Abstract
Weak acids are widely used as food preservatives (e.g., acetic, propionic, benzoic, and sorbic acids), herbicides (e.g., 2,4-dichlorophenoxyacetic acid), and as antimalarial (e.g., artesunic and artemisinic acids), anticancer (e.g., artesunic acid), and immunosuppressive (e.g., mycophenolic acid) drugs, among other possible applications. The understanding of the mechanisms underlying the adaptive response and resistance to these weak acids is a prerequisite to develop more effective strategies to control spoilage yeasts, and the emergence of resistant weeds, drug resistant parasites or cancer cells. Furthermore, the identification of toxicity mechanisms and resistance determinants to weak acid-based pharmaceuticals increases current knowledge on their cytotoxic effects and may lead to the identification of new drug targets. This review integrates current knowledge on the mechanisms of toxicity and tolerance to weak acid stress obtained in the model eukaryote Saccharomyces cerevisiae using genome-wide approaches and more detailed gene-by-gene analysis. The major features of the yeast response to weak acids in general, and the more specific responses and resistance mechanisms towards a specific weak acid or a group of weak acids, depending on the chemical nature of the side chain R group (R-COOH), are highlighted. The involvement of several transcriptional regulatory networks in the genomic response to different weak acids is discussed, focusing on the regulatory pathways controlled by the transcription factors Msn2p/Msn4p, War1p, Haa1p, Rim101p, and Pdr1p/Pdr3p, which are known to orchestrate weak acid stress response in yeast. The extrapolation of the knowledge gathered in yeast to other eukaryotes is also attempted.
Introduction
Weak acids are used in food and chemical industries, in agriculture, and in medicine (Table 1). In the food industry, weak acids are used to control microbial growth (Table 1) (Lund and Eklund, 2000), while in the chemical industry these molecules can be used, as raw materials, for the synthesis of a wide range of products, from plastics to cosmetics and pharmaceuticals, replacing petrochemically derived products that have a more negative environmental impact (reviewed by Abbott et al., 2009). Weak acids are also used as drugs (e.g., artemisinic acid, artesunic acid, mycophenolic acid) or as pesticides (e.g., 2,4-dichlorophenoxyacetic acid, 2,4-D; 2-methyl-4-chlorophenoxyacetic acid, MCPA) (Table 1). The knowledge gathered on the mechanisms underlying Saccharomyces cerevisiae adaptation and resistance to weak acids that are used as food preservatives is expected to improve food and beverage preservation, limiting the activity of weak acid resistant fungi that include S. cerevisiae itself, Zygosaccharomyces bailii, Z. rouxii, and Aspergillus niger (Fleet, 2007). S. cerevisiae has also proven to be an invaluable model eukaryote to study the cytotoxic effects and the cellular responses to weak acids used as pharmaceuticals or pesticides. After more than a decade of postgenomic research, S. cerevisiae turned to be a powerful model system to increase our understanding of the effects and targets of drugs and of underlying resistance mechanisms on more complex and less accessible organisms. Although many of the specific targets of these drugs and pesticides do not exist in yeast, the mechanisms underlying basic cellular processes and chemical stress resistance are apparently conserved among phylogenetically distant organisms, making it possible to extrapolate the knowledge gathered in yeast to higher eukaryotes.
Table 1.
Chemical Structure, Dissociation Constants (pKa), Lipophilic Tendency (Given by the Value of the Logarithm of the Partition Coefficient of the Acid between Octanol and Water—log P), and Main Applications of the Referred Carboxylic Acids
| Acid | Structure | pKa | Log P | Applications | References |
|---|---|---|---|---|---|
| Acetic acid | 4.76 | −0.24 | Preservation of foods (mostly pickles and sauces) and soft drinks. Used in the synthesis of vinylacetate and ethylacetate that are precursors for the chemical synthesis of polymers | Mira et al., 2010; Almeida et al., 2009; Abbot et al., 2007; Mollapour and Piper, 2007; Fernandes et al., 2003; Simões et al., 2006; Kawahata et al., 2006; Tenreiro et al., 2002; Tenreiro et al., 2000; Carmelo et al., 1996; Pampulha and Loureiro-Dias, 1990 | |
| Propionic acid | 4.86 | −0.32 | Preservation of bakery and fresh dairy products | Mira et al., 2009; Gregori et al., 2008; Mollapour and Piper, 2007; Abbot et al., 2007; Fernandes et al., 2003; Simões et al., 2006; Piper et al., 1998,Kren et al., 2003 | |
| Octanoic acid | 4.89 | 3.05 | Preservation of acidic beverages and food products; surface sanitizer in food handling establishmentst and food processing equipment; precursor in the chemical synthesis of esteres used in manufacture of dyes | Viegas et al, 2005; Cabral et al., 2001; Viegas et al., 1998; Viegas et al., 1991; Stevens and Hoffmeyer et al., 1993; Sá-Correia et al., 1989 | |
| Decanoic acid | 4.9 | 4.09 | Food preservative; used in the manufacture of perfumes, dyes and plastics | Alexandre et al., 1996; Stevens and Hoffmeyer et al., 1993; Sá-Correia et al., 1989 | |
| Benzoic acid | 4.2 | 1.71 | Preservation of fruit juices, emulsified sauces, fish products and sugar jams | Piper et al., 1998; Holyoak et al., 1996; Holyoak et al, 1999; Pearce et al., 2001; Hatzixhantis et al., 2003; Simões et al., 2006, Kren et al., 2003 | |
| Sorbic acid | 4.76 | 1.63 | Preservation of cheeses, jams, and fish products | Gregori et al., 2008; Abbot et al., 2007; Makrantoni et al., 2007 Schuller et al., 2004; Mollapour et al., 2004; Mollapour et al., 2006 Hatzixhantis et al., 2003; Kren et al., 2003; Pearce et al., 2001; Piper et al., 1999; Holyoak et al., 1996; Holyoak et al, 1999 | |
| Lactic acid | 3.86 | −0.6 | Sweetner; preservation of beer and diary products; precursor in the synthesis of polyesters and acrylates | Abbot et al., 2008; Kawahata et al., 2006 | |
| α-hop-acids | 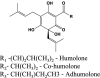 |
Humulone: 5.5 Cohumulone: 4.7 Adhumulone: 5.7 | n.d. | Used in beer production as flavoring and stability agents | Hazelwood et al., 2010 |
| 2,4-D | 2.73 | 2.8 | Auxin-like herbicide used in the control of broadleaf dicothyledon weeds | Teixeira et al., 2007; Teixeira et al., 2006a; Teixeira et al. 2005; Simões et al., 2003; Fernandes et al., 2003; Teixeira and Sá-Correia et al., 2002 | |
| Artesunic acid | 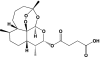 |
4.6 | 4 | Antimalarial and anticancer drug | Alenquer, et al., 2006 |
| Artemisinic acid | 4.34 | 2.9 | Used for the chemical synthesis of the antimalarial drug artemisinine | Ro et al., 2008 | |
| Mycophenolic acid | 4.5 | 3.88 | Immunosupressive drug used for the control of organ rejection in transplants | Desmoucelles et al., 2002; Escobar-Henriques et al., 2001; Saint-Marc et al., 2009 |
A list of references is also provided in which the mechanisms of adaptive response and resistance to these weak acids in yeast cells are addressed.
The accumulation in the growth medium of weak acids produced during yeast metabolism (such as acetic, butyric, and pyruvic acids, among others), together with the high concentrations of ethanol, leads to the growth arrest of fermenting cells and, eventually, to decreased productivity of wine fermentation processes (Gibson et al., 2007; Rasmussen et al., 1995). The production of bioethanol from cellulosic acid-hydrolysates by yeast cells is also limited by the presence of high concentrations of acetic acid and other weak acids (Palmqvist and Hahn-Hägerdal, 2000). Similarly, the large-scale production of weak acids is limited by their accumulation in the growth medium (Abbott et al., 2009). Although S. cerevisiae does not produce large quantities of organic acids, it is considered a very good alternative as a host cell for such processes because it has a lower nutrient requirement and is more tolerant to these acids than the prokaryotes often used (Abbott et al., 2009). The elucidation of the molecular mechanisms underlying S. cerevisiae tolerance to weak acid stress is therefore an essential step for the rational design of fermentation conditions and to guide the engineering of more robust industrial yeast strains.
This review focuses the genomic response and the determinants of tolerance of S. cerevisiae to a number of monocarboxylic weak acids with different chemical structures and lipophilicities, with application in industrial, agricultural, and medical fields (listed in Table 1). Particular emphasis has been given to results obtained from genome-wide approaches, in particular, chemical genomics, transcriptomics, and proteomics analyses, to get a clear-cut picture of the global response to weak acid stress in yeast cells.
