Abstract
As much as 90% of an intravenously (i.v.) injected dose of adenovirus serotype 5 (Ad5) is absorbed and destroyed by liver Kupffer cells. Viruses that escape these cells can then transduce hepatocytes after binding factor X (FX). Given that interactions with FX and Kupffer cells are thought to occur on the Ad5 hexon protein, we replaced its exposed hypervariable regions (HVR) with those from Ad6. When tested in vivo in BALB/c mice and in hamsters, the Ad5/6 chimera mediated >10 times higher transduction in the liver. This effect was not due to changes in FX binding. Rather, Ad5/6 appeared to escape Kupffer cell uptake as evidenced by producing no Kupffer cell death in vivo, not requiring predosing in vivo, and being phagocytosed less efficiently by macrophages in vitro compared to Ad5. When tested as a helper-dependent adenovirus (Ad) vector, Ad5/6 mediated higher luciferase and factor IX transgene expression than either helper-dependent adenoviral 5 (HD-Ad5) or HD-Ad6 vectors. These data suggest that the Ad5/6 hexon-chimera evades Kupffer cells and may have utility for systemic and liver-directed therapies.
Introduction
Adenovirus serotype 5 (Ad5) is widely used for liver gene transfer studies because of its natural tropism for this organ after systemic administration. Recent evidence suggests that this liver tropism is dependent on the major capsid protein, hexon, which can bind host serum coagulation factors to enhance hepatocyte transduction.1 More specifically, it was demonstrated that vitamin K dependent blood factors VII, IX, X, and protein C enhance transduction of Ad5.1,2,3 Factor X (FX) was found to bind Ad5 hexon with particularly high affinity using its γ-carboxyglutamate domain.4 Cryo-electron microscopy and hexon mutagenesis techniques localized FX binding to the hypervariable (HVR) domains of Ad5 hexon.5,6 Adenoviral serotypes possess seven HVR domains that differ widely in sequence, and in part, define them.7 As such, variations in the HVR domain across serotypes and their differences in FX binding affinity generally correlate with the ability of adenovirus (Ad) serotypes to transduce hepatocytes.4
In addition to viral tropism, HVR domains may have a role in sequestration of Ad virions within Kupffer cells. Kupffer cells are resident liver macrophages that sequester 90% of the intravenously (i.v.) injected Ad5 doses, thus representing a major barrier hepatocellular gene transfer.8 Although Kupffer cells comprise only ~7% of liver cells,9 they are thought to represent 80–90% of all macrophages in the body10 and therefore represent a significant whole body pharmacological “sink” for systemically administered Ad5. In addition, Kupffer cells are antigen-presenting macrophages. Therefore, rampant uptake of viral antigens into these cells may amplify immune responses against the vector.
Uptake by Kupffer cells not only depletes circulating Ad5 but it kills these cells themselves.11 Kupffer cells can be destroyed by “predosing” animals first with a high dose of irrelevant Ad5. Four hours later, the therapeutic or reporter-expressing Ad can be injected thereby avoiding destruction by the Kupffer cells. Predosing is remarkably effective and, in the absence of the Kupffer cell sink, virus escapes to other sites as evidenced by its ability to increase hepatocyte expression 40-fold and its ability to increase killing of distant tumor sites.12
In previous work testing systemic delivery of oncolytic Ads, a series of viruses were tested for their ability to mediate this predosing effect.13 Surprisingly, species C Adenovirus serotype 6 (Ad6) demonstrated remarkably less efficient predosing as compared to the extensively studied species C Ad5. This data suggested that Ad5 and Ad6 might have distinctly different interactions with liver cells despite their high sequence identity.
Most of the sequence variation between Ad5 and Ad6 is in the HVR domains of their hexon proteins.14 Given this, we have tested whether this region mediates interactions with liver cells by replacing the HVR domains of Ad5 with those from Ad6. Comparison of this Ad5/6 vector with Ad5 vector after i.v. injection, demonstrates remarkable differences in their pharmacology and interactions with macrophages and Kupffer cells.
Results
Generation of a hexon-chimeric Ad5/6 vector
Given that FX and Kupffer cell interactions are thought occur in the HVR domains of hexon, we replaced the HVR region of Ad5 with that of Ad6. PCR was used to clone the HVR region of Ad6 with terminal ApaI and PstI restriction sites and this was used to replace the corresponding HVR cassette in Ad5. Ad5/6GL expressing the eGFP-luciferase (GL) reporter protein was generated by replacing this region of Ad5GL by red recombination (Supplementary Figure S1). Replacing the Ad5 HVR domain with that of Ad6 significantly decreased the number of charged residues in this region. Ad5 contains 31 negatively charged amino acids (blue; aspartic acid and glutamic acid) and 17 positively charged amino acids (red; arginine, histidine, and lysine). Ad6 displays only 25 negatively and 12 positively charged residues (Supplementary Figure S1a). Both vectors retain the glutamic acid 451 (E451) in HVR7 that is conserved in all FX-binding Ads.6
Ad5/6GL and Ad5GL vectors grew with similar kinetics, although Ad5/6GL yielded approximately fivefold fewer virus particles (vp) and sevenfold fewer plaque forming units after purification (Supplementary Table S1). Western blotting of Ad5GL and Ad5/6GL with serum from Ad5 and Ad5/6-immunized mice demonstrated that these antibodies did not crossreact against each other's hexon proteins (data not shown).
Intravenous injection of mice with Ad5/6GL generates higher liver expression when compared to Ad5GL
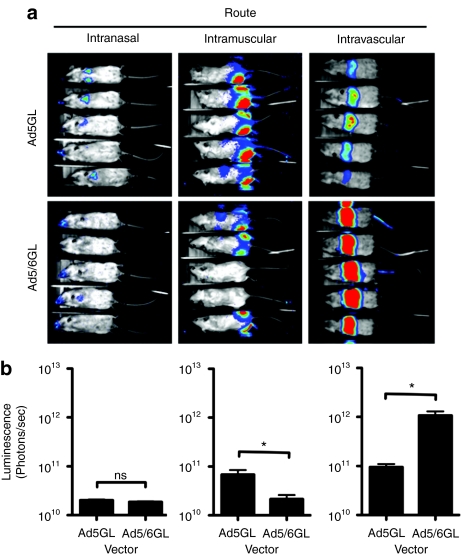
1 × 1010 vp of Ad5GL or Ad5/6GL was injected into female BALB/c mice by three different routes: intramuscular (i.m.), intranasal (i.n.), and i.v. by tail vein (Figure 1). Luciferase imaging 24 hours later demonstrated that both vectors mediated comparable expression when injected i.n. By the i.m. route, Ad5GL generated twofold higher expression than Ad5/6GL. In stark contrast, Ad5/6GL produced >tenfold higher expression in the liver after i.v. injection of equal particle numbers. Considering that the Ad5/6 has a lower infectious unit titer, this tenfold increase likely underestimates Ad5/6 activity. The comparison was repeated four times with several different viral preparations with essentially identical results. That Ad5/6GL demonstrates lower nasal and muscle transduction but higher liver expression indicates a unique biology with respect to systemic delivery and liver infection.
