pRNA-P1 at the last step during regular automated chemical synthesis with coupling efficiencies >90% (Figure 1a). This efficiency is difficult to achieve by other methods. Similarly, P2 can also be labeled with folate by a direct coupling method to create 5′-folate-pRNA P2. Again, the folate-pRNA P1 can self-assemble with pRNA-P2 to form the folate-pRNA monomer, similar to the nonfolate-modified pRNA monomer (Figure 1a).
Folate-pRNA monomeric and dimeric nanoparticles target FR+ cancer cells. Fluorescently labeled P1 or P2 and 5′-folate-P2 or P1 self-assembled to form a fluorescently labeled Cy5-pRNA-folate monomeric nanoparticle that was designed to specifically target the folate receptor (FR+)-bearing cancer cells (Figure 1a). This specific targeting was tested by incubating with KB cells followed by confocal microscopy and flow cytometry analysis. The fluorescence confocal microscopy results demonstrated that ~100% KB cells were positive for Cy5 staining (see the Red Cy5 circle shown in Supplementary Figure S3 indicating cell surface staining at 30-minute incubation time), and inside cells (internalized) with longer incubation time (4 hours). This cell targeting is highly specific for the folate-receptor because the observed binding can be effectively competed out by excess free folate (Supplementary Figure S3). There was no binding signal for incubation with the known FR− cell line (data not shown), and no binding for a control pRNA nanoparticle without folatic acid (data not shown). Flow cytometry analysis also showed 100% staining (Supplementary Figure S3) that is competed out by excess folate. These results together confirmed specific in vitro uptake of the chemically synthesized folate-pRNA nanoparticle by the FR+ cancer cells.
Since folate ligands, upon binding to FR, are internalized through receptor-mediated endocytosis, the trafficking of folate-pRNA nanoparticle (red) upon binding to receptors was then investigated. To help assess this, the Cy5-folate-pRNA nanoparticle was incubated with the KB cells transfected with GFP-Rho-B fusion protein plasmid, followed by confocal examination. GFP-Rho-B is expressed exclusively in endosomes (early and mid-stage) and displays a green fluorescence. Four hours postincubation, the Cy5-pRNA nanoparticle was found to be colocalized with GFP-Rho-B in endosomes in KB cells (yellow) as expected (Supplementary Figure S3). Similarly, the Cy5-pRNA-Ba′/folate-pRNA-Ab′ dimeric nanoparticle can also bind and become internalized by FR+ cancer cells (data not shown), as measured by the same method used for the monomer above.
Folate-pRNA nanoparticles specifically target FR+ human xenograft tumors in immunocompromised mice upon systemic exposure
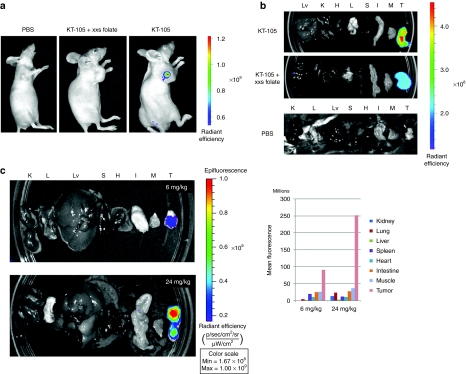
It was hypothesized that a ligand-labeled pRNA nanoparticle would have beneficial tissue distribution and the advantage of self-delivery to the disease tissues of interest upon systemic administration. This assumption was based on the following rationales: first, the particles contained a specific ligand that could target only disease tissue; second, it was relatively small in size and hydrophilic, consequently it would not be trapped in the liver and spleen by RES; third, it was sufficiently large not to be filtered out by or accumulated in the kidneys. To confirm this, the fluorescence-labeled folate-pRNA nanoparticle (AlexaFluor647-folate-pRNA) was systemically delivered to nude mice bearing either HeLa or KB xenograft tumors. The AlexaFluor647 was chosen as a near-infrared fluorescent dye for its brightness, photostability, and far-red emission spectrum that does not overlap with autofluorescence levels in tissue to be conjugated to pRNA (5′-P2; see above) by direct coupling during the last step of P2 synthesis. Twenty-four hours postadministration, the animals were imaged using IVIS Lumina station. Whole-body imaging was first carried out and then ex vivo imaging of major organ in order to fully assess the fluorescence signals. The results demonstrated that the fluorescence was mostly concentrated in the tumors, in contrast to the small amount of fluorescence in other normal mouse tissues (Figure 3a,b). This property can be an important advantage of the pRNA nanoparticle for more efficient delivery with fewer side effects, in contrast to other nanodelivery technologies.43 As a negative control, a group of mice was predosed with 10 mg/kg of folic acid administered intraperitonealy 10 minutes before injection of AlexaFluor647-folate-pRNA through the tail vein. Interestingly, both whole body and organ imaging analyses revealed significantly reduced fluorescence intensity in the tumors (Figure 3a,b), compared to the mice that were not dosed with folic acid. These results clearly demonstrated the specific tumor targeting in vivo by folate-pRNA nanoparticles.
Figure 3.