Toxicity of, and Adaptive response to, Weak Acids in Yeast: An Overview
The antimicrobial potential of carboxylic acids is essentially determined by their chemical properties and, in particular, by their hydrophobicity, volatility, and pKa. At external pH below the weak acid pKa value, the lipophilic undissociated form of the acid (RCOOH) predominates and may permeate the plasma membrane by simple diffusion (Fig. 1A). It has recently been described that acetic acid can also enter the yeast cells by a process of facilitated diffusion, mediated by the aquaglyceroporin Fps1p (Mollapour and Piper, 2007). Once in the near-neutral cytosol, the chemical dissociation of the weak acid occurs leading to the release of protons (H+) and of the respective counterion (RCOO−). Due to their electric charge, these ions are not able to cross the hydrophobic lipid plasma membrane bilayer and accumulate in the cell interior (Fig. 1A). Therefore, the antimicrobial activity of weak acids at low pH relies on the effects of the undissociated acid form. Depending on the medium pH and the weak acid pKa, the concentration of the toxic form may reach a value close to the total acid concentration. As they are difficult to separate, the antimicrobial effect of the undissociated form of weak acids and the effect of low pH itself are frequently confused. When a strong acid is used as the acidulant a high concentration of protons is generated in the external environment that may affect the cell wall structure and alter the conformation of proteins protruding from the plasma membrane (Booth and Statford, 2003). However, because protons diffuse poorly across the plasma membrane, the inhibitory potential of low pH is much lower than the one exerted by weak acids that dissociate directly in the cytosol (Carmelo et al., 1996). Moreover, besides affecting internal pH homeostasis, weak acids also have an impact on the lipid organization and function of cellular membranes, consistent with their strong propensity to become more inhibitory as they become more hydrophobic (Fernandes et al., 2005; Piper et al., 1998; Stratford and Anslow 1996). As a consequence of this membrane deleterious effect, the nonspecific permeabilization of plasma and vacuolar membranes is registered in weak acid-challenged cells (Sá-Correia et al., 1989; Stevens and Hofemyer, 1993; Teixeira et al., 2005), as well as the perturbation of the function of membrane embedded proteins. The loss of plasma membrane integrity increases cell permeability to ions and other small metabolites leading to the stimulation of passive diffusion of protons from the exterior to the cytosol, this contributing to the reduction of internal pH in weak acid-challenged cells (Sá-Correia et al., 1989; Stevens and Hofemyer, 1993; Teixeira et al., 2005) and to the dissipation of the electrochemical potential maintained across this membrane, a driving force for secondary transport (Fig. 1A).
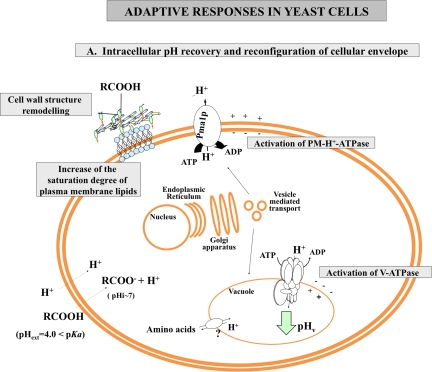
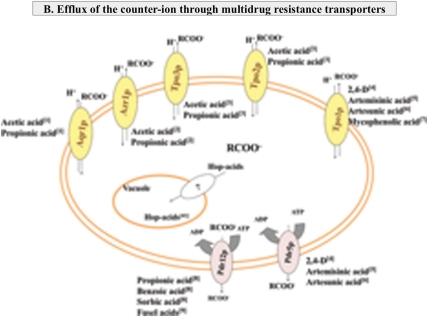
FIG. 1.
Mechanistic model for the adaptive yeast response to weak acid-induced stress. (A) Stimulation of the activity of H+-ATPases present in the plasma and vacuolar membranes contribute to the recovery of internal pH (pHi) to more physiological values and for metabolite compartmentalization in acid-stressed cells. The reconfiguration of cell wall structure and plasma membrane lipid composition may reduce the diffusion rate of undissociated weak acids and reduce the weak acid-induced plasma membrane damage. (B) Detoxification through multidrug resistance (MDR) transporters of the ATP-binding cassette (ABC) (in yellow) and Major Facilitator Superfamily (MFS) (in pink) is required to reduce the internal concentration of the weak acid counterion. The substrates established for each MDR transporter are indicated based on the information available in the literature. 1) Tenreiro et al., 2000; 2) Tenreiro et al., 2002; 3) Fernandes et al., 2003; 4) Teixeira and Sá-Correia, 2002; 5) Ro et al., 2008; 6) Alenquer et al., 2006; 7) Desmoucelles et al., 2002; 8) Piper et al., 1998; 9) Hazelwood et al., 2006.
When an inhibitory concentration of a weak acid is added to an exponentially growing yeast culture, growth arrest occurs and the cell population enters in a period of growth latency. During this lag phase, a reduction of cell viability is often registered but, depending on the level of weak acid-induced stress and on the severity of growth inhibition, exponential growth may be resumed after a more or less extended period of time. However, when these adapted cells are reinoculated into a fresh-growth medium at an identical pH and supplemented with the same weak acid concentration, no delay in cell growth is observed (Cabral et al., 2001; Teixeira and Sá-Correia, 2002; Viegas et al., 1998). During the referred adaptation period a number of molecular responses are activated these being detailed in the following sections.
Intracellular acidification and pH recovery
Intracellular acidification, caused by the dissociation of a weak acid in the neutral cytosol and by increased H+ influx rate prompted by the acid-induced membrane permeabilization, leads to decreased DNA and RNA synthesis rate, inhibited metabolic activity and disruption of the proton gradient maintained across the plasma membrane (Booth and Statford, 2003; Holyoak et al., 1996; Krebs et al., 1983; Pampulha and Loureiro-Dias, 1990). To avoid the dissipation of plasma membrane potential and to maintain the internal pH within physiological values, yeast cells rely on the stimulation of the activity of the plasma membrane H+-ATPase (PM-H+-ATPase), Pma1p, which couples ATP hydrolysis to proton extrusion (Fig. 1A). PM-H+-ATPase activity is increased in response to octanoic acid (Viegas and Sá-Correia, 1991), sorbic acid (Holyoak et al., 1996), decanoic acid (Alexandre et al., 1996), acetic or succinic acids (Carmelo et al., 1997), and 2,4-D (Fernandes et al., 2003), presumably to counteract intracellular acidification and the dissipation of plasma membrane potential induced by these lipophilic weak acids (Fig. 1A). Mounting evidences, particularly those coming from chemical genomic analysis, support the idea that the H+-ATPase present in the vacuolar membrane, the V-ATPase, is also crucial for pHi homeostasis under weak acid stress (Desmoucelles et al., 2002; Fernandes et al., 2003; Kawahata et al., 2006; Makrantoni et al., 2007; Mira et al., 2009; Mollapour et al., 2004; Schuller et al., 2004), its activity also being suggested and proved to increase in response to various weak acids (Carmelo et al., 1997; Fernandes et al., 2003; Makrantoni et al., 2007). By sequestering protons into the lumen of the vacuole, V-ATPase activity contributes to the recovery of cytosolic pH and to counteract the acid-induced dissipation of the transmembrane potential across the vacuolar membrane (Fig. 1A). V-ATPase function is also crucial for the normal function of other physiological processes (reviewed by Kane, 2006) which, based on the results gathered from large-scale chemical genomics screenings (see below), are required for maximal tolerance to weak acid stress, such as endocytosis, targeting of newly synthesized lysosomal enzymes, protein processing, intracellular trafficking, and Pma1p sorting to the plasma membrane (Fernandes et al., 2003; Mira et al., 2009; Mollapour et al., 2004). An expression proteomic analysis has demonstrated that vacuolar compartimentalization of amino acids occurring in cells stressed with the acid herbicide 2,4-D is dependent on a functional V-ATPase (Teixeira et al., 2005), suggesting that this proton pump may also play a role in the compartmentalization of metabolites in response to weak acid stress (Fig. 1A).
Detoxification through multidrug resistance transporters
The accumulation of weak acid counterions in the cell interior may lead to an increase in turgor pressure, oxidative stress, protein aggregation, lipid peroxidation, inhibition of membrane trafficking, and perturbation of plasma and vacuolar membranes spatial organization (reviewed by Piper et al., 2001; Teixeira et al., 2007). The reduction of the intracellular pool of the weak acid counterion(s) is therefore essential, and several specific transporters have been implicated in yeast tolerance to different weak acids, in particular, those transporters related to multidrug resistance (MDR). Among them, Pdr12p, a plasma membrane transporter of the ATP-binding cassette (ABC) superfamily has been more thoroughly characterised, the results obtained demonstrating its involvement in the active efflux of propionate, sorbate, or benzoate anions (Holyoak et al., 1999; Piper et al., 1998) (Fig. 1B). On the other hand, Pdr12p was found to play no detectable role in the active expulsion of counterions of more lipophilic acids such as octanoic acid, decanoic acid, artesunic acid, or 2,4-D (Alenquer et al., 2006; Hatzixanthis et al., 2003; Piper et al., 1998; Teixeira and Sá-Correia, 2002), or of the more hydrophilic acetate anion (Piper et al., 1998). Interestingly, Pdr12p was recently implicated in the active extrusion of fusel acids produced during amino acid catabolism, this being the proposed physiological function for this transporter in the yeast cell (Hazelwood et al., 2006). PDR5, encoding a close homologue of Pdr12p and a well-known efflux pump required for resistance to a very wide range of xenobiotic compunds (Paumi et al., 2009), was proposed to be involved in the expulsion of the counterions of the highly lipophilic 2,4-D and of artesunic and artemisinic acids, based on the higher susceptibility of Δpdr5 cells to these weak acids compared to wild-type cells (Alenquer et al., 2006; Ro et al., 2008; Teixeira and Sá-Correia, 2002) (Fig. 1B). In contrast, the elimination of PDR5 gene did not increased yeast susceptibility to acetic, propionic, and benzoic acids (our unpublished results). No other ABC transporters were so far implicated in weak acid tolerance, as revealed by gene-by-gene and large-scale global phenotypic screenings (Desmoucelles et al., 2002; Mira et al., 2009; Mollapour et al., 2004; Piper et al., 1998; Schuller et al., 2004). The expression of a number of genes encoding drug:H+ antiporters belonging to the major facilitator superfamily (MFS) have also been found to contribute for yeast tolerance to different weak acids: AQR1 and AZR1 genes are required for maximal tolerance to acetic and propionic acids (Tenreiro et al., 2000, 2002); TPO1 gene was found to be a major determinant of resistance to 2,4-D, artesunic and mycophenolic acids (Alenquer et al., 2006; Desmoucelles et al., 2002; Teixeira and Sá-Correia, 2002) and the TPO1 close homologues TPO2 and TPO3, were found to provide protection against acetic, propionic, and benzoic acids (Fernandes et al., 2005) (Fig. 1B). Although usually described as drug pumps, there are evidences supporting the concept that these MFS–MDR transporters do have a natural substrate, the drug transport occurring only fortuitously or opportunistically, this hypothesis being consistent with the described promiscuous activity of these transporters in providing protection against a large number of structurally and chemically unrelated drugs (reviewed by Sá-Correia et al., 2009). The role of MFS–MDR transporters in reducing the intracellular concentration of different weak acid counterions might also result from the transport of endogenous metabolite(s) that may affect the partition of the weak acid between the exterior and the intracellular environment, through an alteration of intracellular pH and/or plasma membrane potential (Sá-Correia et al., 2009). The increased resistance of S. cerevisiae to α-hop-acids, a group of weak acids largely used in brewing industries (Table 1), was very recently correlated with the ability of the cells to accumulate these molecules into the vacuole, although the transporter(s) mediating this phenomenon were not identified (Hazelwood et al., 2010) (Fig. 1B). To date, it remains to be identified an export system for lactate but the existence of such system has been anticipated (Abbott et al., 2009).