Figure 1.
In vivo expression of unmodified and chimeric adenoviruses when injected via different routes. BALB/c mice were injected with 1 × 1010 virus particles of either Ad5GL or Ad5/6GL by different routes. (a) Mice were imaged 24 hours later using a Lumazone imager: intranasal, 10-minute 1 × 1 binning, 1,315–1,389 gray values; intramuscular, 10-minute 2 × 2 binning, 712–1,300 gray values; intravenous, 1 minute 1 × 1 binning, 1,296–2,000 gray values. Shown are representative white light images overlaid with luciferase expression in pseudo-color. (b) Quantification of luciferase expression in mice injected via different routes after 24 hours. N = 5. *P < 0.05; ns = not significant. Means ± SEM.
FX binds to Ad5/6GL and is crucial for hexon-mediated transduction of Ad5/6GL in vitro
The binding of Ad5 hexon to vitamin K dependent blood factors, particularly FX, has been shown to traffic virions to hepatocytes in vivo.2,5 Hexon protein was purified from Ad5GL or Ad5/6GL infected cell lysates, immobilized on biosensor chips, and tested for binding to human FX using Surface Plasmon Resonance (Supplementary Table S2). Consistent with previous work,4 Ad5 hexon bound FX with high affinity (KD = 1.68 × 10−9 mol/l). In contrast, FX affinity for Ad5/6 hexon was tenfold lower (KD = 1.63 × 10−8 mol/l). Closer inspection revealed that association (Ka) of unmodified and chimeric hexon with FX was similar, but the latter appeared to dissociate (Kd) more rapidly. These results were consistent with the binding kinetics of Ad6 to FX (Supplementary Table S2). As previously shown, immobilization of FX on the chip instead of hexon increased the KD by approximately tenfold.4
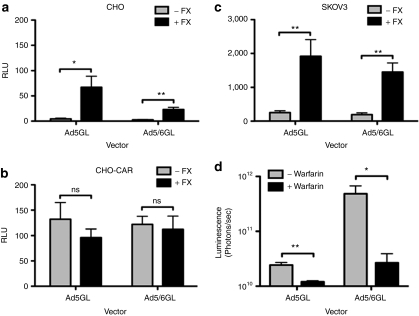
To assess functional FX binding in vitro, Ad5GL and Ad5/6GL were tested for their ability to transduce cells that are nonpermissive for Ad5 infection. SKOV3 and CHO cell lines have low levels of CAR, the coxsackie and adenovirus receptor,13 but in the presence of FX, Ad5 is able to transduce these cells.4,6 Both viruses were incubated on cells in the presence or absence of physiological levels of FX (10 µg/ml) for 3 hours at 37 °C, and were assayed for transduction by luciferase activity 24 hours later (Figure 2a,c). Transgene expression by both viruses increased significantly in the presence of FX. When tested on CHO cells expressing CAR, the FX effect was masked (Figure 2b).
Figure 2.
Ad5/6GL and Ad5GL interact with blood factors comparably. (a) CHO, (b) CHO cells transiently transfected with coxsackie-adenovirus receptor (CAR), or (c) SKOV3 were infected with unmodified or chimeric adenovirus (Ad) vectors (multiplicity of infection 1,000) ± FX (10 µg/ml). A luciferase assay was performed after 24 hours to determine transgene expression. N = 6. (d) BALB/c mice were injected subcutaneously with warfarin (133 µg in 100 µl peanut oil) 3 and 1 day before intravenous injection of 1 × 1010 virus particles (vp) unmodified or chimeric adenoviruses. Mice were imaged 24 hours later using a Lumazone imager and quantified for luciferase transgene expression; 3 minutes 2 × 2 binning. N = 5. *P < 0.05; **P < 0.005; ns = not significant. Means ± SEM.
Vitamin K dependent blood clotting factors are crucial for liver transduction by Ad5/6GL in vivo
Warfarin inhibits vitamin-K mediated γ-carboxylation of blood factors such as FX, decreasing their ability to retarget Ad5 to hepatocytes.12,15 To test if blood factors mediated Ad5/6GL liver transduction in vivo, BALB/c mice were treated with warfarin prior to i.v. injection of both viruses (Figure 2d). For both, warfarinization dramatically decreased luciferase expression in the liver. The fold decrease between mice after warfarinization was twofold for Ad5 and 18-fold for Ad5/6.
Ad5/6GL is less efficiently phagocytosed by macrophages in vitro
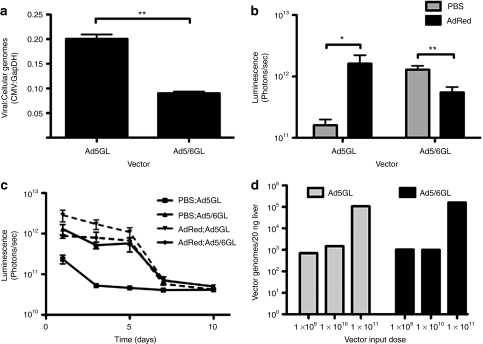
Phagocytic Kupffer cells are a considerable sink for i.v. injected Ad5 particles. To test whether the Ad6 HVRs changed recognition of Ad virions by phagocytic cells, the viruses were incubated with RAW 264.7 murine monocyte-macrophage cells in vitro. Although we determined low levels of expression in vitro (data not shown) these cells are thought to predominantly phagocytose rather than endocytose Ad.16 Given this, phagocytosis was assessed by incubating the cells with 10,000 vp/cell of Ad5 or Ad5/6 and after 24 hours, viral genomes were quantified by real-time PCR (Figure 3a). This analysis revealed that ~50% less Ad5/6GL viral genomes were taken up into RAW macrophages when compared to unmodified Ad5GL.
Figure 3.
Ad5/6GL is taken up into macrophages less than unmodified Ad5GL. (a) adenovirus (Ad) vectors were added to RAW 267.4 cells (multiplicity of infection 10,000). Cells were harvested 24 hours later and genomic DNA was extracted. Real-time PCR was used to quantify viral and cellular genomes using cytomegalovirus (CMV) and GAPDH specific primers, respectively. Data are normalized to cellular genomes. N = 4. BALB/c mice were predosed intravenous (i.v.) with 3 × 1010 virus particles (vp) in 100 µl in phosphate-buffered saline (PBS). After 4 hours, the mice were injected with 1 × 1010 vp in PBS of unmodified or chimeric Ad vector. Mice were imaged and luciferase expression was quantified (b) 24 hours after Ad vector injection and (c) over time. N = 5. (d) Varying doses of Ad vectors were injected i.v. into mice. After 1 hour, mice were euthanized and 20 ng of tissue from the large liver lobe was harvested. Total DNA was extracted and real-time PCR was used to quantify viral genomes using CMV primers. *P < 0.05; **P < 0.005; ns = not significant. Means ± SEM.