The folate-packaging RNA (pRNA) nanoparticle targets FR+ tumors upon systemic administration. (a) HeLa xenograft tumor-bearing nude mice were injected with 15 nmol (~24 mg/kg) of KT-105 (Folate-AlexaFluor647-labeled pRNA nanoparticle) through the tail-vein (right). The control mice were injected with either phosphate-buffered saline (PBS) (left) or with folate (intraperitoneal (i.p.), 10 mg/kg) 10 minutes before KT-105 injection (middle). The mice were euthanized 24 hours after injection and whole-body imaging was conducted using IVIS Lumina station. (b) Following whole-body imaging, the mice were dissected and the major organ were isolated for imaging. H, heart; I, intestine; K, kidney; L, lung; Lv, liver; M, muscle; S, spleen; T, tumor. (c) KB xenograft tumor-bearing nude mice were injected with 15 nmol (24 mg/kg) (lower panel) or 3.75 nmol (upper panel) (~6 mg/kg) of KT-105 through the tail vein. And organ were isolated and imaged as described above.
Next, the dose effect on the accumulation of pRNA in the tumor tissue was tested. The results demonstrated that a higher dose level caused increased fluorescence intensity in the tumor. The mean fluorescence increased 2.5-fold when the dose was increased from 6 to 24 mg/kg (Figure 3c). This dose-dependent accumulation of the fluorescent-pRNA nanoparticle suggests that the amount of pRNA nanoparticles delivered to target tissues is controllable by the amount of administrated dose and thus suitable for delivery of therapeutics and diagnostics, an important feature of pharmaceutical entities.
PK analysis of 2′-F-pRNA nanoparticle
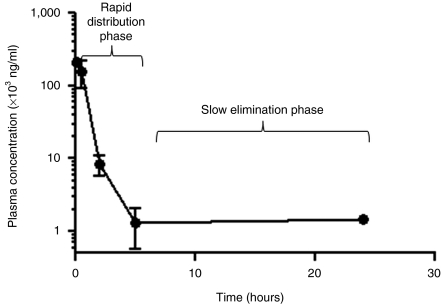
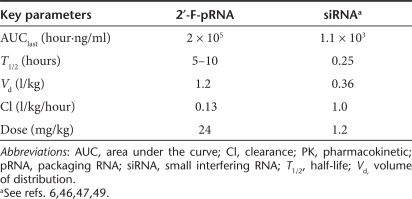
PK parameters describe the fate and/or behavior of a drug in the in vivo environment after administration. Two of the key factors that may affect the PK profile are metabolic stability and clearance (e.g., renal filtration and elimination by RES). The 2′-F modified pRNA nanoparticles have significantly enhanced metabolic stability. The dimensions of the monomeric pRNA nanoparticles are >11 nm, which is above the threshold of kidney filtration. The hydrophilic nature, in addition to the optimum size, reduces RES-mediated clearance. These facts should significantly improve the PK profiles of pRNA nanoparticles by extending T1/2 and reducing the elimination rate. A PK analysis was carried out for the folate-2′-F-pRNA nanoparticle in tumor-bearing mice upon systemic administration (a single intravenous injection), in which P1 was fluorescently labeled (AlexaFluor647) at the 5′-end (Figure 1a). Fluorescence labeling was to ensure sensitive detection and quantification of nanoparticles in plasma sample, as compared to the low sensitivity of UV detection/quantification. The labeled folate-2′-F-pRNA was injected via the tail vein at time 0, followed by blood collection at different time points postadministration (5 minutes, 30 minutes, 2 hours, 5 hours, and 24 hours). The fluorescent nanoparticle concentration in serum was then determined using capillary gel electrophoresis. The choice of using capillary gel electrophoresis was to ensure the quantification of the whole nanoparticle at corresponding elution time, instead of fragmented/degraded pRNA molecules. The plasma concentration–time plot demonstrated a typical two-phase kinetics with an initial rapid distribution phase, followed by a relatively slow elimination phase (Figure 4). The secondary PK parameters were calculated using a noncompartmental model, as shown in Table 1. Our results demonstrated significantly extended terminal-phase T1/2 of 5–10 hours and the increased systemic exposure of AUClast of 2.0 × 105 hour·ng/ml when dosed at 24 mg/kg. The normalized volume of distribution Vd (1.2 l/kg) suggests that a significant fraction seemed to be distributed to peripheral tissues (outside vascular or extravasation), particularly in tumor (as shown from biodistribution results). The relatively small clearance (Cl) value (significantly below the kidney filtration rate45) suggests that the nanoparticles were not efficiently filtered out by the kidneys. In addition, an apparent dose proportionality of exposure was also observed, suggesting no saturation of the elimination pathways (data not shown). This preferred PK of pRNA nanoparticle is consistent with our hypothesis based on the characteristic physicochemical properties (size, hydrophilicity, and stability) and metabolic stability. In summary, pRNA is rapidly distributed and slowly eliminated, in contrast to the rapid elimination of small interfering RNA (reported PK parameters in the literature are shown in Table 1).46,47,48,49,50
Figure 4.
Plasma concentration of packaging RNA (pRNA) nanoparticle upon systemic administration. Tumor-bearing mice (n = 3) were intravenously (i.v.) injected with 15 nmol of KT-105 (Folate-AlexaFluor647 pRNA nanoparticle). Blood was collected at different time points postadministration (5 minutes, 30 minutes, 2 hours, 5 hours, and 24 hours) through the lateral saphenous vein. The serum was isolated and the serum concentration of the nanoparticle was determined using capillary gel electrophoresis (CGE). The semi-log plot of the plasma concentration versus time is shown.
Table 1. Secondary PK parameters of pRNA nanoparticles.