Weak acid-induced remodelling of the cellular envelope
The active expulsion of weak acid anions from the cell interior would be energetically expensive and futile if the undissociated acid could reenter the cells at a similar rate. Therefore, the restriction of the passive diffusion and, consequently, of the reentrance of the undissociated form of these weak acids, should be an essential step in yeast adaptation to these lipophilic compounds. One of the mechanisms proposed to reduce the diffusion rate of weak acids is the reinforcement of cell wall structure to decrease its porosity (Fig. 1A). Indeed, this mechanism was first described to occur in response to the acid herbicide 2,4-D, mediated by the glycosylphosphatidylinositol cell wall protein (GPI-CWP) Spi1p (Simões et al., 2003), and extended to weak acid food preservatives (Kapteyn et al., 2001; Simões et al., 2006). Significantly, the Spi1p-dependent remodelling of cell wall structure was also demonstrated to reduce weak acid-induced plasma membrane damage (Simões et al., 2006). The strong upregulation of SPI1 and of other genes involved in cell wall structure is observed in cells challenged with different weak acids (Abbott et al., 2008; Hazelwood et al., 2010; Kapteyn et al., 2001; Mira et al., 2009, 2010; Ro et al., 2008; Schuller et al., 2004; Teixeira et al., 2006a), sustaining the idea that the reinforcement of cell wall structure is a general response mechanism to weak acid-induced toxicity. Despite that, the cell wall-related genes contributing for tolerance are not the same for all weak acids (Kawahata et al., 2006; Mira et al., 2009; Mollapour et al., 2004; Schuller et al., 2004), suggesting that the molecular players underlying weak acid-induced alteration of cell wall structure may differ depending on the weak acid structure. For example, Spi1p, was identified as a key player in the remodelling of the cell wall structure in response to 2,4-D and benzoic acid, but had no detectable role in the acetic acid-induced alteration of cell wall structure (Simões et al., 2003, 2006). In the case of acetic acid, it was described that the removal of the aquaglyceroporin Fps1p from the plasma membrane, as the result of a strong increase in the endocytosis rate of this protein, prevents the facilitated diffusion of undissociated acetic acid through this channel thereby reducing the internal concentration of acetic acid (Mollapour and Piper, 2007).
Plasma membrane lipid composition is also altered in decanoic acid- or 2,4-D- challenged cells, compared to cells cultivated in the absence of these weak acids, an increase in the ratio of saturated:unsaturated membrane fatty acids being reported in these stressed cells (Alexandre et al., 1996; Viegas et al., 2005). Such alteration contributes to the adjustment of membrane fluidity and it is also likely to protect membrane lipids from oxygen-derived free radical attack induced by weak acids (Viegas et al., 2005) (Fig. 1A). Consistent with such response, the levels of OLE1 gene transcripts, encoding the sole Δ9 desaturase that catalyzes the conversion of saturated to unsaturated fatty acids, is reduced in 2,4-D- (Viegas et al., 2005), sorbic acid- (Schuller et al., 2004), and artemisinic acid-stressed cells (Ro et al., 2008).
Sensing and response to nutrient and energy limitation
Based on results from transcriptomic and proteomic analyses, the TOR (Target-of-Rapamycin) pathway, a regulatory system dedicated to the yeast response to nutrient starvation, was suggested to be activated in response to 2,4-D (Teixeira et al., 2006a) and acetic acid (Almeida et al., 2009). Such mechanism was proposed to counteract the nutrient limitation condition imposed by these weak acids and is consistent with the reduced concentration of amino acids registered inside 2,4-D and acetic acid-stressed cells (Almeida et al., 2009; Teixeira et al., 2005). The upregulation of genes and/or proteins involved in the uptake and biosynthesis of amino acids was also observed in cells cultivated in the presence of acetic acid (Almeida et al., 2009; Mira et al., 2010), propionic acid (Mira et al., 2009), sorbic acid (de Nobel et al., 2001) and 2,4-D (Teixeira et al., 2005, 2006a).
The dramatic alteration of carbohydrate metabolism is also a feature of weak acid-stressed yeast cells, as suggested by the upregulation of a number of genes and proteins encoding enzymes of glycolysis and of the Krebs cycle in response to acetic acid (Almeida et al., 2009; Mira et al., 2010), propionic acid (Mira et al., 2009), sorbic acid (Abbott et al., 2007; de Nobel et al., 2001; Schuller et al., 2004), benzoic acid (Abbott et al., 2007), lactic acid (Abbott et al., 2008) and 2,4-D (Teixeira et al., 2005, 2006a). Such response was proposed to compensate the severe depletion of ATP observed in these weak acid-stressed cells (Holyoak et al., 1996; Krebs et al., 1983; Warth, 1991) caused, at least, by the inhibition of the activity of glycolytic enzymes (Holyoak et al., 1996; Pampulha and Loureiro-Dias, 1990; Pearce et al., 2001). The activation of energy-consuming defence mechanisms, specially those involving major ATP consuming processes such as the PM-H+-ATPase, V-H+-ATPase, and ABC drug pumps, should also contribute to enhance energy depletion in weak acid-challenged cells. Not surprisingly, a large number of genes involved in ATP synthesis were identified as determinants of resistance to multiple weak acids, in particular to acetic, propionic, lactic, and sorbic acids (Kawahata et al., 2006; Mira et al., 2009; Mollapour et al., 2004; our unpublished results).
Tolerance to citric, propionic, acetic, and lactic acids and to α-hop-acids was also found to involve the expression of genes related to the uptake of potassium, calcium, iron, and zinc (Abbott et al., 2008; Hazelwood et al., 2010; Lawrence et al., 2004; Mira et al., 2009; our unpublished results) suggesting that these ions availability is crucial for tolerance to weak acid stress. The ability of the negatively charged counterion to chelate one or more metal cations, depending on the anion–metal stability constants, has the potential to limit metal availability in the growth medium and therefore to prevent growth in weak acid-supplemented growth media (Abbott et al., 2008; Hazelwood et al., 2010; Lawrence et al., 2004).
Genome-Wide Requirements for Maximal Tolerance to Weak Acids in Yeast
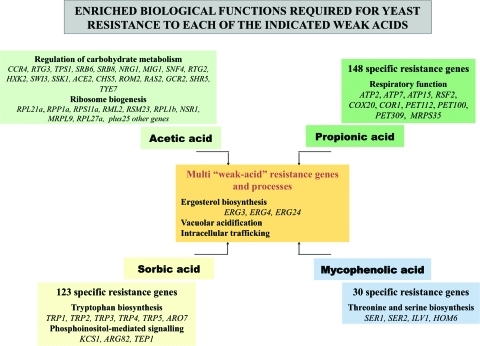
The availability of two strain collections containing thousands of deletion mutants lacking every nonessential yeast gene (haploid collection), or one of the two copies of each essential gene (diploid collection), has opened the door to the fields of Chemical Genomics and Phenomics in this experimental eukaryotic model. Based on these tools, the biological targets and the mechanisms of resistance to a wide range of xenobiotic compounds have been being characterised in yeast cells. Some of these global phenotypic screenings have been performed focused on the inhibitory action of various weak acids including propionic acid (Mira et al., 2009), mycophenolic acid (Desmoucelles et al., 2002), sorbic acid (Mollapour et al., 2004), and acetic acid (Kawahata et al., 2006; our unpublished results). The determinants of resistance to these four weak acids, identified through chemical genomic screenings, are compared in Figure 2. Vacuolar acidification, intracellular trafficking, and ergosterol biosynthesis are cellular processes required for general weak acid tolerance (Fig. 2), consistent with the conclusions gathered in a large-scale study where 400 structurally unrelated chemicals were tested to identify the genes/processes essential for multidrug resistance in yeast (Hillenmeyer et al., 2008). However, with the exception of ERG3, ERG4, and ERG24 genes, the genes associated to these biological functions that are required for resistance to each one of the different weak acids do not coincide (Fig. 2). Many genes whose expression confers resistance to a single weak acid were also identified, presumably representing more specific biological targets of each weak acid. We have clustered the 165 yeast genes that specifically provide resistance to sorbic acid (Mollapour et al., 2004), based on their biological function (according to the MIPS functional catalog), and the frequency of each class was compared in this dataset and in the genome to search for the enriched functional classes (those with an associated p-value below 0.01). Based on this statistical analysis, specific sorbic acid resistance genes are involved in “tryptophan biosynthesis” and in “phosphoinositol-mediated signal transduction” (Fig. 2), consistent with the specific inhibitory effect that this weak acid has over the major tryptophan permease, Tat2p (Liu et al., 2004) and with the activation of phosphoinositoide signaling that was registered in sorbic acid-challenged cells (Mollapour et al., 2006). A similar analysis was carried out using the 30 genes that specifically provide protection toward mycophenolic acid, these resistance genes being enriched in those involved in serine and threonine biosynthesis (Fig. 2). Mycophenolic acid is a known inhibitor of inosine monophosphate dehydrogenase, the enzyme that controls the rate of synthesis of guanine monophosphate in the purine biosynthetic pathway. Significantly, yeast susceptibility to mycophenolic acid was found to be fully bypassed upon guanine addition, suggesting that the toxic effect of this weak acid in yeast cells is very specific (Escobar-Henriques et al., 2001). Nonetheless, the existence of secondary targets for mycophenolic acid other than guanine metabolism has also been acknowledged (Escobar-Henriques et al., 2001). Interestingly, threonine metabolism was one of the biological processes more severely affected by the purine nucleotide imbalance in mycophenolic acid-challenged cells (Saint-Marc et al., 2009). The list of genes specific for acetic acid tolerance was enriched in those associated to the regulation of carbohydrate metabolism (in particular, to glucose sensing and signaling) and to ribosome biogenesis (our unpublished results) (Fig. 2). Interestingly, extensive degradation of ribosomal RNA is observed in acetic acid-stressed cells as a result of the activation of apoptotic mechanisms (Mroczek and Kufel, 2008). In the case of propionic acid, the specific resistance genes were enriched in those involved in respiratory function (Fig. 2), remaining to be established whether there is a link between propionic acid resistance and respiration.