Ad5/6GL evades Kupffer cells for increased liver expression in vivo
The in vitro RAW cell data suggested that swapping the Ad5 HVR for Ad6 HVR reduced phagocytosis by macrophage cells. To explore this in vivo, mice were predosed by i.v. injection of 3 × 1010 vp of Ad5-dsRed to destroy Kupffer cells and prevent trapping of subsequent doses of Ad.11 Four hours later, predosed or control mice were injected i.v. with either Ad5GL or Ad5/6GL and luciferase expression was measured over the following 10 days (Figure 3b,d). Consistent with previous data, predosing with Ad5 increased subsequent transduction by Ad5GL by tenfold.12,13 In contrast, predosing did not increase liver expression by Ad5/6GL. Liver tissue from mice injected with varying doses of Ad vectors was harvested analyzed by real-time PCR for viral genomes. Despite dramatic changes in liver expression, numbers of viral genomes 1 hour (Figure 3c) and 24 hours (data not shown) between the two vectors were similar.
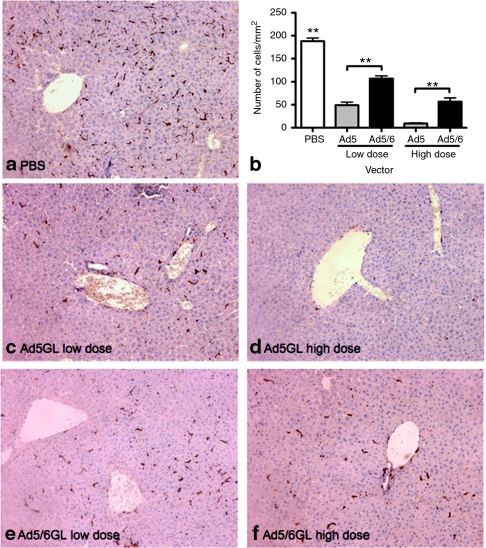
Elimination of Kupffer cells by predosing above suggested that Ad5 was being trapped by Kupffer cells while Ad5/6 is not. Since it has been previously shown that Ad5 subsequently causes extensive Kupffer cell depletion, we hypothesized that Ad5/6 would induce less Kupffer cell death. To test this, mice were injected i.v. with phosphate-buffered saline (PBS), a low dose (1 × 1010 vp) or a high dose (3 × 1010 vp) of either Ad5 or Ad5/6. Six hours later, the large liver lobes of the liver were harvested and immunohistochemistry for F4/80 was performed to detect Kupffer cells (Figure 4). Six hours after injection, the high dose of Ad5 caused a 20-fold reduction in F4/80+ cells while Ad5/6 caused only a threefold decrease (Figure 4b). These data suggested that in the hours after injection, Ad5/6 causes less loss of Kupffer cells from the liver than Ad5.
Figure 4.
Ad5/6 depletes Kupffer cells less than Ad5. BALB/c mice were injected intravenous (i.v.) with phosphate-buffered saline, a low dose [1 × 1010 virus particles (vp)], or a high dose (3 × 1010 vp) of adenovirus (Ad) vectors. Six hours later, the large lobe of the liver was harvested, paraffin embedded and sectioned (4 µm thick). (a,c–f) Immunohistochemical staining of Kupffer cells (anti-F4/80 macrophage marker). (b) Quantification of F4/80 positive cells 6 hours after i.v. injection of Ad vectors. **P < 0.005; ns = not significant. Means ± SEM.
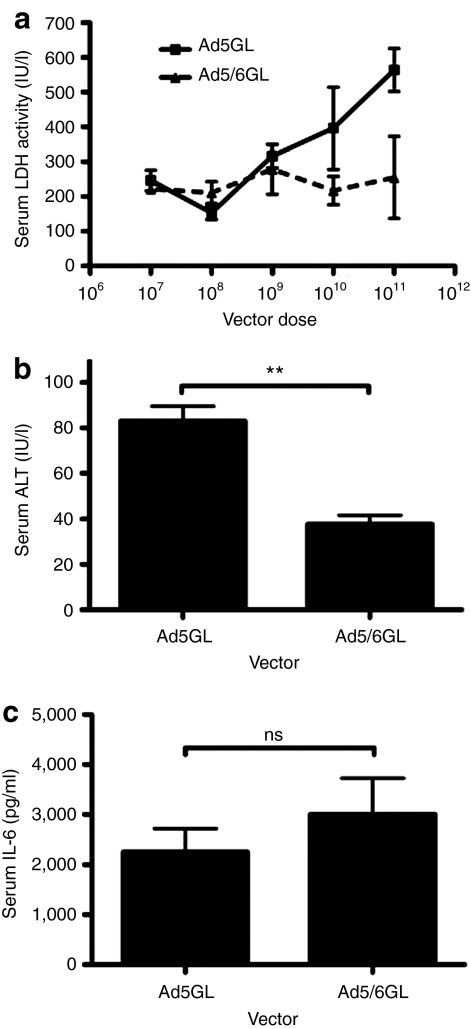
When Kupffer cells are killed by Ad5, they release lactate dehydrogenase (LDH) into the blood within 30 minutes of virus injection.11 To test this, Ad5 and Ad5/6 were injected at increasing doses into mice and LDH levels were measured in serum harvested 30 minutes later (Figure 5a). Under these conditions, LDH levels rose with increasing doses of Ad5GL virus. In contrast, even the highest dose of Ad5/6GL did not increase LDH levels, indicating that Kupffer cells remained viable at least at 30 minutes. The threefold loss of F4/80+ cells observed in Figure 4 at 6 hours suggests that Ad5/6 may cause lower toxicity than Ad5 and that this may manifest later than 30 minutes after injection.
Figure 5.
Ad5/6 demonstrates reduced toxicity in vivo. (a) BALB/c mice were injected intravenous with up to 1 × 1011 virus particles (vp) of adenovirus (Ad) vectors. Serum was harvested 30 minutes later and analyzed for lactate dehydrogenase levels. N = 3. BALB/c mice were injected intravenously with 2.5 × 1011 vp of Ad vectors. (b) Serum was harvested at 72 hours and assayed for alanine aminotransferase (ALT) levels. N = 3. (c) Serum was harvested at 6 hours and assayed for interleukin-6 levels (IL-6). N = 3. **P < 0.005; ns = not significant. Means ± SEM.