Toxicological evaluation of pRNA nanoparticles
One of the most important criteria for using pRNA nanoparticles as a nanodelivery platform is its safety profile. To this end, two toxicological assessments of pRNA were performed in mice. The rationale for choosing mice was to be consistent with both PK and tumor targeting studies. First, three groups of four immunocompetent C57B/6 mice each were injected via the tail-vein with phosphate-buffered saline (PBS) (group 1), pRNA at 30 mg/kg (group 2), and pRNA at 10 mg/kg (group 3), respectively. Three hours postadministration, the plasma samples from the treated animals were analyzed for mouse TNF-α and interleukin-6 induction by enzyme-linked immunosorbent assay. The results showed no induction of TNF-α and interleukin-6 at the tested dose levels (data not shown).
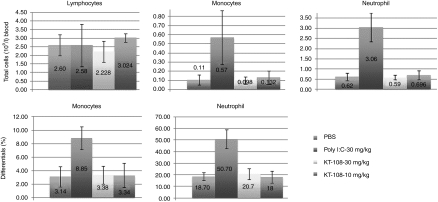
In the second study, four groups of five C57B/6 mice each were repeatedly administered pRNA nanoparticles once every 48 hours by tail vein injection for 1 week (four injections total). Each group was injected with PBS (group 1); 30 mg/kg poly I:C (group 2); 30 mg/kg pRNA (group 3); or 10 mg/kg pRNA (group 4), respectively. The animals were then examined for a variety of toxicological parameters. All the animals survived the entire dosing period without noticeable clinical signs or body weight changes. There were no gross pathology differences between the pRNA and PBS groups. There were slight enlargements in the spleen in the poly I:C treated group as compared to the PBS group (Supplementary Figure S4). There were no enlargements in the liver, kidneys, and other organs for any of the treated groups as compared to the PBS control group (Supplementary Figure S4). Clinical pathology analysis indicated no obvious change in the pRNA-treated groups as compared to the PBS control group, whereas there were statistically significant increases in both the total number of white blood cells, monocytes, and neutrophils in the poly I:C-treated group, along with an increase in the differentials of monocytes and neutrophils (Figure 5) correlating to the enlarged spleen (see above). These changes in the poly I:C-treated animals are consistent with the well-known immunostimulatory nature of poly I:C. Clinical chemistry results demonstrated normal parameters in all the treated animals. There was no change in the liver enzyme levels, correlating with the absence of liver enlargement in these animals (except that the alanine transaminase levels of animal # 1588 were above normal range, but this seems to be unrelated to pRNA because it was the only anomaly among all 10 animals and was without a dose correlation). All these data demonstrated that pRNA is safe, in contrast to poly I:C at the same dose level (30 mg/kg).
Figure 5.
White blood cells counts in mice after packaging RNA (pRNA) nanoparticles injection. Clinical pathology analysis was conducted for the C57B/6 mice upon 1-week repeat intravenous (i.v.)-administrations of pRNA versus polyinosinic:polycytidylic acid (poly I:C) at the indicated doses. Total cell counts (left) and differentials (right) were displayed.
Discussion
RNA nanoparticles have several important features (for review, see ref. 5). First, RNA base pairing is strong (ΔG of UC/AG: −2.4 kcal/mol versus ΔG of dTdC/dAdG −1.5 kcal/mol), enabling a more stable 2° structure. In contrast to DNA, RNA can fold into well-defined and stable 3° structures. For example, kissing–loop interactions can be two- to threefolds more stable than the RNA duplex of the same sequence.29,30 Second, RNA, as an informational molecule, can be fabricated in different sizes, structures, and functions, which are all coded in its sequences. Consequently, there is a potential for rationally engineering of RNA 3° structures that can lead to useful functional modules (e.g., RNAi trigger and self-folding RNA scaffolds mimicking ribozymes and aptamers). Third, since RNA modularity is hierarchically manifested at chemical, structural, and supramolecular levels, a desired RNA nanostructure with multiple substructures and functions, in principle, can be predesigned and constructed by a bottom-up process using programmable basic building blocks. Fourth, scalable total synthetic chemistry procedures are now available for industrial manufacturing and precise modifications of these predesigned building blocks. In addition, two more attributes of RNA molecules make them potentially attractive pharmaceutical products: (i) RNA is completely biocompatible, biodegradable, and nonimmunogenic, and (ii) modified RNA is resistant to degradation by ribonucleases without altering the original structure/functions in many cases.18
Despite these important features in theory, RNA nanotechnology faces practical challenges because of its extreme flexibility and variability. Each RNA nucleotide has 8° of freedom, and there are competitions between the favorable stacking interaction and the nonfavorable conformational entropy. RNA folds in a complex process that involves divalent-ion-mediated electrostatic interactions, conformational entropy, multistate RNA folding kinetics, and noncanonical interactions.29,30 A slight change in primary sequences can result in unpredictable new structures. Therefore, by the currently available tools, although it is theoretically possible to design RNA nanoparticles with the certain above features, it is very challenging to reliably predict how this nanoparticle might behave in reality. However, nature has provided us with RNA nanoscaffolds that are evolutionally selected to possess these features. Bacterial phage phi29 pRNA may be one of such naturally occurring molecules.1,4,6,23 Our present studies indeed demonstrated that chemically synthesized bipartite pRNA is tightly folded and highly stable. It can be bottom-up self-assembled into a functional nanoparticle from fully chemically synthesized basic building blocks. The entire process is highly scalable and its high modularity accommodates potentially broad applications.