FIG. 2.
Yeast genes that were found to confer resistance to acetic, propionic, sorbic, and mycophenolic acids, based on the results of (Desmoucelles et al., 2002; Mira et al., 2009; Mollapour et al., 2004; and our unpublished results) were selected and clustered according to their biological function using the MIPS functional catalog. The classes considered enriched in the different gene lists (associated p-value below 0.01) are shown.
Genome-Wide Transcriptional Response to Weak Acid Stress in Yeast and Underlying Regulatory Networks
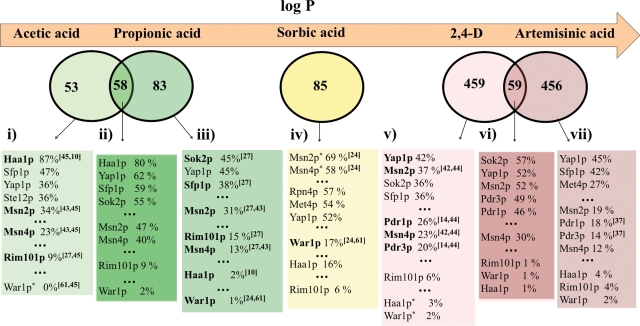
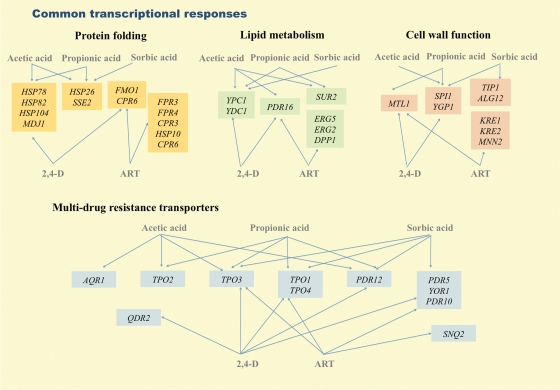
The analysis of the genomic expression programs activated in response to different weak acids support the concept that yeast exhibits specific responses determined by the chemical structure of the weak acid, in particular, by its lipophilicity (Abbott et al., 2007; Mira et al., 2009, 2010; Schuller et al., 2004; Teixeira et al., 2006a). Indeed, the comparison of the genes upregulated in response to acetic acid (Mira et al., 2010), propionic acid (Mira et al., 2009), sorbic acid (Schuller et al., 2004), 2,4-D (Teixeira et al., 2006a), and artemisinic acid (Ro et al., 2008), five differently lipophilic weak acids, reveals that the genes activated in response to acetic and propionic acids or in response to 2,4-D and artemisinic acids overlap at a significant extent, consistent with the close hydrophobicity of each pair of weak acids (Fig. 4). Only one gene, TPO3, encoding a multidrug resistance transporter, was found to be induced in response to all weak acids, indicating that the existence of a general weak acid response is rather limited (results not shown). A previous transcriptomic analysis carried out in the presence of acetic, propionic, benzoic, and sorbic acids came to a similar conclusion (Abbott et al., 2007). It is evident from our analysis that although the genes activated in response to all weak acids do not coincide, they are involved in the same biological functions, this being the case of those genes involved in protein folding, lipid metabolism, cell wall function, and multidrug resistance (Fig. 3). For example, the multidrug resistance transporters PDR5, YOR1, PDR10, and SNQ2 genes are only induced by the more lipophilic weak acids sorbic and artemisinic acids and 2,4-D, whereas TPO2, also encoding a multidrug resistance transporter, is only induced in response to the more hydrophilic acetic and propionic acids (Fig. 3). The fact that the results considered in this comparison have been obtained in different experimental conditions might contribute for the observed differences, however, the specificities of each group of upregulated genes may also reflect the activation of different regulatory pathways that may respond differently to each weak acid. Indeed, PDR5, YOR1, PDR10, and SNQ2 genes are documented targets of Pdr1p/Pdr3p transcription factors that respond to 2,4-D, sorbic and artemisinic acids (Alenquer et al., 2006; Teixeira and Sá-Correia, 2002) but are dispensable for response to acetic and propionic acids (Piper et al., 1998; our unpublished results). Similarly, TPO2 is one of the genes most strongly upregulated by Haa1p, which plays a major role in response to more hydrophilic acetic and propionic acids but has only a negligible role in response to more lipophilic weak acids (Fernandes et al., 2005; Mira et al., 2010). Another interesting feature observable when comparing the results from transcriptomics and chemogenomics analysis, shown in Figure 2 and Figure 3, is the fact that some of the genes (e.g., AZR1) that confer resistance to weak acids are not transcriptionally activated by them, while some of the upregulated genes (e.g., YOR1 and PDR10) are not required to increase yeast tolerance toward weak acid stress. Such phenomenon has been registered before, and suggests that a broad transcriptional response may be more cost-effective than the existence of a large number of specific regulatory components (Mira et al., 2009; Schuller et al., 2004; Teixeira et al., 2006a).
FIG. 4.
Clustering of the genes upregulated in response to acetic, propionic, sorbic, and artemisinic acids (Mira et al., 2009, 2010; Ro et al., 2008; Schuller et al., 2004) or to 2,4-D (Teixeira et al., 2006a) with their documented regulators, according to the information deposited in the YEASTRACT database (March 2010), plus data coming from our recent work (Mira et al., 2010). Based on the level of overlapping, the responsive genes were clustered in three groups: acetic and propionic acid-induced genes, sorbic acid-induced genes and artemisinic acid- and 2,4-D-induced genes. YEASTRACT database was used to cluster, with their documented regulators, the genes that: (1) are activated in cells exposed to acetic acid but not in the presence of propionic acid stress; (2) are upregulated upon exposure to acetic or propionic acid; (3) are activated in cells exposed to propionic acid but not upon acetic acid challenge; (4) are upregulated in yeast cells exposed to sorbic acid; (5) are activated in cells exposed to 2,4D but not in the presence of artemisinic acid stress; (6) are upregulated upon exposure to 2,4-D or artemisinic acid; (7) are activated in cells exposed to artemisinic acid but not upon 2,4-D challenge. The transcription factors were organized by decreasing percentage of documented targets in each dataset. The transcription factors that provide protection toward a given weak acid are distinguished (in bold) from those known to be dispensable for tolerance (indicated with *).
FIG. 3.
Biological functions found to be represented in the yeast transcriptome-wide response to all weak acids analysed. For each biological function, the specific genes responding to the indicated weak acids are highlighted. ART, Artemisinic acid.