Ad5/6GL reduces toxicity
Ad5/6 appears to cause less Kupffer cell death which might impact the release of inflammatory mediators by these phagocytic cells and reduce liver damage. To test this, high doses of each vector (2.5 × 1011 vp) were injected i.v. into BALB/c mice and blood was harvested at 72 hours later to measure hepatocyte damage by release of alanine aminotransferase (ALT) into the blood. While both viruses produced relatively low liver damage (Figure 5b), ALT levels were significantly lower by Ad5/6GL than Ad5GL. Changes in Kupffer cell uptake and systemic cell transduction might also modify the innate immune response to the viruses. To test this, blood was also drawn 6 hours after injection of the high dose viruses and interleukin-6 (IL-6) levels were assayed (Figure 5c). In this case, IL-6 levels were similar suggesting that systemic innate immune responses are not overtly different between Ad5 and the Ad5/6 chimera.
Ad5/6GL does not increase liver expression in C57BL/6 mice
C57BL/6 mice are a fortuitous negative control for Kupffer effects on Ad. When C57BL/6 mice are injected with equal doses of Ad5, gene expression is significantly higher than in BALB/c mice. This effect is due in part to 20 times as many Ad genomes being sequestered in Kupffer cells in BALB/c mice than in C57BL/6 mice.17 Given this, we compared Ad5 and Ad5/6 in C57BL/6 mice after i.v. injection (Figure 6a). In these mice, the Ad5/6 chimera showed no enhanced luciferase activity consistent with reduced Kupffer cell uptake in these mice.
Figure 6.
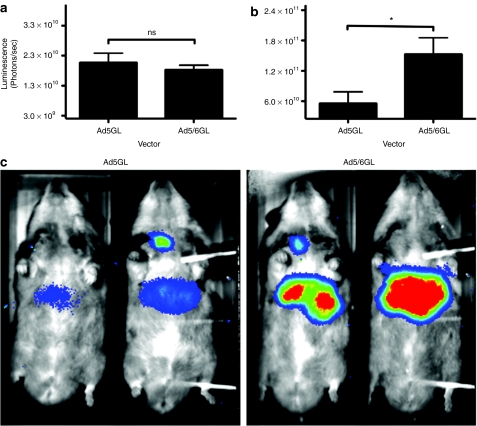
Effect of Ad5/6GL in C57Bl/6 mice and Syrian hamsters. 1 × 1010 virus particles (vp) of Ad5GL or Ad5/6GL was injected intravenously into C57BL/6 mice and intrajugularly into Syrian hamsters. (a) Mice were imaged for luciferase expression after 24 hours for 3 minutes at 2 × 2 binning (N = 10). (b,c) Hamsters were imaged after 4 days for 10 minutes at 2 × 2 binning (N = 4). Shown are (c) white light images overlaid with luciferase expression in pseudo-color from 688 to 1400 gray values and (b) quantification of luciferase expression. *P = 0.05; ns = not significant. Means ± SEM.
Ad5/6GL increases liver expression in hamsters
Syrian hamsters are thought to be an optimal model for Ad5 biology, since these animals support Ad5 infection better than mice.18 To determine whether Ad5/6 activity was a peculiarity of BALB/c mice, 1 × 1010 vp of Ad5 and Ad5/6 were injected intrajugularly into Syrian hamsters (Figure 6b,c). Consistent with the BALB/c model, Ad5/6GL generated threefold greater luciferase expression in the liver compared to Ad5GL. This data suggests that Kupffer cell uptake of Ad5 and improved transduction by Ad5/6 is a common effect across different species.
Helper-dependent Ad5, Ad6, and Ad5/6 vectors
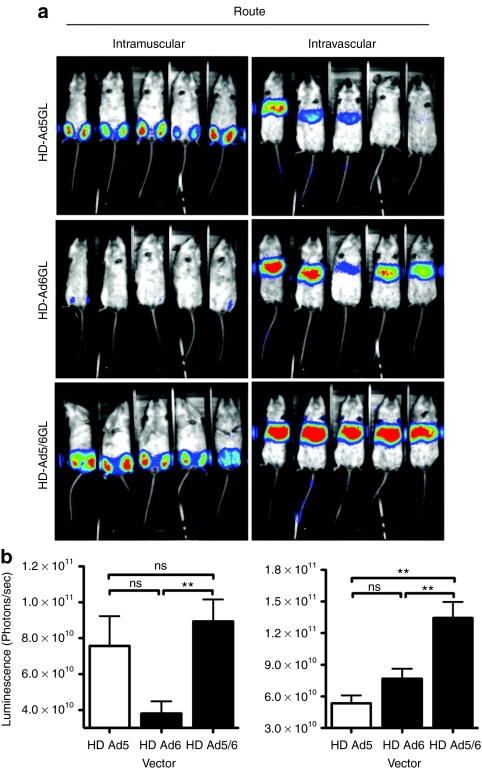
Helper-dependent adenoviral (HD-Ad) vectors have improved safety profiles and extended expression when compared to replication defective (RD) Ad vectors.19,20,21 To test whether the hexon chimera modified HD-Ad vector biology, an HD-Ad5 vector expressing eGFP-luciferase was packaged by Ad5, Ad5/6, and Ad6 helper viruses.22 In vitro comparison of transduction of A549 cells by HD-Ad5, Ad5/6, and Ad6 by luciferase assay confirmed that all three helper dependent vectors transduced A549 cells to a similar degree, although transduction was fourfold less than that the first generation Ad5 and Ad5/6 vectors (data not shown). HD-Ads were injected i.v. into BALB/c mice and imaged 24 hours later (Figure 7). Consistent with the RD vectors, HD-Ad5/6GL expressed 2.5-fold more luciferase in the liver than HD-Ad5. Interestingly, HD-Ad5/6 also demonstrated ~1.7-fold greater luciferase expression than HD-Ad6.
Figure 7.
In vivo expression of unmodified and chimeric helper dependent vectors. BALB/c mice were injected with 1 × 1010 virus particles (vp) of either HD-Ad5GL or HD-Ad5/6GL by different routes. Mice were imaged 24 hours later using a Lumazone imager for 3 minutes at 1 × 1 binning, 1,320–1,600 gray values. Shown are (a) white light images overlaid with luciferase expression in pseudo-color. (b) Quantification of luciferase expression. Intramuscular, left and intravascular, right. N = 5. **P < 0.005; ns = not significant. Means ± SEM.
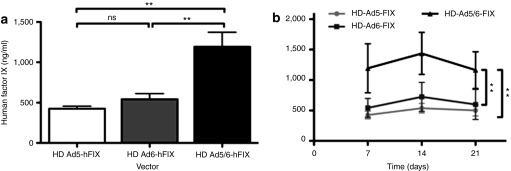
To test whether the chimera might have utility for liver gene therapy for hemophilia, a helper-dependent vector expressing human Factor IX (hFIX) transgene from a hepatocyte-specific promoter was also packaged with Ad5, Ad5/6, and Ad6 helper viruses. When injected into BALB/c mice, HD-Ad5/6-hFIX produced approximately threefold and approximately twofold higher levels of serum hFIX after 1 week when compared to both HD-Ad5 and HD-Ad6, respectively (Figure 8a). This statistical trend continues for at least 3 weeks (Figure 8b). These data suggest that the Ad5/6 chimeric virus may have utility for long-acting HD-Ad-based liver-directed gene therapy.