A successful nanodelivery system needs to have a suitable pharmacological profile of safety, PKs, and tissue distribution, coupled with a specific disease-targeting mechanism. Nanoparticles with such profiles usually have several key characteristics: (i) optimal particle size (10–100 nm) to avoid kidney or RES-mediated clearance, (ii) biocompatibility derived from optimal physicochemical properties such as surface charge and hydrophobicity and rigidity of the particle,51 (iii) thermodynamic and metabolic stability, (iv) non- or minimal cytotoxicity, and lack of induction of IFN response. From our accumulated data, pRNA nanoparticle has all these characteristics. To our knowledge, it is the first RNA nanoparticle to have been comprehensively examined pharmacologically in vivo, and demonstrated to be safe with favorable PK and biodistribution profiles, as well as self-delivery to tumor tissues by specific targeting mechanism.
The current studies demonstrated two rather important pharmacological characteristics of pRNA nanoparticle. First, it did not induce the IFN response, even for pRNA nanoparticles with a long 5′/3′ duplex region, modified or nonmodified. In particular, pRNA nanoparticles did not cause the TLR3 pathway activation in the TLR3-overexpressing reporter cell line. One possible interpretation could be that it has a tightly folded structure with multiple loops, resulting in little interaction with TLR3, and/or it has a larger particle size than linear duplex RNA molecules. However, whether this is true remains to be tested. Second, pRNA nanoparticle accumulates in negligible amounts in liver, spleen, and lung, in contrast to almost all known nanoparticles. Although the true underlying mechanism is yet to be investigated, this could represent a great advantage of pRNA nanoparticles over existing nanotechnologies for therapeutic delivery, where pRNA can minimize organ toxicities and provide better distribution to the targeted disease tissues. These unique characteristics of pRNA could be a direct result of its physicochemical properties(hydrophilicity, negative surface charge, stable tertiary structure, and optimal particle size).51 We believe that the sufficiently small size of the monomeric pRNA nanoparticle enables it to pass through the smallest blood vessels and deliver to a wide spectrum of peripheral tissues. A tumor targeting pRNA nanoparticle thus should not only target the primary tumor site where the vasculature is leaky, but also metastatic disease in remote tissues.
Another advantage of pRNA nanoparticle is its robustness for precise functionalization. As demonstrated in this report, folate and fluorescence dyes can be stoichiometrically incorporated at the precise locations as desired. Our previous studies have also shown other types of ligand such as RNA aptamers be incorporated into pRNA.52 Aptamers as functional modules could be of particular importance, because they can be produced using the similar chemical process and have some similar biocompartibility profiles as RNA nanoparticles. One of the concerns over the attachment of aptamers to RNA nanoparticle has been the possible disruption of structures/functions of either the nanoparticle or the aptamer, or both. However, we have not seen this disruption in several aptamers that we have tested so far: CD4-aptamer-pRNA chimeric nanoparticle,16,17 and PSMA-aptamer (S. Abdelmawla, P. Guo, and Q.-X. Li et al., unpublished results), which is consistent with our hypothesis that the unique stability of pRNA supports its diverse functionalization.
We have previously also demonstrated that the 5′/3′ double helix region of pRNA could accommodate changes in the sequence to act as RNAi trigger, leading to RNAi activity, which we called “pRNAi.”16 With the preferred pharmacological profiles, we predict that a targeted 2′-F-pRNAi nanoparticle would likely cause in vivo silencing in the targeted tissues upon systemic administration. We anticipate that pRNAi would be a superior RNAi therapeutic over the naked small interfering RNA because of the larger particle size and better PK profile of pRNA, and also over many polymer and liposomal based small interfering RNA delivery platforms because of the better safety profile and tissue distribution profile. At present, we are investigating the pRNAi nanoparticle both in vitro and in vivo as an RNAi delivery platform for therapeutic applications. Besides RNAi trigger, pRNA nanoparticle can also carry other conventional therapeutic payloads, such chemotherapeutic agents, which could also result in therapeutics superior to the free drugs in both safety and activity.
Another type of application for pRNA nanoparticles is its possible use for imaging for targeted disease tissues as demonstrated in this study. In addition to fluorescent dyes, other reporters as radioactive isotopes or contrasting agents can also be incorporated for a variety of imaging applications, including PET and NMR. We anticipate that such a system can lead to potential superior imaging properties over small molecule imaging agents due to advantageous PK and biodistribution profiles. In particular, tumor targeting pRNA nanoparticles carrying therapeutic payloads and imaging agents can facilitate noninvasive diagnostic imaging of tumors and monitor whether the therapeutic payloads are delivered to the tumor sites. It can also facilitate personalized treatment by imaging tumors and metastatic sites for treatment justification and assessment of tumor response.
Materials and Methods
Chemical synthesis of 2′-F-pRNA
Chemical synthesis of bipartite pRNA: The 2′-F-pRNA monomer molecule was first chemically synthesized as two pieces with an approximate length of ~55 nt of P1 and ~60 nt of P2 (depending on the particular pRNA monomer) by Trilink Biotechnologies (San Diego, CA). The first oligonucleotide (P1) has a 3′-OH group. The second oligonucleotide (P2) has a 5′-phosphate group (Figure 1a). To assemble the pRNA monomer, simply mix and/or anneal P1 and P2 at an equal molar ratio at the desired concentration. In order to obtain 5′-folate, 5′-Cy5, or 5′-AlexaFluor647 pRNA on P1 or P2, folate-CEP, Cy5-CEP, or AlexaFluor-CEP were synthesized and used in the last coupling step during synthesis of P1 and P2.