The exploitation of the data emerging from transcriptomic analysis of weak acid-stressed cells with bioinformatic tools, in particular, with those available in the YEASTRACT database, allowing clustering of genes with their documented regulators (Teixeira et al., 2006b) opens the door to the identification of novel transcription factors that may underlie yeast transcriptional response to weak acid stress. So far, four regulatory pathways, dependent on the transcription factors Haa1p (Fernandes et al., 2005; Mira et al., 2010), Rim101p (Mira et al., 2009), Msn2p/Msn4p (Schuller et al., 2004), and War1p (Kren et al., 2003; Schuller et al., 2004), were identified as mediating yeast response to weak acid stress. Pdr1p and Pdr3p, the major transcription factors controlling multidrug resistance in yeast (Gulshan and Moye-Rowley, 2007), are also known to be key players in the control of yeast transcriptional response to artemisinic acid (Ro et al., 2008) and to 2,4-D (Teixeira et al., 2006a; Teixeira and Sá-Correia, 2002). The role of these signaling systems in mediating yeast-adaptive response and tolerance to weak acid stress is detailed in the following subsections of this article (see below). The YEASTRACT database was used to cluster transcription factors with genes that: (1) are activated in cells exposed to acetic acid but not in the presence of propionic acid stress (acetic acid-specific); (2) are upregulated upon exposure to both acetic and propionic acids; (3) are activated in cells exposed to propionic acid but not under acetic acid challenge (propionic acid-specific); (4) are upregulated in yeast cells exposed to sorbic acid; (5) are activated in cells exposed to 2,4-D but not in the presence of artemisinic acid stress (2,4-D specific); (6) are upregulated upon exposure to both 2,4-D and artemisinic acid; (7) are activated in cells exposed to artemisinic acid but not upon 2,4-D challenge (artemisinic acid-specific). Such division of the datasets was performed to distinguish, when possible, the regulators required that control more specific transcriptional response (e.g., responses to a given weak acid or a group of weak acids) from those that might have a broader effect. Besides Msn2p and Msn4p transcription factors whose role in response to weak acid stress is discussed below, Yap1p and Sfp1p transcription factors also emerge from this analysis as having high percentage of documented targets among the genes induced by all weak acids under examination (Fig. 4). Yap1p is a key player in the control of transcriptional response to oxidative stress in yeast cells (Fernandes et al., 1997) and Sfp1p is involved in the transcriptional regulation of ribosomal genes in response to nutrient starvation and stress (Marion et al., 2004). Although it is not established whether Yap1p and Sfp1p do have a role in the control of the genome-wide transcriptional response to the weak acids under examination, it is interesting that all these molecules were suggested to have a pro-oxidant effect in yeast cells (Piper, 1999; Ro et al., 2008; Teixeira et al., 2004) and to induce general nutrient limitation (Almeida et al., 2009; Mira et al., 2010; Teixeira et al., 2005, 2006a). Noteworthy, the elimination of YAP1 gene increased yeast susceptibility to 2,4-D (our unpublished results) but had no relevant effect on tolerance to acetic, propionic, and sorbic acids (Piper et al., 1998; our unpublished results). Similarly, Sfp1p was only described as a determinant of resistance to propionic acid (Mira et al., 2009). Sok2p, a transcription factor known to be involved in the regulation of stress response in yeast cells (Shenhar and Kassir, 2001), was also found to have high percentages of documented targets in all datasets (Fig. 4). Interestingly, Sok2p has a high percentage of documented targets among the genes specifically activated in response to propionic acid, (Fig. 3), this correlating with the identification of this transcription factor as a determinant of resistance to this weak acid (Mira et al., 2009). Altogether these observations suggest that Msn2p/Msn4p, and eventually Yap1p, Sfp1p, and Sok2p, may be involved in the control of a “general weak acid transcriptional response.” Another interesting observation drawn from the analysis of results shown in Figure 4 is the remarkably high percentage of documented Haa1p targets among the genes specifically induced by acetic acid, compared to the one registered with the genes induced in response to sorbic and artemisinic acids or 2,4-D, which suggests that the Haa1p-dependent regulatory system is mostly dedicated to yeast response to acetic acid stress. This idea is also reinforced by the fact that the predominance of Haa1p in the dataset of “acetic and propionic acid-induced genes” is attributed to its involvement in response to acetic acid stress as this transcription factor had no relevant role in the regulation of the genes specifically induced by propionic acid (Fig. 4). The existence of a similar specific transcriptional response for other weak acids under examination was not evident from our analysis (Fig. 4).
The Msn2p/Msn4-regulon
Msn2p and Msn4p are two homologous transcription factors whose biological role is the control of yeast transcriptional response to environmental stress (Gasch et al., 2000). Indeed, the participation of Msn2p and Msn4p in the transcriptional response to a number of weak acids has been confirmed both by microarray (Schuller et al., 2004) and gene-by-gene approaches (Teixeira et al., 2006a; Simões et al., 2003, 2006). Most of the genes upregulated by Msn2p and Msn4p in response to weak acid stress encode proteins of the environmental stress response such as molecular chaperones (e.g., Hsp26p, Sse2p), enzymes of the carbohydrate metabolism (e.g., Hxk1p, Gpd1p), and of the antioxidant defense system (e.g., Ctt1p, Gpx1p) (Schuller et al., 2004; Simões et al., 2006). Interestingly, many of the genes transcriptionally activated by Msn2p/Msn4p under weak acid stress do not have in their promoter region a binding site for these transcription factors (5′-CCCCT-3′), suggesting that the effect of the Msn2p/Msn4p-system is mediated by the hierarchical control of downstream signaling pathways. It is noteworthy that the important role of Msn2p and Msn4p as regulators of a high percentage of the weak acid-induced genes does not always correlate with a protective effect of these transcription factors towards these same weak acids (Fig. 4). For example, Msn2p and Msn4p are implicated in yeast tolerance to 2,4-D, acetic, and propionic acids (Mira et al., 2009; Simões et al., 2003, 2006), but are dispensable for sorbic acid tolerance (Schuller et al., 2004) (Fig. 4). The induction of Msn2p/Msn4-regulon in response to multiple weak acids provides the cells with the necessary flexibility to respond to a wide range of challenges, although the physiological response brought by this activation does not always result in an increased resistance to the weak acid.
The Haa1p-regulon
The first biological function attributed to the Haa1p transcription factor, first identified based on its homology with a set of copper-activated transcription factors (Keller et al., 2001), was in yeast tolerance to acetic and propionic acids (Fernandes et al., 2005). The protective effect exerted by the expression of the HAA1 gene was found to decrease markedly as the lipophilicity of the weak acid increases, attaining a maximal level against acetic acid (Fernandes et al., 2005). Remarkably, this pattern of differential protection correlates with the level of activation mediated by Haa1p in the genomic expression responses to differently lipophilic weak acids, as evidenced by the results shown in Figure 4. A number of the genes activated directly or indirectly by Haa1p in response to acetic acid were found to exert protection against this weak acid (Mira et al., 2010), the most significant protective effect being obtained with the expression of SAP30 and HRK1 genes (Mira et al., 2010). SAP30 encodes a subunit of the Rpd3L histone deacetylase complex, while HRK1 encodes a protein kinase belonging to a family of kinases dedicated to the posttranslational regulation of plasma membrane transporters (Goossens et al., 2000). The Rpd3L complex was recently demonstrated to be involved in the control of the transcriptional response to environmental stress mediated by the Msn2p/Msn4p transcription factors (Alejandro-Osorio et al., 2009), thus being possible that this complex may also be required for the control of the activity of transcription factors involved in response to acetic acid stress (Mira et al., 2010). The deletion of HRK1 gene led to an increased accumulation of radiolabeled acetic acid into yeast cells cultivated in the presence of this weak acid (Mira et al., 2010), a similar phenotype to the one observed upon HAA1 deletion (Fernandes et al., 2005), which seems to indicate the involvement of Haa1p and Hrk1p in the reduction of intracellular acetate concentration. Other Haa1p-regulated genes that also confer resistance to acetic acid include the drug:H+ antiporters Tpo2p and Tpo3p, proposed to mediate acetate export (Fernandes et al., 2005), and the cell wall-related protein Ygp1p that may contribute to the acid-induced remodelling of cell wall structure and thus prevent the reentry of acetate at the same rate as the expulsion of this counterion (Fernandes et al., 2005).
The War1p-regulon
The elimination of the WAR1 gene leads to hypersusceptibility phenotypes toward moderately lipophilic weak acids (propionic acid, sorbic acid, benzoic acid, and some fusel acids) (Hazelwood et al., 2006; Kren et al., 2003; Schuller et al., 2004). Consistent with the idea that the main function of War1p is the regulation of PDR12 transcription, the susceptibility phenotypes of Δwar1 cells are similar to those exhibited by the Δpdr12 mutant (Hazelwood et al., 2006; Kren et al., 2003; Piper et al., 1998; Schuller et al., 2004). A set of other War1p-regulated genes in response to sorbic acid were identified, based on mRNA profiling but, apart from PDR12, none of these genes provides protection against this weak acid (Schuller et al., 2004). Recent results indicate that War1p activity is controlled upon the direct binding of weak acid counterions, this interaction eliciting the affinity of this transcription factor for the promoter region of its target genes (Gregori et al., 2008). In particular, the ability of sorbate anion to induce a conformational modification in War1p structure was demonstrated, this resulting in the potent transcriptional activation of PDR12 gene in sorbic acid-challenged cells (Gregori et al., 2008). Notably, the percentage of War1p documented target genes is much higher among the sorbic acid-induced genes than within the group of genes activated in response to acetic acid, propionic acid, artemisinic acid, or to 2,4-D (Fig. 4).
The Pdr1p/Pdr3p-regulon
Pdr1p and Pdr3p transcription factors are the key players in the control of pleiotropic drug resistance in yeast (reviewed by Gulshan and Moye-Rowley, 2007). The elimination of PDR1 and, less significantly of PDR3, renders the yeast cells more susceptible to the more lipophilic weak acids such as artemisinic acid, artesunic acid, and 2,4-D (Alenquer et al., 2006; Ro et al., 2008; Teixeira and Sá-Correia, 2002) but has no effect in tolerance to acetic, propionic, or benzoic acids (Piper et al., 1998; our unpublished results). The molecular mechanisms mediated by Pdr1p/Pdr3p that leads to increased resistance toward 2,4-D and artesunic acid were found to involve the multidrug resistance transporters Pdr5p and Tpo1p, presumably required for the active expulsion of these cytotoxic compounds (Alenquer et al., 2006; Teixeira and Sá-Correia, 2002). Although many Pdr1p/Pdr3p target genes are upregulated in response to artemisinic acid (Ro et al., 2008) (Fig. 4), suggesting a mechanistic action similar to the one described under 2,4-D stress, neither the role of Pdr1p/Pdr3p nor of their target genes in yeast tolerance to artemisinic acid was examined. Remarkably, the activation of the Pdr1p/Pdr3p-dependent regulatory system was recently associated to the perturbation of plasma membrane lipid homeostasis (Schuller et al., 2007), which is consistent with the much higher percentage of documented targets of Pdr1p and Pdr3p registered among the genes activated in response to the more lipophilic artemisinic acid and 2,4-D than within the genes induced in response to acetic, propionic, or sorbic acids (Fig. 4). Additionally, Pdr1p was also recently demonstrated to be activated upon binding of drugs (Thakur et al., 2008), suggesting that various weak acid counterions might also directly modulate the activity of this transcription factor, as described for War1p (Gregori et al., 2008).