Figure 8.
Expression of hFIX from intravenous (i.v.) injection of helper dependent adenovirus (Ad) vectors. Helper dependent (HD) vectors were injected intravenously into BALB/c mice at 2.5 × 1010 virus particles (vp)/mouse. Serum was harvested on (a) day 7 and (b) up to day 21. Serum was analyzed for human Factor IX by enzyme linked imunosorbent assay. N = 5 for Ad5 and Ad5/6; N = 4 for Ad6. **P < 0.005; ns = not significant. Means ± SEM.
Discussion
It is widely known that Ad5 rapidly localizes to the liver after systemic administration, thus highlighting its potential use as a liver gene therapy vector. Ad5 hexon binds circulating FX with high affinity, which then acts as a “molecular bridge” for binding to hepatocytes.4,5,23 Another species C adenovirus, Ad6, was reported to have a similar affinity for FX,4 but a potentially different interaction with Kupffer cells.13 We genetically replaced the Ad5 HVR cassette with that of Ad6 in order to study the role of this region in liver transduction.
When injected i.v. into BALB/c mice, Ad5/6 exhibited a tenfold increase in liver expression compared to Ad5, but lower expression when injected i.m. or i.n. In vitro functional assays confirmed the role of vitamin K dependent blood factors in both Ad5 and Ad5/6 hepatocyte transduction, and in vivo ablation of blood factors with warfarin reduced liver transduction for both vectors. This implies the necessity of FX binding for liver tropism, although despite the injection of warfarin, Ad5/6 expression remained higher than that of Ad5. Moreover, surface plasmon resonance (SPR) analysis revealed that Ad6 and Ad5/6 had lower affinity of for FX compared to Ad5. These results indicate that the interaction of Ad5/6GL with blood factors is just sufficient for cell binding and liver tropism, but is unlikely to be the underlying cause of increased liver transduction.
Within the liver, Kupffer cells take up the majority of systemically injected Ad5 via scavenger receptors.8,24 Scavenger receptors are thought to recognize charge clusters,25 it is therefore interesting that 20% of the amino acids in Ad5 HVR loops is either negatively charged (D, E) or positively charged (H, K, R). For example, HVR1 has a net negative charge of 13. Conversely, Ad6 HVR1 has net negative charge of only eight or five fewer than Ad5. When all 720 hexons are considered in aggregate, we hypothesized that this 40% decrease in charge could significantly decrease recognition of Ad5/6 by Kupffer cells.
Consistent with this, Ad5/6 was phagocytosed 50% less efficiently than Ad5 by macrophages in vitro. In vivo, we show by three assays that Ad5/6 interacts with Kupffer cells to a lower degree than Ad5. First, eliminating Kupffer cells by predosing before Ad vector injection conferred no increase in liver transduction for Ad5/6, while Ad5 received a marked boost. Second, we show by immunohistochemistry that a high dose of Ad5 depletes Kupffer cells dramatically by 20-fold, while Ad5/6 decreases Kupffer cells by only threefold. Finally, we show Ad5 killed Kupffer cells in a dose-dependent fashion as measured by LDH release. On the other hand, Ad5/6 generated constant levels of LDH even at high doses, although increased variability in LDH measurements at higher doses may indicate some Kupffer cell killing. These data indicate that the Ad6 HVRs appear to interact to a significantly lesser degree with Kupffer cells in vivo.
Ad5 and Ad5/6 were compared in RD vectors, but the effect of RD-Ad6 was not measured due to the lack of a RD Ad6 vector containing the luciferase transgene. Instead, the Ad5, 6, and 5/6 were compared in HD-Ad vectors. A luciferase assay showed that HD-Ad vectors showed at least twofold decreased transduction in vitro (data not shown). Nevertheless, HD-Ad5/6 demonstrated significantly increased liver expression compared to Ad5, although the fold increase was not as large. It was interesting to observe that Ad5/6 was also better at transducing liver than Ad6. This may have resulted from the longer fiber shaft of the Ad5 fiber, which is generally more efficient compared to the shorter fiber of Ad6.14,26 Fiber shaft length may also explain why HD-Ad6 exhibited such poor expression in muscle compared to HD-Ad5 or HD-Ad5/6.
Ad5/6 showed markedly increased expression in BALB/c mice and Syrian hamsters. Interestingly, Ad5/6 had no benefit in C57BL/6 mice. Recently, Vigant et al. also found differences in behavior between hexon-modified Ad5 vectors in these two mice strains.27 Snoeys et al. determined that the overall number of Kupffer cells between the two mouse strains is not significantly different, but consistent with our data, BALB/c mice take up approximately sixfold more Ad DNA in their nonparenchymal cells than C57BL/6 mice, although both mice take up large quantities of virus.17 When analyzed by cell type, they showed that BALB/c Kupffer cells took up ~20 times more Ad DNA than C57BL/6 cells after an i.v. injection of the vector. Since endothelial cells also express scavenger receptors,28,29 fundamental differences in the location, types, and/or density of these receptors between animals may also explain why Ad5 and Ad5/6 expressed similarly in C57BL/6 mice. Variations in scavenging and sequestration of vectors beyond Kupffer cells (i.e., in hepatocytes endothelial cells, blood cells, etc.) may also explain why the ALT hepatotoxicity levels were twofold greater in Ad5 compared to Ad5/6.
When adenoviruses from species B, C, and D were tested for Kupffer uptake and hepatocyte infection, the species B and D viruses appeared to evade both cell types.13 While this makes them potentially excellent for systemic therapies that need to avoid the liver, it also obviates their use for efficient liver-directed gene therapy. Conversely, species C viruses are able to infect hepatocytes, but are also absorbed by Kupffer cells to varying degrees. The Ad5/6 chimera appears to bridge these two biologies by evading Kupffer cells while still being able to efficiently infect liver hepatocytes. Evading Kupffer cells and macrophages is likely generally advantageous for improving virus pharmacology for liver and systemic therapy. Since dendritic cells also express scavenger receptors,30,31 evading phagocytosis by any antigen presenting cell may reduce adaptive responses against Ad and its transgene products.
Materials and Methods
Cell culture. Human embryonic kidney cells (293) were purchased from Microbix (Toronto, Ontario, Canada). A549 Human Lung Carcinoma cells (ATCC CCL-185; ATCC, Manassas, VA) and RAW 264.7 murine monocyte-macrophage cells (ATCC CRL-2278) were purchased from the American Type Culture Collection (ATCC). All cells were maintained in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal calf serum (Gibco, Carlsbad, CA) and penicillin/streptomycin at 100 U/ml (Gibco).