Enzymatic ligation of P1 and P2 to form a single full-length pRNA monomer: First, P1 and P2 oligonucleotides were dissolved in 1× T4-RNA ligase buffer (New England BioLabs, Ipswich, MA) at 50 µmol/l final concentration for each oligo. The oligonucleotides were annealed by heating to 90 °C for 10 minutes followed by slow cooling to room temperature over the course of 45 minutes. To the annealed oligonucleotides solution, T4-RNA ligase I (1.2 U/µl) (New England BioLabs) was added and the reaction carried out at 37 °C overnight to form the pRNA monomer. The ligated product was extracted using phenol:chloroform followed by ethanol precipitation and subsequent 10%/8 mol/l urea PAGE purification. The purified product was quantified by UV absorbance at 260 nm using a NanoDrop ND-1000 spectrophotometer (Nanodrop, Wilimington, DE).
Cellular uptake of pRNA nanoparticle by confocol microscopy and flow cytometry. Fluorescently labeled-pRNA nanoparticles were used for cellular uptake studies. Essentially, two labeling methods were used. First, direct coupling of fluorescence at 5′ end of the oligonucleotides was used as described above. Second, the pRNA was labeled with fluorescein using the Label IT kit and purified according to the manufacturer's instructions (Invitrogen, Eugene, OR). KB cells were grown in a folate-free RPMI-1640 medium (Invitrogen) supplemented with 10% fetal bovine serum. For the cellular uptake experiment, the cells were cultured in 8-well-chambered slides (LabTek; Nunc, Roskilde, Denmark) at a density of 1 × 104 cells/well. Twenty-four hours later, the culture media was replaced with fresh 200 µl folate-free media, and the fluorescently labeled folate-pRNA nanoparticles were added at a final concentration of 200 nmol/l, either in the absence or in the presence of 200-fold excess of free-folic acid. After incubation of the cells at 37 °C for 30 minutes, confocal microscopy was conducted. Images were recorded with an inverted Nikon A1 confocal microscope using a ×40 objective. Images were processed using NIS software provided by Nikon (Mellville, NJ).
IFN response assays in vitro
Assess induction of IFN-response genes: To examine the induction of OAS1&2, MX1, and IFITM1, KB cells were transfected with pRNAi (50 nmol/l) and poly I:C (1 µg/ml), respectively using the Fugene-HD transfection reagent (Roche, Promega, Madison, WI). After 24 hours, total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA), and reverse transcriptase-PCR was conducted using the primer sets provided by the “IFNγ qRT-primers” kit (InvivoGen, San Diego, CA) in conjunction with the one-step reverse transcriptase-PCR kit (Invitrogen) according to the manufacturer's instructions. Target gene expression was normalized to GAPDH expression. All reactions were done in triplicates and with no reverse transcriptase or no template as negative controls.
To examine the expression of TLR-3, -7, -9, human peripheral mononuclear cells were incubated for 24 hours with 2′-F modified pRNA and poly I:C as a positive control. The expression levels of human TLR receptors were analyzed by semiquantitative reverse transcriptase-PCR using TLR reverse transcriptase-Primer Set (Invivogen), as described above.
Examination of TNF-α induction in the mouse RAW-246 cell line: Mouse macrophage cell line, RAW-246 (ATCC, Manassas, VA), was plated at a density of 30,000 cells/well in 200 µl Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum in 96-well plate. The cells were incubated with pRNA nanoparticles at final concentrations of 400, 100, 4 µg/ml for 3 hours at 37 °C. Aliquots (20 µl) from the culture medium were taken and analyzed using the enzyme-linked immunosorbent assay kit for murine TNF-α, per the manufacturer's instruction (Antigenix America, Huntington, NY).
Examination of human TLR-3 pathway activation: HEK-Blue-hTLR3 (Invivogen) cells were set up at a density of 300,000 cells/ml in a test medium containing Dulbecco's modified Eagle's medium, 4.5 g/l glucose, 100 µg/ml Normocin, 2 mmol/l -glutamine, 10% (vol/vol) heat-inactivated fetal bovine serum. The cell suspension (180 µl) was mixed with a 20-µl pRNA solution and plated in each well of a 96-well plate with final concentrations of 3,000, 1,000, and 300 ng/ml, followed by overnight incubation at 37 °C. Aliquots (20 µl) from the cell supernatant were mixed with a 180 µl QUANTI-Blue detection medium (Invivogen) and incubated at 37 °C for 3 hours. The levels of SEAP were determined by measuring the UV absorbance at 620–655 nm.
PK analysis in mice. HeLa tumor bearing Balb/c nude male mice (three mice total) (see below) were each injected with 600 µg of AlexaFluor647-labeled pRNA monomer (24 mg/kg) through the tail vein. Blood was collected (up to 20 µl) through the lateral saphenous vein at 5 minutes, 30 minutes, 2 hours, 5 hours, and 24 hours time points. Blood samples were collected in BD Vacutainer SST Serum Separation Tubes. The tubes were inverted five times and left for 30 minutes at room temperature (to allow the blood to clot), followed by centrifugation at 1,000–1,300 g for 10 minutes. Two micro liter supernatant serums were mixed with 1 µl of proteinase-K and 37 µl of water. The mixture was incubated at 37 °C for 30 minutes before loading onto the capillary gel electrophoresis machine. The pRNA in the blood was quantified by capillary gel electrophoresis through measurement of the fluorescence intensity using a P/ACE MDQ capillary electrophoresis system with a 635 nm laser suitable for AlexaFluor647 fluorophores (Beckman Coulter, Fullerton, CA). The capillaries used were 32 cm in length and 100 µm in diameter. Samples were loaded by voltage injection for 40 seconds at 4 kV and allowed to run for 25 minutes at 7.8 kV. The gel was replaced after every run. The buffer and the nondenaturing gel were purchased from Beckman Coulter (dsDNA 1000 Kit). The electropherograms produced were integrated using the software “32 Karat” version 7 provided by Beckman Coulter. The pRNA concentrations were calculated from a standard curve. The serum concentration profiles were fitted with an IV bolus noncompartmental model using the Kinetica program (Fisher Scientific, Pittsburgh, PA), and the key secondary PK parameters were deduced. The PK studies were performed at Purdue animal facility per approved protocol.