The Rim101p pathway
The Rim101 pathway was recently implicated in yeast adaptive response to propionic acid (Mira et al., 2009) extending the biological role of this pathway beyond its involvement in alkaline pH response, in structuring the cell wall and in mediating tolerance to high concentrations of sodium and lithium (reviewed by Penalva et al., 2008). Based on the comparison of the transcriptomes of wild-type and Δrim101 cells, Rim101p was found to regulate a small subset of the genes transcriptionally activated in response to propionic acid, including several determinants of resistance to this weak acid: KNH1, encoding a protein involved in β-1,6-glucan synthesis; CWP1, encoding a cell wall mannoprotein linked to β-1,3- and β-1,6-glucan heteropolymer; BAG7, encoding a GTPase-activating protein involved in the stimulation of β-1,3-glucan synthesis through Rho1p; YIL029c, encoding a protein of uncharacterized function (Mira et al., 2009). Consistent with its role in the upregulation of genes related to cell wall function, the expression of RIM101 protects the cell against lyticase activity both in the presence or absence of propionic acid (Mira et al., 2009). A link between the Rim101p pathway and the homeostasis of internal pH was also uncovered the Δrim101 cells exhibiting a lower cytosolic pH and an impaired vacuolar acidification under propionic acid stress (Mira et al., 2009).
Weak Acid Resistance in Other Eukaryotes: Lessons from S. cerevisiae
The remarkably high resistance of the food spoilage yeast species Zygosaccharomyces bailii to weak acids food preservatives (e.g., acetic acid, benzoic acid. and sorbic acid) was related with its ability to use these weak acids as carbon sources, even in the presence of glucose (Mollapour and Piper, 2001; Sousa et al., 1998; Stratford et al., 2007). Interestingly, no induction of a Pdr12p-like protein was observed in Z. bailii cells challenged with sorbic acid (Papadimitriou et al., 2007), strongly indicating that the key role played by the Pdr12p–War1p axis in S. cerevisiae resistance is not operating in this species. The recent release of the genome sequence of Z. rouxii (http://cbi.labri.fr/Genolevures), an osmotolerant food spoilage yeast that is also resistant to weak acid preservation (Fleet, 2007; Martorell et al., 2007), provides an important experimental platform to extrapolate the knowledge gathered in S. cerevisiae to both Z. rouxii and Z. bailii, because the genome sequence of this last yeast is not available. Interestingly, robust homologues of Rim101p and Haa1p transcription factors are found in the Z. rouxii genome (ORFs ZYRO0G07964g and ZYRO0F04862g, respectively), and most of the genes found to be regulated by these two transcription factors in S. cerevisiae are also found conserved in this species. It is therefore likely that Rim101p- and Haa1p-like signalling pathways are functionally active in Z. rouxii and in Z. bailii, eventually mediating tolerance to these weak acids in these spoilage yeasts. Other relevant spoilage fungi resistant to weak acids include Aspergillus species and in particular, Aspergillus niger (Fleet, 2007). The sorbic acid-resistance phenotype of this filamentous fungus was attributed to its capability to convert the weak acid into less harmful compounds (Plumridge et al., 2008), but little is known about the mechanisms underlying acetic acid and propionic acid resistance in this organism. Although no relevant homologue of Haa1p was found in A. niger, a functional Rim101p pathway is known to be active in this organism (reviewed by Penalva et al., 2008).
Short-chain weak acids, in particular acetic acid, are known to elicit the fungicidal action of fluconazole towards Candida albicans under conditions where the fungicide alone is not effective (Moosa et al., 2004). Therefore, the understanding of weak acid resistance mechanisms in pathogenic Candida species may contribute to delineate strategies to overcome antifungal drug resistance. The existence of functional Msn2p/Msn4p-like pathways have been described both in C. albicans and in C. glabrata, these pathways being controlled, respectively, by CaMnl1p (an orphan Msn2-like protein), CaMsn4p, CgMsn2p, and CgMsn4p (Nicholls et al., 2004; Ramsdale et al., 2008; Roetzer et al., 2008). Like in S. cerevisiae, CgMsn2p and CgMsn4p are essential for the transcriptional response of C. glabrata to a number of environmental stresses including osmotic stress, heat shock, and oxidative stress (Roetzer et al., 2008); however, these transcription factors are largely dispensable for response and tolerance of this yeast to propionic and sorbic acids (Mundy and Cormack, 2009; Roetzer et al., 2008). Differently, CaMnl1p and CaMsn4p are not required for the transcriptional regulation of C. albicans environmental response to stress (Nicholls et al., 2004), but CaMnl1p was recently demonstrated to be a main player in the control of the genome-wide transcriptional alterations induced by acetic acid (Ramsdale et al., 2008). Robust homologues of Haa1p and Rim101p are present in Candida glabrata (CAGL0L09339g and CAGL0E03762g, respectively), remaining to be examined whether these genes have a role in the response of this pathogenic yeast to weak acids. In C. albicans, no significant homologue of Haa1p is found, but a functional Rim101p-dependent pathway is present in this fungus (reviewed by Penalva et al., 2008) as well as an orthologue of War1p, CaWar1p, that mediates the resistance of this fungus to sorbic and propionic acids (Lebel et al., 2006; Mundy and Cormack, 2009). Although general evidences suggest that stress signalling pathways may have diverged significantly in Candida species from those active in the budding yeast (Nicholls et al., 2004; Ramsdale et al., 2008), the information gathered from the elucidation of the weak acid-sensing systems in S. cerevisiae is expected to guide the identification of corresponding regulatory systems in these pathogenic yeasts.
The knowledge gathered in S. cerevisiae on the response and resistance to the weak acid herbicide 2,4-D has paved the way to gain insights into 2,4-D resistance in the plant model Arabidopsis thaliana (Cabrito et al., 2009; Teixeira et al., 2007). The A. thaliana AtPdr9p conferred resistance to 2,4-D in plant (Ito and Gray, 2006), similar to the beneficial effect exerted by the expression of its close homologue Pdr5p during cultivation of yeast cells in the presence of this herbicide (Teixeira and Sá-Correia, 2002). More recently, the heterologous expression of A. thaliana ORF At5g13750, encoding a homologue of Tpo1p, in yeast cells decreased the inhibition of cell growth in the presence of 2,4-D (Cabrito et al., 2009). The extrapolation and exploitation of the knowledge obtained in S. cerevisiae to the plant model A. thaliana may help to control the acquired resistance to 2,4-D of specific weed species or to allow the rational design of crops with increased tolerance to this herbicide.
The emergence of Plasmodium falciparum-resistant strains to artemisinine and to its derivatives, including artesunic acid, has already been described (Dondorp et al., 2009), compromising the usefulness of these drugs as therapeutics in the near future. Interestingly, close homologues of Pdr5p that confer resistance to artesunic acid in yeast are found in P. falciparum (Alenquer et al., 2006), although the eventual role played by these proteins in the resistance of the parasite to artemisinine and artesunic acid remain to be scrutinized. Artesunic acid is also active against a large number of tumor cell lines that can also develop resistance to this drug as the result of the activity of multidrug resistance transporters (Efferth et al., 2003). One of such transporters is the p-glycoprotein, a human ABC transporter related to yeast Pdr5p (Paumi et al., 2009). Interestingly, it was also recently demonstrated the involvement of the MFS transporter TETRAN, the human orthologue of Tpo1p, in drug resistance (Mima et al., 2007), suggesting that results involving Tpo1p in artesunic acid resistance in yeast may have a parallel in human cells.
Concluding Remarks and Future Perspectives
Functional genomics has deeply modified the understanding of yeast global response to weak acid-induced stress. Chemical Genomic studies opened the door to the identification, at a genome-wide scale, of the key players conferring resistance to weak acids (Desmoucelles et al., 2002; Mira et al., 2009; Mollapour et al., 2004; Schuller et al., 2004) (Fig. 2). On the other hand, extensive transcriptomic profilings have characterized a number of transcriptional regulatory networks active in yeast response to weak acid stress (Mira et al., 2009, 2010; Saint-Marc et al., 2009; Schuller et al., 2004; Teixeira et al., 2006a) and contributed to the understanding of the highly dynamic nature of the weak acid-sensing regulatory networks by defining how these systems crosstalk in response to different weak acids (Fig. 4). However, the identification of many other transcription factors as determinants of resistance to specific groups of weak acids, specially as the result of global phenotypic screenings, strongly suggests that the size and the complexity of such regulatory network is much higher than the simple model that can be established so far. The use of expression proteomics to study yeast response to weak acid stress has mostly confirmed the results suggested by transcriptomics analysis (Almeida et al., 2009; de Nobel et al., 2001; Lawrence et al., 2004; Makrantoni et al., 2007; Teixeira et al., 2005). Nevertheless, with the emergence of new experimental approaches that permit mapping of posttranslational modifications, the novel branches of proteomics (phosphoproteomics, glycoproteomics, acetylation proteomics, among others) are expected to add an additional layer of knowledge for the elucidation of the molecular mechanisms underlying adaptation and tolerance to weak acid stress in yeast cells. Considering that many yeast stress responsive transcription factors are activated upon phosphorylation, phosphoproteomics may provide a link between transcriptional regulatory networks and the upstream kinase-mediated signaling pathways. Metabolomics is still a relatively unexplored field to provide clues into the mechanisms of yeast response to weak acid stress. However, because metabolites are the end products of cellular regulatory processes and their levels can be regarded as the ultimate response of biological systems to an environmental change, metabolic footprinting may provide an essential knowledge for the full understanding of the impact of weak acids toxicity in the overall physiology of the yeast cell. Although there is still much to be done, the integration of all the knowledge gathered at the different genomic levels has already provided a clear-cut picture of the yeast response to various weak acids, an information that considering the wide range of applications of these molecules may have a great impact in industry, agriculture and human health. Future research on the field will require the further development of computational tools aiming the integration of data coming from the different genome-wide approaches and the collaborative activity of multidisciplinary teams with expertise in biological sciences, functional genomics, and bioinformatics.