Adenoviruses. Ad5GL and Ad5dsRed: First-generation, replication-defective (E1 and E3 deleted) Ad5 vectors carrying enhanced green fluorescent protein and luciferase fusion gene (GL) or dsRed were produced with the AdEasy system (Qbiogene, Irvine, CA/MP Biomedicals, Solon, OH) in 293 cells as described previously.16
Ad6: Wild type Ad6 (ATCC).
Ad5/6GL: Ad6 viral genomic DNA was purified using a PureLink Viral RNA/DNA Mini Kit (Invitrogen, Carlsbad, CA) and the HVR cassette was amplified by PCR (F primer 5′-TACTTTGACATCCGCGGCGTGCTGG-3′ and R primer 5′-GTGGGGCCATGGGGAAGAAGGTGGCG-3′), cut with ApaI and PstI and cloned into the Ad5GL backbone. 293 cells were transfected with the full-length recombined construct using Polyfect reagent (Qiagen, Valencia, CA). All viruses were purified on two consecutive CsCl gradient centrifugations, desalted using Econopac 10-DG chromatography columns (Bio-Rad, Hercules, CA), resuspended in 0.5 mol/l sucrose in KPBS and stored at −80 °C. Viral particle concentration was determined by A260 measurements. Viral plaque forming units and TCID50 was determined by serial dilution of virus on 293 cells. Typical yields for Ad5 and Ad5/6 were 3.4 × 1012 and 6.4 × 1011 vp/ml. Particle/plaque forming units (TCID50) ratios were 108 and 404 for Ad5GL and Ad5/6GL, respectively.
Helper-dependent adenoviral vectors: The chimeric helper virus AdNG177 contains the Ad6 HVR region in an otherwise serotype 5 genome and was constructed by replacing the 6710 bp Sfi I fragment in the serotype 5 helper virus AdNG16332 with the 6815 bp Sfi I fragment from Ad5/6GL. HDAd5/6-GL was produced by coinfecting 116 cells with AdNG177 and HD5-GL22 as described in detail elsewhere.33 HDAd5/6-hFIX was produced by coinfecting 116 cells with AdNG177 and HDD21.7E4PEPCK-hFIX-WL34 as described in detail elsewhere.33 All vector particle numbers were determined by absorbance at 260 nm and confirmed by real-time PCR. Transduction of different vectors was compared by luciferase assay.
Animals. Female BALB/c, C57BL/6 or male Syrian hamsters were purchased from Harlan Sprague-Dawley (Indianapolis, IN). They were housed in the Mayo Clinic Animal Facility under the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC) guidelines with animal use protocols approved by the Mayo Clinic Animal Use and Care Committee. All animal experiments were carried out according to the provisions of the Animal Welfare Act, PHS Animal Welfare Policy, the principles of the NIH Guide for the Care and Use of Laboratory Animals, and the policies and procedures of Mayo Clinic. Blood was collected from the cheek in BD Microtainer tubes and serum was separated by centrifugation for 1 minutes at 16,000g.
Ad injections. All viruses were diluted in PBS but were brought up in different volumes depending on the route of injection. For mice: i.n. (10 µl in left nare), i.m. (50 µl total, 25 µl per hind leg muscle), and i.v. (100 µl in the tail vein). In hamsters: i.m (200 µl total, 100 µl per hind leg muscle), and i.j. (intrajugular; 200 µl in the jugular). For predosing, mice were first injected i.v. with 3 × 1010 vp of Ad5dsRed in 100 µl of PBS 4 hours before i.v. injection of 1 × 1010 vp of Ad5GL or Ad5/6GL. For toxicity experiments, 2.5 × 1011 vp was injected i.v. For LDH assay, up to 1 × 1011 vp was injected i.v. For FIX experiments, 2.5 × 1010 vp was injected i.v. For all other experiments, mice were injected with a total viral dose of 1 × 1010 vp.
Warfarinization. Mice were injected subcutaneously with 133 µg warfarin in 100 µl peanut oil both 3 days and 1 day prior to i.v. injection of Ad.
In vivo bioluminescence imaging. Mice and hamsters were anesthetized with ketamine/xylazine and injected intraperitoneally with 100 µl (for mice) or 200 µl (for hamsters) of -luciferin (20 mg/ml; Molecular Imaging Products, Bend, OR). Animals were imaged on the Lumazone Imagine System (Photometrics, Roper Scientific, Tucson, AZ) for 1–10 minutes with 1 × 1 or 2 × 2 binning using no filters or photomultiplication. Lumazone imaging software was used to determine total light intensity per mouse (photons/second) for data analysis.
SPR analyses. FX protein was purchased from Hematologic Technologies (Essex Junction, VT). Ad5GL and Ad5/6GL hexon was purified from supernatants of CsCl virus preparations. Hexon protein was further purified and buffer exchanged on a 10G and then G-75 Sephadex column (Bio-Rad) into Hank's buffered salt solution. Hexon and FX protein was immobilized onto research grade CM5 sensor chips using the Biacore 2000 instrument and the amine coupling reagents (Biacore, Pittsburgh, PA). All data was collected in replicates of three for each concentration of analyte.
In vitro luciferase assay. To detect luciferase expression in vitro, CHO, CHO-CAR, SKOV3, and RAW 264.7 cells were plated to 80–90% confluency in 96-well plates and infected with varying multiplicities of infection of Ad vectors. After a 24 hours incubation at 37 °C, cells were lysed with cold passive lysis buffer (Promega, Madison, WI). Luciferase assay reagent was added and relative luminescence units was measured with the Beckman Coulter DTX 880 Multimode Detector system.
Uptake of virus into RAW 264.7. RAW 264.7 cells were seeded to 85–90% confluency in 24 well plates and virus was applied to cells at an multiplicity of infection 10,000 for 24 hours. Media and noncell bound virus was removed and cells were harvested. DNA was extracted using the DNeasy Blood and Tissue kit according to the manufacturer's protocol (Qiagen) with an RNase A digestion. Real-time PCR was performed as described below for both viral [cytomegalovirus (CMV)] and cellular (GAPDH) genomic targets. Reactions were performed in triplicate with uninfected cellular DNA as the negative control.
In vivo vector genome quantification. Mice were injected with varying doses of Ad vectors. After 1 or 24 hours, animals were euthanized and 20 ng of the large liver lobe was harvested. DNA was extracted using the DNeasy Blood and Tissue kit according to the manufacturer's protocol (Qiagen) with an RNase A digestion. Real-time PCR was performed as described below for both viral (CMV) genomic targets. Reactions were performed in triplicate with a PBS injected mouse as the negative control.