Toxicological evaluation in mice. For the cytokine induction study, three groups of four immunocompetent female C57B/6 mice (Harlan, Indianapolis, IN) aged 10 weeks were injected via tail vein with PBS, Ba′-2′-F-sur-pRNAi (KT-108) at 30 mg/kg and KT-108 at 10 mg/kg. The blood was collected by cardiac puncture 3 hours postinjection. The plasma was prepared and analyzed for mouse TNF-α and interleukin-6 using enzyme-linked immunosorbent assay (Antigenix America). For a 7-day repeat dose study, four groups of five mice were intravenously injected (tail vein) with: (i) PBS, (ii) poly I:C at 30 mg/kg (Sigma-Aldrich, St Louis, MO), (iii) KT-108 at 30 mg/kg, and (iv) KT-108 at 10 mg/kg. The mice were treated once every 48 hours for 7 days (four doses total). The mice were monitored for clinical signs and body weights. On day 7, 3 hours after the fourth injection, the mice were euthanized per protocol and blood samples were collected by cardiac puncture for standard panel clinical chemistry (including PT, aPTT) and clinical pathology analysis. The gross pathology and organ weights were recorded (including spleen, lymph nodes, liver, and kidneys). The toxicology study was conducted at Explora BioLab per approved protocol (San Diego, CA).
Tumor targeting and biodistribution by imaging in xenograft tumor models. For biodistribution and tumor targeting studies, 6-week-old male nude mice (nu/nu) were purchased from NCI, Frederick, MD and maintained on a folate-free diet for a total of 2 weeks before the start of the experiment. The mice were injected with different cancer cells (KB cells ~3 × 106 cells/mouse, HeLa cells ~1 × 107 cells/mouse in 40% matrigel in a folate-free RPMI-1640 medium. When the tumors reached ~500 mm3, the mice were injected intravenously through the tail vein with a single dose of 15 nmol (24 mg/kg) of KT-105 (FA-AlexaFluor647 labeled-pRNA). The control mice were injected with 10 mg/kg of free folate intraperitonealy 10 minutes before the KT-105 injection. The mice were euthanized by CO2 asphyxiation 24 hours postinjection, and whole-body imaging was conducted using an IVIS Lumina station (Bindley Bioscience Center at Purdue University, West Lafayette, IN). After whole-body imaging, the mice were dissected and the major organs and tumors were collected and laid out for organ imaging (tumors, liver, spleen, heart, lung, intestine, kidney, and skeletal muscle). The tumor targeting studies were performed at Purdue animal facility per approved protocol.
SUPPLEMENTARY MATERIAL Figure S1. CGE analysis of monomeric and dimeric pRNA nanoparticles. Figure S2. Chemically synthesized 2′F-pRNA nanoparticle are metabolically stable. The different forms of nanoparticeles were incubated with different concentrations of sera or with RNase A (1U, 30 min) for the indicated periods at 37°C, followed by phenol:chloroform extraction/ethanol precipitation, and 10%/8 M urea PAGE (visualized by Sybr® Gold). Figure S3. Cellular uptake of pRNA nanoparticles with various folate conjugation. A-I: KB cells were incubated with FA-pRNA nanoparticle containing folate at the 5'-end of P1. The pRNA was labeled with fluoresceine using Silencer ® siRNA labeling kit (Ambion). The cells were incubated with the pRNA (200 nM) in absence or presence of excess folate (200 fold) for 30 min., followed by flow analysis (A-C), or confocal microscopy (D-I). J-M: KB cells were transfected with GFP-Rho-B-expressing cassette to label early and late endosomes (green). 24 h later, KB cells were incubated with 200 nM Cy5-FA-pRNA nanoparticle (red) for the indicated periods followed by confocal microscopy. Cy5 was conjugated to the 5'-end of P1 and folate was conjugated on 5'-end of P2. The co-localization of pRNA with the endosomes is shown in yellow. A positive control panel (M) is shown where chloroquine (chQ) was added 4 hours post incubation to induce endosomal release. Figure S4. The organ weights of pRNA-treated mice. Selected organs from the C57B/6 mice after 1 week of repeat systemic administration of pRNA nanoparticles at a dose of 30 mg/kg were weighted and displayed.
Acknowledgments
The authors thank Drs Pierce and Bergstrom for scientific advice and editing of the manuscript. The authors also thank Dr Taylor at Bindley Imaging Center, and Dr Zhang for useful advices. This work was partially supported by Indiana 21st century fund and NIH SBIR1R43GM087081-01 to Kylin Therapeutics, Inc. as well as NIH EB003730 and CA151648 to P.G. The majority of the work was conducted at Bindley Bioscience Center, Discovery Park, Purdue University.
Supplementary Material
CGE analysis of monomeric and dimeric pRNA nanoparticles.
Chemically synthesized 2′F-pRNA nanoparticle are metabolically stable. The different forms of nanoparticeles were incubated with different concentrations of sera or with RNase A (1U, 30 min) for the indicated periods at 37°C, followed by phenol:chloroform extraction/ethanol precipitation, and 10%/8 M urea PAGE (visualized by Sybr® Gold).