Acknowledgments
Research carried out in our laboratory and described in this review article was financially supported by FEDER, Fundação para a Ciência e Tecnologia (FCT), and POCTI and PTDC programs (contracts PTDC/AGR-ALI/102608/2008, PTDC/BIO/66151/2006, PTDC/EIA/67722/2006, POCTI/AGR/45347/2002, POCTI/ BIO/38115/2001, and a postdoctoral grant, SFRH/BPD/46982/2008, to N.P.M.)
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Abbott D.A. Knijnenburg T.A. de Poorter L.M. Reinders M,J. Pronk J.T. Van Maris A.J. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 2007;7:819–833. doi: 10.1111/j.1567-1364.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- Abbott D.A. Suir E. Van Maris A.J.A. Pronk J.T. Physiological and transcriptional responses to high concentrations of lactic acid in anaerobic chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microb. 2008;74:5759–5768. doi: 10.1128/AEM.01030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott D.A. Zelle R.M. Pronk J.T. Van Maris A.J. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 2009;9:1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- Alejandro-Osorio A.L. Huebert D.J. Porcaro D.T. Sonntag M.E. Nillasithanukroh S. Will J.L., et al. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 2009;10:R57. doi: 10.1186/gb-2009-10-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenquer M. Tenreiro S. Sá-Correia I. Adaptive response to the antimalarial drug artesunate in yeast involves Pdr1p/Pdr3p-mediated transcriptional activation of the resistance determinants TPO1 and PDR5. FEMS Yeast Res. 2006;6:1130–1139. doi: 10.1111/j.1567-1364.2006.00095.x. [DOI] [PubMed] [Google Scholar]
- Alexandre H. Mathieu B. Charpentier C. Alteration in membrane fluidity and lipid composition, and modulation of H(+)-ATPase activity in Saccharomyces cerevisiae caused by decanoic acid. Microbiology. 1996;142(Pt 3):469–475. doi: 10.1099/13500872-142-3-469. [DOI] [PubMed] [Google Scholar]
- Almeida B. Ohlmeier S. Almeida A.J. Madeo F. Leão C. Rodrigues F., et al. Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics. 2009;9:720–732. doi: 10.1002/pmic.200700816. [DOI] [PubMed] [Google Scholar]
- Booth I.R. Statford N. Acidulants and low pH. In: Russel N, editor; Gould G, editor. Food Preservatives. Kluwer Academic/Plenum Publishers; New York: 2003. pp. 25–48. [Google Scholar]
- Cabral M.G. Viegas C.A. Sá-Correia I. Mechanisms underlying the acquisition of resistance to octanoic-acid-induced-death following exposure of Saccharomyces cerevisiae to mild stress imposed by octanoic acid or ethanol. Arch Microbiol. 2001;175:301–307. doi: 10.1007/s002030100269. [DOI] [PubMed] [Google Scholar]
- Cabrito T.R. Teixeira M.C. Duarte A.A. Duque P. Sá-Correia I. Heterologous expression of a Tpo1 homolog from Arabidopsis thaliana confers resistance to the herbicide 2,4-D and other chemical stresses in yeast. Appl Microbiol Biotechnol. 2009;84:927–936. doi: 10.1007/s00253-009-2025-5. [DOI] [PubMed] [Google Scholar]
- Carmelo V. Bogaerts P. Sá-Correia I. Activity of plasma membrane H+-ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at optimal and low pH. Arch Microbiol. 1996;166:315–320. doi: 10.1007/s002030050389. [DOI] [PubMed] [Google Scholar]
- Carmelo V. Santos H. Sá-Correia I. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim Biophys Acta. 1997;1325:63–70. doi: 10.1016/s0005-2736(96)00245-3. [DOI] [PubMed] [Google Scholar]
- de Nobel H. Lawrie L. Brul S. Klis F. Davis M. Alloush H., et al. Parallel and comparative analysis of the proteome and transcriptome of sorbic acid-stressed Saccharomyces cerevisiae. Yeast. 2001;18:1413–1428. doi: 10.1002/yea.793. [DOI] [PubMed] [Google Scholar]
- Desmoucelles C. Pinson B. Saint-Marc C. Daignan-Fornier B. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J Biol Chem. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- Dondorp A.M. Nosten F. Yi P. Das D. Phyo A.P. Tarning J., et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efferth T. Sauerbrey A. Olbrich A. Gebhart E. Rauch P. Weber H.O., et al. Molecular modes of action of artesunate in tumor cell lines. Mol Pharmacol. 2003;64:382–394. doi: 10.1124/mol.64.2.382. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques M. Balguerie A. Monribot C. Boucherie H. Daignan-Fornier B. Proteome analysis and morphological studies reveal multiple effects of the immunosuppressive drug mycophenolic acid specifically resulting from guanylic nucleotide depletion. J Biol Chem. 2001;276:46237–46242. doi: 10.1074/jbc.M103416200. [DOI] [PubMed] [Google Scholar]
- Fernandes A.R. Durao P.J. Santos P.M. Sá-Correia I. Activation and significance of vacuolar H+-ATPase in Saccharomyces cerevisiae adaptation and resistance to the herbicide 2,4-dichlorophenoxyacetic acid. Biochem Biophys Res Commun. 2003;312:1317–1324. doi: 10.1016/j.bbrc.2003.11.072. [DOI] [PubMed] [Google Scholar]
- Fernandes A.R. Mira N.P. Vargas R.C. Canelhas I. Sá-Correia I. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Commun. 2005;337:95–103. doi: 10.1016/j.bbrc.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Fernandes L. Rodrigues-Pousada C. Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet G.H. Yeasts in foods and beverages: impact on product quality and safety. Curr Opin Biotechnol. 2007;18:170–175. doi: 10.1016/j.copbio.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Gasch A.P. Spellman P.T. Kao C.M. Carmel-Harel O. Eisen M.B. Storz G., et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B.R. Lawrence F.M. Leclaire J.P.R. Powell C.D. Smart K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev. 2007;31:535–569. doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Goossens A. de la Fuente N. Forment J. Serrano R. Portillo F. Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol Cell Biol. 2000;20:7654–7661. doi: 10.1128/mcb.20.20.7654-7661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori C. Schuller C. Frohner I.E. Ammerer G. Kuchler K. Weak organic acids trigger conformational changes of the yeast transcription factor War1 in vivo to elicit stress adaptation. J Biol Chem. 2008;283:25752–25764. doi: 10.1074/jbc.M803095200. [DOI] [PubMed] [Google Scholar]
- Gulshan K. Moye-Rowley W.S. Multidrug resistance in fungi. Eukaryotic Cell. 2007;6:1933–1942. doi: 10.1128/EC.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzixanthis K. Mollapour M. Seymour I. Bauer B.E. Krapf G. Schuller C., et al. Moderately lipophilic carboxylate compounds are the selective inducers of the Saccharomyces cerevisiae Pdr12p ATP-binding cassette transporter. Yeast. 2003;20:575–585. doi: 10.1002/yea.981. [DOI] [PubMed] [Google Scholar]
- Hazelwood L.A. Tai S.L. Boer V.M. de Winde J.H. Pronk J.T. Daran J.M. A new physiological role for Pdr12p in Saccharomyces cerevisiae: export of aromatic and branched-chain organic acids produced in amino acid catabolism. FEMS Yeast Res. 2006;6:937–945. doi: 10.1111/j.1567-1364.2006.00094.x. [DOI] [PubMed] [Google Scholar]
- Hazelwood L.A. Walsh M.C. Pronk J.T. Daran J.M. Involvement of vacuolar sequestration and active transport in tolerance of Saccharomyces cerevisiae to hop iso-alpha-acids. Appl Environ Microbiol. 2010;76:318–328. doi: 10.1128/AEM.01457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E. Fung E. Wildenhain J. Pierce S.E. Hoon S. Lee W., et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak C.D. Bracey D. Piper P.W. Kuchler K. Coote P.J. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J Bacteriol. 1999;181:4644–4652. doi: 10.1128/jb.181.15.4644-4652.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak C.D. Stratford M. McMullin Z. Cole M.B. Crimmins K. Brown A.J, et al. Activity of the plasma membrane H(+)-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl Environ Microb. 1996;62:3158–3164. doi: 10.1128/aem.62.9.3158-3164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H. Gray W.M. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 2006;142:63–74. doi: 10.1104/pp.106.084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P.M. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J.C. Ter Riet B. Vink E. Blad S. De Nobel H. Van den Ende H., et al. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol Microbiol. 2001;39:469–479. doi: 10.1046/j.1365-2958.2001.02242.x. [DOI] [PubMed] [Google Scholar]
- Kawahata M. Masaki K. Fujii T. Iefujii H. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 2006;6:924–936. doi: 10.1111/j.1567-1364.2006.00089.x. [DOI] [PubMed] [Google Scholar]
- Keller G. Ray E. Brown P.O. Winge D.R. Haa1, a protein homologous to the copper-regulated transcription factor Ace1, is a novel transcriptional activator. J Biol Chem. 2001;276:38697–38702. doi: 10.1074/jbc.M107131200. [DOI] [PubMed] [Google Scholar]
- Krebs H.A. Wiggins D. Stubbs M. Sols A. Bedoya F. Studies on the mechanism of the antifungal action of benzoate. Biochem J. 1983;214:657–663. doi: 10.1042/bj2140657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren A. Mamnun Y.M. Bauer B.E. Schuller C. Wolfger H. Hatzixanthis K., et al. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol Cell Biol. 2003;23:1775–1785. doi: 10.1128/MCB.23.5.1775-1785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.L. Botting C.H. Antrobus R. Coote P.J. Evidence of a new role for the high-osmolarity glycerol mitogen-activated protein kinase pathway in yeast: regulating adaptation to citric acid stress. Mol Cell Biol. 2004;24:3307–3323. doi: 10.1128/MCB.24.8.3307-3323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel K. Macpherson S. Turcotte B. New tools for phenotypic analysis in Candida albicans: the WAR1 gene confers resistance to sorbate. Yeast. 2006;23:249–259. doi: 10.1002/yea.1346. [DOI] [PubMed] [Google Scholar]