Quantitative real-time PCR. Concentration of DNA samples was determined by Abs260 and then diluted to 20 ng/µl. Real-time PCR was performed on the DNA in an Applied Biosystems Prism 7900HT sequence detection system with SDS 2.3 software. Each well contained 10 µl Sybr Green (Applied Biosystems, Warrington, UK), 1 µl H2O, 2 µl of 3 µmol/l F Primer, 2µl of 3 µmol/l R Primer, and 5 µl sample (i.e., 100 ng DNA/well). The parameters used were one cycle of 95 °C for 15 minutes, and 40 cycles of 95 °C for 15 seconds, 55 °C for 1 minute, and 72 °C for 30 seconds. To normalize data, PCR was done for both the cellular housekeeping gene GAPDH (using previously published F primer 5′-CGTGTTCCTACCCCCAATGT-3′ and R primer 5′-TGTCATCATACTTGGCAGGTTTCT-3′) and viral CMV gene (F primer 5′-CAAGTGTATCATATGCCAAGTACGCCCC-3′ and R primer 5′-CCCCGTGAGTCAAACCGCTATCCACGCC-3′).16 Viral DNA was normalized to cellular DNA by dividing CMV genomes by GAPDH genomes.
Immunohistochemistry. BALB/c mice were injected i.v. with PBS or Ad vectors at a low dose (1 × 1010 vp) or at a high dose (3 × 1010 vp). After 6 hours, the large liver lobe was harvested and incubated over night at room temperature in formalin. Tissue was embedded in paraffin and cut to 4 µm sections. These were probed with rat anti-F4/80 primary (MCA497EL; AbD Serotec, Raleigh, NC) at a 1:200 dilution and ah horseradish peroxidase-conjugated anti-rat secondary, with DAB substrate. Quantifying positively stained cells: Images of five fields of view for each mouse section were taken. The ImageJ software was used to convert them to binary images, and the “Threshold” feature was used to separate out positively stained cells from background nuclei. “Analyze particles” was used to count numbers of stained regions (25-infinity).
Determination of serum IL-6, ALT, and hFIX levels. Serum was collected at 6 hours after injection and IL-6 levels were quantified using an OptEIA ELISA kit (BD Biosciences, San Diego, CA) according to the manufacturer's protocol. The amount of IL-6 was determined by measuring absorbance at 450 nm and compared to a standard curve with known IL-6 concentration. Serum was collected at 72 hours and ALT was quantified using the Alanine Aminotransferase kit (Biotron Diagnostics, Hemet, CA) according to the manufacturer's protocol. Serum was collected 7 days after injection and hFIX levels were analyzed by ELISA (Enzyme Research Laboratories, South Bend, IN) according to the manufacturer's protocol.
LDH assay. Serum was collected from BALB/c mice 30 minutes after i.v. injection with varying doses of Ad vector. LDH levels were measured within 24 hours using the Liquid Reagent kit (Pointe Scientific, Canton, MI) and decreasing all volumes by fivefold due to small serum collections. LDH activity was measured by incubation at 37 °C and reading the absorption at 340 nm with a spectrophotometer.
Data analysis. Graphs and statistical analyses were performed using Prism Graphical software. Analysis between two and three groups was determined by unpaired t-test, and one-way ANOVA (Newman–Keuls post-test), respectively.
SUPPLEMENTARY MATERIAL Figure S1. Design and characterization of adenoviral constructs. (a) Comparison of Ad5 and Ad6 HVR cassette sequence between the AvrII and PstI cloning restriction sites. Blue circles = negatively amino acids (asp, glu); Red triangles = positively charged amino acids (arg, his, lys). (b) Schematic of replication defective Ad5/6GL chimera with Ad6 HVR cassette in Ad5 backbone. eGFPLuciferase is inserted into the E1 region under a CMV promoter. ITR = Inverted terminal repeat. Means + SEM. Table S1. Ad5/6GL grows to lower particle and infectious unit titers compared to Ad5GL. Table S2. Ad5/6 hexon has lower affinity for Factor X (FX) compared to Ad5 hexon.
Acknowledgments
We would like to thank Mary Barry for her helpful technical assistance. This work was supported by a grant to M.A.B. from NIH (R01-CA136945) and a grant to P.N. from NIH (R01-DK067324). We thank the Mayo Clinic Cancer Center for the use of the TACMA Shared Resource, which provided immunohistological services. Mayo Clinic Cancer Center is supported in part by an NCI Cancer Center Support Grant SP30 CA1503-37. We also thank Mayo Clinic Department of Histology and Pathology for tissue embedding services.
Supplementary Material
Design and characterization of adenoviral constructs. (a) Comparison of Ad5 and Ad6 HVR cassette sequence between the AvrII and PstI cloning restriction sites. Blue circles = negatively amino acids (asp, glu); Red triangles = positively charged amino acids (arg, his, lys). (b) Schematic of replication defective Ad5/6GL chimera with Ad6 HVR cassette in Ad5 backbone. eGFPLuciferase is inserted into the E1 region under a CMV promoter. ITR = Inverted terminal repeat. Means + SEM.
Ad5/6GL grows to lower particle and infectious unit titers compared to Ad5GL.
Ad5/6 hexon has lower affinity for Factor X (FX) compared to Ad5 hexon.