Cellular uptake of pRNA nanoparticles with various folate conjugation. A-I: KB cells were incubated with FA-pRNA nanoparticle containing folate at the 5'-end of P1. The pRNA was labeled with fluoresceine using Silencer ® siRNA labeling kit (Ambion). The cells were incubated with the pRNA (200 nM) in absence or presence of excess folate (200 fold) for 30 min., followed by flow analysis (A-C), or confocal microscopy (D-I). J-M: KB cells were transfected with GFP-Rho-B-expressing cassette to label early and late endosomes (green). 24 h later, KB cells were incubated with 200 nM Cy5-FA-pRNA nanoparticle (red) for the indicated periods followed by confocal microscopy. Cy5 was conjugated to the 5'-end of P1 and folate was conjugated on 5'-end of P2. The co-localization of pRNA with the endosomes is shown in yellow. A positive control panel (M) is shown where chloroquine (chQ) was added 4 hours post incubation to induce endosomal release.
The organ weights of pRNA-treated mice. Selected organs from the C57B/6 mice after 1 week of repeat systemic administration of pRNA nanoparticles at a dose of 30 mg/kg were weighted and displayed.
REFERENCES
- Guo P, Zhang C, Chen C, Garver K., and, Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray CT, Major F.et al. (1998Function of hexameric RNA in packaging of bacteriophage phi 29 DNA in vitro Mol Cell 2141–147. [DOI] [PubMed] [Google Scholar]
- Jaeger L., and, Leontis NB. Tecto-RNA: One-Dimensional Self-Assembly through Tertiary Interactions This work was carried out in Strasbourg with the support of grants to N.B.L. from the NIH (1R15 GM55898) and the NIH Fogarty Institute (1-F06-TW02251-01) and the support of the CNRS to L.J. The authors wish to thank Eric Westhof for his support and encouragement of this work. Angew Chem Int Ed Engl. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Shu D, Moll WD, Deng Z, Mao C., and, Guo P. Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J Nanosci Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM., and, Cullis PR. Drug delivery systems: entering the mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- Guo P, Coban O, Snead NM, Trebley J, Hoeprich S, Guo S.et al. (2010Engineering RNA for targeted siRNA delivery and medical application Adv Drug Deliv Rev 62650–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo PX, Erickson S., and, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Shu D, Zhang H, Jin J., and, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zhang H., and, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. Structure and function of phi29 hexameric RNA that drives the viral DNA packaging motor: review. Prog Nucleic Acid Res Mol Biol. 2002;72:415–472. doi: 10.1016/s0079-6603(02)72076-x. [DOI] [PubMed] [Google Scholar]
- Hoeprich S., and, Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA (pRNA) monomer, dimer, and hexamer of Phi29 DNA packaging motor. J Biol Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- Chen C, Sheng S, Shao Z., and, Guo P. A dimer as a building block in assembling RNA. A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- Mat-Arip Y, Garver K, Chen C, Sheng S, Shao Z., and, Guo P. Three-dimensional interaction of Phi29 pRNA dimer probed by chemical modification interference, cryo-AFM, and cross-linking. J Biol Chem. 2001;276:32575–32584. doi: 10.1074/jbc.M100045200. [DOI] [PubMed] [Google Scholar]
- Guo S, Tschammer N, Mohammed S., and, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled A, Guo S, Li F., and, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J, Guo, S, Cinier, M, Shlyakhtenko, L, Shu, Y, Chen, C.et al. (2011Fabrication of stable and RNase- resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging ACS Nano 5237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Lee C-S., and, Guo P. The proximate 5' and 3' ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage 29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- Garver K., and, Guo P. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage phi29. RNA. 1997;3:1068–1079. [PMC free article] [PubMed] [Google Scholar]
- Reid RJ, Bodley JW., and, Anderson D. Characterization of the prohead-pRNA interaction of bacteriophage phi 29. J Biol Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- Chen C, Zhang C., and, Guo P. Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Huang LP, Hoeprich S., and, Guo P. Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices. J Nanosci Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- Guo S, Huang F., and, Guo P. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther. 2006;13:814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Trottier M., and, Guo PX. Circularly permuted viral pRNA active and specific in the packaging of bacteriophage 29 DNA. Virology. 1995;207:442–451. doi: 10.1006/viro.1995.1103. [DOI] [PubMed] [Google Scholar]
- Hoeprich S, Zhou Q, Guo S, Shu D, Qi G, Wang Y.et al. (2003Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus Gene Ther 101258–1267. [DOI] [PubMed] [Google Scholar]
- Li L, Liu J, Diao Z, Shu D, Guo P., and, Shen G. Evaluation of specific delivery of chimeric phi29 pRNA/siRNA nanoparticles to multiple tumor cells. Mol Biosyst. 2009;5:1361–1368. doi: 10.1039/b903428e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y, Cinier, M., and, Guo P.2011Bottom-up assembly of therapeutic pRNA-siRNA nanoparticles using two RNA fragments Mol Therthis issue. Doi:10.1038/mt.2011.23. [DOI] [PMC free article] [PubMed]