REFERENCES
- Parker AL, Waddington SN, Nicol CG, Shayakhmetov DM, Buckley SM, Denby L.et al. (2006Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes Blood 1082554–2561. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov DM, Gaggar A, Ni S, Li ZY., and, Lieber A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J Virol. 2005;79:7478–7491. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofherr SE, Shashkova EV, Weaver EA, Khare R., and, Barry MA. Modification of adenoviral vectors with polyethylene glycol modulates in vivo tissue tropism and gene expression. Mol Ther. 2008;16:1276–1282. doi: 10.1038/mt.2008.86. [DOI] [PubMed] [Google Scholar]
- Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H.et al. (2008Adenovirus serotype 5 hexon mediates liver gene transfer Cell 132397–409. [DOI] [PubMed] [Google Scholar]
- Kalyuzhniy O, Di Paolo NC, Silvestry M, Hofherr SE, Barry MA, Stewart PL.et al. (2008Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo Proc Natl Acad Sci USA 1055483–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba R, Bradshaw AC, Parker AL, Bhella D, Waddington SN, Nicklin SA.et al. (2009Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer Blood 114965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K, Matsuzaki Y, Hongo S, Ohmi A, Okamoto M, Nishimura H.et al. (2009Stability of the seven hexon hypervariable region sequences of adenovirus types 1-6 isolated in Yamagata, Japan between 1988 and 2007 Virus Res 14032–39. [DOI] [PubMed] [Google Scholar]
- Alemany R, Suzuki K., and, Curiel DT. Blood clearance rates of adenovirus type 5 in mice. J Gen Virol. 2000;81 Pt 11:2605–2609. doi: 10.1099/0022-1317-81-11-2605. [DOI] [PubMed] [Google Scholar]
- Jacobs F, Wisse E., and, De Geest B. The role of liver sinusoidal cells in hepatocyte-directed gene transfer. Am J Pathol. 2010;176:14–21. doi: 10.2353/ajpath.2010.090136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilzer M, Roggel F., and, Gerbes AL. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006;26:1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- Manickan E, Smith JS, Tian J, Eggerman TL, Lozier JN, Muller J.et al. (2006Rapid Kupffer cell death after intravenous injection of adenovirus vectors Mol Ther 13108–117. [DOI] [PubMed] [Google Scholar]
- Shashkova EV, Doronin K, Senac JS., and, Barry MA. Macrophage depletion combined with anticoagulant therapy increases therapeutic window of systemic treatment with oncolytic adenovirus. Cancer Res. 2008;68:5896–5904. doi: 10.1158/0008-5472.CAN-08-0488. [DOI] [PubMed] [Google Scholar]
- Shashkova EV, May SM., and, Barry MA. Characterization of human adenovirus serotypes 5, 6, 11, and 35 as anticancer agents. Virology. 2009;394:311–320. doi: 10.1016/j.virol.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov DM., and, Lieber A. Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol. 2000;74:10274–10286. doi: 10.1128/jvi.74.22.10274-10286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SN, Parker AL, Havenga M, Nicklin SA, Buckley SM, McVey JH.et al. (2007Targeting of adenovirus serotype 5 (Ad5) and 5/47 pseudotyped vectors in vivo: fundamental involvement of coagulation factors and redundancy of CAR binding by Ad5 J Virol 819568–9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok H, Palmer DJ, Ng P., and, Barry MA. Evaluation of polyethylene glycol modification of first-generation and helper-dependent adenoviral vectors to reduce innate immune responses. Mol Ther. 2005;11:66–79. doi: 10.1016/j.ymthe.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Snoeys J, Mertens G, Lievens J, van Berkel T, Collen D, Biessen EA.et al. (2006Lipid emulsions potently increase transgene expression in hepatocytes after adenoviral transfer Mol Ther 1398–107. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Spencer JF., and, Wold WS. Use of the Syrian hamster as an animal model for oncolytic adenovirus vectors. Methods Mol Med. 2007;130:169–183. doi: 10.1385/1-59745-166-5:169. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Cotter MJ, Zaiss AK, White LR, Liu Q, Chan T.et al. (2004Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo J Virol 785966–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Mack LM, Kelly R, Ontell M, Kochanek S., and, Clemens PR. Persistence in muscle of an adenoviral vector that lacks all viral genes. Proc Natl Acad Sci USA. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N, O'Neal W, Rice K, Leland M, Kaplan J, Piedra PA.et al. (1999Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons Proc Natl Acad Sci USA 9612816–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver EA, Nehete PN, Buchl SS, Senac JS, Palmer D, Ng P.et al. (2009Comparison of replication-competent, first generation, and helper-dependent adenoviral vaccines PLoS ONE 4e5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AH, Mcvey JH, Waddington SN, Di Paolo NC., and, Shayakhmetov DM. The influence of blood on in vivo adenovirus bio-distribution and transduction. Mol Ther. 2007;15:1410–1416. doi: 10.1038/sj.mt.6300206. [DOI] [PubMed] [Google Scholar]
- Haisma HJ, Boesjes M, Beerens AM, van der Strate BW, Curiel DT, Plüddemann A.et al. (2009Scavenger receptor A: a new route for adenovirus 5 Mol Pharm 6366–374. [DOI] [PubMed] [Google Scholar]
- Xu Z, Tian J, Smith JS., and, Byrnes AP. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J Virol. 2008;82:11705–11713. doi: 10.1128/JVI.01320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver EA, Hillestad ML, Khare R, Palmer D, Ng P., and, Barry MA. Characterization of species C human adenovirus serotype 6 (Ad6) Virology. 2011;412:19–27. doi: 10.1016/j.virol.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigant F, Descamps D, Jullienne B, Esselin S, Connault E, Opolon P.et al. (2008Substitution of hexon hypervariable region 5 of adenovirus serotype 5 abrogates blood factor binding and limits gene transfer to liver Mol Ther 161474–1480. [DOI] [PubMed] [Google Scholar]
- Adachi H., and, Tsujimoto M. Endothelial scavenger receptors. Prog Lipid Res. 2006;45:379–404. doi: 10.1016/j.plipres.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Ishii J, Adachi H, Shibata N, Arai H., and, Tsujimoto M. Scavenger receptor expressed by endothelial cells (SREC)-I interacts with protein phosphatase 1alpha in L cells to induce neurite-like outgrowth. Biochem Biophys Res Commun. 2007;360:269–274. doi: 10.1016/j.bbrc.2007.06.047. [DOI] [PubMed] [Google Scholar]
- Amiel E, Nicholson-Dykstra S, Walters JJ, Higgs H., and, Berwin B. Scavenger receptor-A functions in phagocytosis of E. coli by bone marrow dendritic cells. Exp Cell Res. 2007;313:1438–1448. doi: 10.1016/j.yexcr.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JO, Park HY, Xu Q, Park JI, Zvyagintseva T, Stonik VA.et al. (2009Ligand of scavenger receptor class A indirectly induces maturation of human blood dendritic cells via production of tumor necrosis factor-alpha Blood 1135839–5847. [DOI] [PubMed] [Google Scholar]
- Palmer DJ., and, Ng P. Physical and infectious titers of helper-dependent adenoviral vectors: a method of direct comparison to the adenovirus reference material. Mol Ther. 2004;10:792–798. doi: 10.1016/j.ymthe.2004.06.1013. [DOI] [PubMed] [Google Scholar]
- Palmer D., and, Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Palmer DJ, Mane V, Finegold M, Beaudet AL., and, Ng P. Increased hepatic transduction with reduced systemic dissemination and proinflammatory cytokines following hydrodynamic injection of helper-dependent adenoviral vectors. Mol Ther. 2005;12:99–106. doi: 10.1016/j.ymthe.2005.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design and characterization of adenoviral constructs. (a) Comparison of Ad5 and Ad6 HVR cassette sequence between the AvrII and PstI cloning restriction sites. Blue circles = negatively amino acids (asp, glu); Red triangles = positively charged amino acids (arg, his, lys). (b) Schematic of replication defective Ad5/6GL chimera with Ad6 HVR cassette in Ad5 backbone. eGFPLuciferase is inserted into the E1 region under a CMV promoter. ITR = Inverted terminal repeat. Means + SEM.
Ad5/6GL grows to lower particle and infectious unit titers compared to Ad5GL.
Ad5/6 hexon has lower affinity for Factor X (FX) compared to Ad5 hexon.