- Chen SJ. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu Rev Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkovitz E, Sapiro G, Tannenbaum A., and, Williams LD. Statistical analysis of RNA backbone. IEEE/ACM Trans Comput Biol Bioinform. 2006;3:33–46. doi: 10.1109/TCBB.2006.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurschat WC, Müller J, Wombacher R., and, Helm M. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishigaki K, Taguchi K, Kinoshita Y, Aita T., and, Husimi Y. Y-ligation: an efficient method for ligating single-stranded DNAs and RNAs with T4 RNA ligase. Mol Divers. 1998;4:187–190. doi: 10.1023/a:1009644028931. [DOI] [PubMed] [Google Scholar]
- Stark MR, Pleiss JA, Deras M, Scaringe SA., and, Rader SD. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA. 2006;12:2014–2019. doi: 10.1261/rna.93506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumport RI., and, Uhlenbeck OC. T4 RNA ligase as a nucleic acid synthesis and modification reagent. Gene Amplif Anal. 1981;2:313–345. [PubMed] [Google Scholar]
- Kühn R, Schwenk F, Aguet M., and, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Kim SY, Li J, Bentsman G, Brooks AI., and, Volsky DJ. Microarray analysis of changes in cellular gene expression induced by productive infection of primary human astrocytes: implications for HAD. J Neuroimmunology. 157:17–26. doi: 10.1016/j.jneuroim.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Huang S, Wei W., and, Yun Y. Upregulation of TLR7 and TLR3 gene expression in the lung of respiratory syncytial virus infected mice. Wei Sheng Wu Xue Bao. 2009;49:239–245. [PubMed] [Google Scholar]
- Groskreutz DJ, Monick MM, Powers LS, Yarovinsky TO, Look DC., and, Hunninghake GW. Respiratory syncytial virus induces TLR3 protein and protein kinase R, leading to increased double-stranded RNA responsiveness in airway epithelial cells. J Immunol. 2006;176:1733–1740. doi: 10.4049/jimmunol.176.3.1733. [DOI] [PubMed] [Google Scholar]
- Kamen BA., and, Caston JD. Properties of a folate binding protein (FBP) isolated from porcine kidney. Biochem Pharmacol. 1986;35:2323–2329. doi: 10.1016/0006-2952(86)90458-2. [DOI] [PubMed] [Google Scholar]
- Leamon CP, Reddy JA, Vlahov IR, Westrick E, Dawson A, Dorton R.et al. (2007Preclinical antitumor activity of a novel folate-targeted dual drug conjugate Mol Pharm 4659–667. [DOI] [PubMed] [Google Scholar]
- Reddy JA, Westrick E, Santhapuram HK, Howard SJ, Miller ML, Vetzel M.et al. (2007Folate receptor-specific antitumor activity of EC131, a folate-maytansinoid conjugate Cancer Res 676376–6382. [DOI] [PubMed] [Google Scholar]
- Reddy JA, Dorton R, Westrick E, Dawson A, Smith T, Xu LC.et al. (2007Preclinical evaluation of EC145, a folate-vinca alkaloid conjugate Cancer Res 674434–4442. [DOI] [PubMed] [Google Scholar]
- Turk MJ, Waters DJ., and, Low PS. Folate-conjugated liposomes preferentially target macrophages associated with ovarian carcinoma. Cancer Lett. 2004;213:165–172. doi: 10.1016/j.canlet.2003.12.028. [DOI] [PubMed] [Google Scholar]
- Paulos CM, Turk MJ, Breur GJ., and, Low PS. Folate receptor-mediated targeting of therapeutic and imaging agents to activated macrophages in rheumatoid arthritis. Adv Drug Deliv Rev. 2004;56:1205–1217. doi: 10.1016/j.addr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Kwon, Y. Kluwer Academic, New York; 2001. Handbook of Essential Pharmacokinetics, Pharmacodynamics, and Drug Metabolism for Industrial Scientists. [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W.et al. (2005Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs Nat Biotechnol 231002–1007. [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B.et al. (2005Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication Hepatology 411349–1356. [DOI] [PubMed] [Google Scholar]
- Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR.et al. (2009Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors Nat Biotechnol 27839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SD, Chen YC, Hackett MJ., and, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil SE. Nanoparticle therapeutics: a personal perspective. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:264–271. doi: 10.1002/wnan.6. [DOI] [PubMed] [Google Scholar]
- Zhou J., and, Rossi JJ. The therapeutic potential of cell-internalizing aptamers. Curr Top Med Chem. 2009;9:1144–1157. doi: 10.2174/156802609789630893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CGE analysis of monomeric and dimeric pRNA nanoparticles.
Chemically synthesized 2′F-pRNA nanoparticle are metabolically stable. The different forms of nanoparticeles were incubated with different concentrations of sera or with RNase A (1U, 30 min) for the indicated periods at 37°C, followed by phenol:chloroform extraction/ethanol precipitation, and 10%/8 M urea PAGE (visualized by Sybr® Gold).
Cellular uptake of pRNA nanoparticles with various folate conjugation. A-I: KB cells were incubated with FA-pRNA nanoparticle containing folate at the 5'-end of P1. The pRNA was labeled with fluoresceine using Silencer ® siRNA labeling kit (Ambion). The cells were incubated with the pRNA (200 nM) in absence or presence of excess folate (200 fold) for 30 min., followed by flow analysis (A-C), or confocal microscopy (D-I). J-M: KB cells were transfected with GFP-Rho-B-expressing cassette to label early and late endosomes (green). 24 h later, KB cells were incubated with 200 nM Cy5-FA-pRNA nanoparticle (red) for the indicated periods followed by confocal microscopy. Cy5 was conjugated to the 5'-end of P1 and folate was conjugated on 5'-end of P2. The co-localization of pRNA with the endosomes is shown in yellow. A positive control panel (M) is shown where chloroquine (chQ) was added 4 hours post incubation to induce endosomal release.
The organ weights of pRNA-treated mice. Selected organs from the C57B/6 mice after 1 week of repeat systemic administration of pRNA nanoparticles at a dose of 30 mg/kg were weighted and displayed.