- Liu M. Brusilow W.S. Needleman R. Activity of the yeast Tat2p tryptophan permease is sensitive to the anti-tumor agent 4-phenylbutyrate. Curr Genet. 2004;46:256–268. doi: 10.1007/s00294-004-0531-7. [DOI] [PubMed] [Google Scholar]
- Lund B. Eklund T. Control of pH and use of organic acids. In: Lund B, editor; Baird-Parker T, editor; Gould G, editor. The Microbiological Safety and Quality of Food. Springer; Berlin: 2000. [Google Scholar]
- Makrantoni V. Dennison P. Stark M.J. Coote P.J. A novel role for the yeast protein kinase Dbf2p in vacuolar H+-ATPase function and sorbic acid stress tolerance. Microbiology. 2007;153:4016–4026. doi: 10.1099/mic.0.2007/010298-0. [DOI] [PubMed] [Google Scholar]
- Marion R.M. Regev A. Segal E. Barash Y. Koller D. Friedman N., et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell P. Stratford M. Steels H. Fernandez-Espinar M.T. Querol A. Physiological characterization of spoilage strains of Zygosaccharomyces bailii and Zygosaccharomyces rouxii isolated from high sugar environments. Int J Food Microbiol. 2007;114:234–242. doi: 10.1016/j.ijfoodmicro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Mima S. Ushijima H. Hwang H.J. Tsutsumi S. Makise M. Yamaguchi Y., et al. Identification of the TPO1 gene in yeast, and its human orthologue TETRAN, which cause resistance to NSAIDs. FEBS Lett. 2007;581:1457–1463. doi: 10.1016/j.febslet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Mira N.P. Lourenco A.B. Fernandes A.R. Becker J.D. Sá-Correia I. The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res. 2009;9:202–216. doi: 10.1111/j.1567-1364.2008.00473.x. [DOI] [PubMed] [Google Scholar]
- Mira N.P. Becker J. Sá-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS. 2010;14 doi: 10.1089/omi.2010.0048. (This issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M. Piper P.W. The ZbYME2 gene from the food spoilage yeast Zygosaccharomyces bailii confers not only YME2 functions in Saccharomyces cerevisiae, but also the capacity for catabolism of sorbate and benzoate, two major weak organic acid preservatives. Mol Microbiol. 2001;42:919–930. doi: 10.1046/j.1365-2958.2001.02686.x. [DOI] [PubMed] [Google Scholar]
- Mollapour M. Piper P.W. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol. 2007;27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M. Fong D. Balakrishnan K. Harris N. Thompson S. Schuller C., et al. Screening the yeast deletant mutant collection for hypersensitivity and hyper-resistance to sorbate, a weak organic acid food preservative. Yeast. 2004;21:927–946. doi: 10.1002/yea.1141. [DOI] [PubMed] [Google Scholar]
- Mollapour M. Phelan J.P. Millson S.H. Piper P.W. Cooke F.T. Weak acid and alkali stress regulate phosphatidylinositol bisphosphate synthesis in Saccharomyces cerevisiae. Biochem J. 2006;395:73–80. doi: 10.1042/BJ20051765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa M.-Y.S. Sobel J.D. Elhais H. Du W. Akins R.A. Fungicidal activity of fluconazole against Candida albicans in a synthetic vagina-simulative medium. Antimicrob Agents Chemotherap. 2004;48:161–167. doi: 10.1128/AAC.48.1.161-167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek S. Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res. 2008;36:2874–2888. doi: 10.1093/nar/gkm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R.D. Cormack B. Expression of Candida glabrata adhesins after exposure to chemical preservatives. J Infect Dis. 2009;199:1891–1898. doi: 10.1086/599120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S. Straffon M. Enjalbert B. Nantel A. Macaskill S. Whiteway M., et al. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell. 2004;3:1111–1123. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist E. Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates. I: inhibition and detoxification. Bioresource Technol. 2000;74:17–24. [Google Scholar]
- Pampulha M.E. Loureiro-Dias M.C. Activity of glycolytic enzymes of Saccharomyces cerevisiae in the presence of acetic acid. Appl Microbiol Biotechnol. 1990;34:375–380. [Google Scholar]
- Papadimitriou M.N. Resende C. Kuchler K. Brul S. High Pdr12 levels in spoilage yeast (Saccharomyces cerevisiae) correlate directly with sorbic acid levels in the culture medium but are not sufficient to provide cells with acquired resistance to the food preservative. Int J Food Microbiol. 2007;113:173–179. doi: 10.1016/j.ijfoodmicro.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Paumi C.M. Chuk M. Snider J. Stagljar I. Michaelis S. ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol Mol Biol Rev. 2009;73:577–593. doi: 10.1128/MMBR.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce A.K. Booth I.R. Brown A.J. Genetic manipulation of 6-phosphofructo-1-kinase and fructose 2,6-bisphosphate levels affects the extent to which benzoic acid inhibits the growth of Saccharomyces cerevisiae. Microbiology. 2001;147:403–410. doi: 10.1099/00221287-147-2-403. [DOI] [PubMed] [Google Scholar]
- Penalva M.A. Tilburn J. Bignell E. Arst H.N., Jr Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Piper P. Mahe Y. Thompson S. Pandjaitan R. Holyoak C. Egner R., et al. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17:4257–4265. doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. Calderon C.O. Hatzixanthis K. Mollapour M. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology. 2001;147:2635–2642. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]
- Piper P.W. Yeast superoxide dismutase mutants reveal a pro-oxidant action of weak organic acid food preservatives. Free Radic Biol Med. 1999;27:1219–1227. doi: 10.1016/s0891-5849(99)00147-1. [DOI] [PubMed] [Google Scholar]
- Plumridge A. Stratford M. Lowe K.C. Archer D.B. The weak-acid preservative sorbic acid is decarboxylated and detoxified by a phenylacrylic acid decarboxylase, PadA1, in the spoilage mold Aspergillus niger. Appl Environ Microbiol. 2008;74:550–552. doi: 10.1128/AEM.02105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdale M. Selway L. Stead D. Walker J. Yin Z. Nicholls S.M., et al. MNL1 regulates weak acid-induced stress responses of the fungal pathogen Candida albicans. Mol Biol Cell. 2008;19:4393–4403. doi: 10.1091/mbc.E07-09-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J.E. Schultz E. Snyder R.E. Jones R.S. Smith C.R. Acetic-acid as a causative agent in producing stuck fermentations. Am J Enol Viticult. 1995;46:278–280. [Google Scholar]
- Ro D.K. Ouellet M. Paradise E.M. Burd H. Eng D. Paddon C.J., et al. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008;8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A. Gregori C. Jennings A.M. Quintin J. Ferrandon D. Butler G., et al. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol Microbiol. 2008;69:603–620. doi: 10.1111/j.1365-2958.2008.06301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Correia I. Salgueiro S.P. Viegas C.A. Novais J.M. Leakage induced by ethanol and octanoic and decanoic acids in Saccharomyces cerevisiae. Yeast. 1989;5:S123–S127. [Google Scholar]
- Sá-Correia I. Dos Santos S.C. Teixeira M.C. Cabrito T.R. Mira N.P. Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 2009;17:22–31. doi: 10.1016/j.tim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Saint-Marc C. Pinson B. Coulpier F. Jourdren L. Lisova O. Daignan-Fornier B. Phenotypic consequences of purine nucleotide imbalance in Saccharomyces cerevisiae. Genetics. 2009;183:529–538. doi: 10.1534/genetics.109.105858. 1SI–7SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller C. Mamnun Y.M. Mollapour M. Krapf G. Schuster M. Bauer B.E., et al. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller C. Mamnun Y.M. Wolfger H. Rockwell N. Thorner J. Kuchler K. Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4932–4944. doi: 10.1091/mbc.E07-06-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenhar G. Kassir Y. A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:1603–1612. doi: 10.1128/MCB.21.5.1603-1612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões T. Teixeira M.C. Fernandes A.R. Sá-Correia I. Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes: important role of SPI1. Appl Environ Microbiol. 2003;69:4019–4028. doi: 10.1128/AEM.69.7.4019-4028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões T. Mira N.P. Fernandes A.R. Sá-Correia I. The SPI1 gene, encoding a glycosylphosphatidylinositol (GPI)-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak acids food preservatives. Appl Environ Microb. 2006;72:7168–7175. doi: 10.1128/AEM.01476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M.J. Rodrigues F. Corte-Real M. Leão C. Mechanisms underlying the transport and intracellular metabolism of acetic acid in the presence of glucose in the yeast Zygosaccharomyces bailii. Microbiology. 1998;144(Pt 3):665–670. doi: 10.1099/00221287-144-3-665. [DOI] [PubMed] [Google Scholar]
- Stevens S. Hofemyer J.-H.S. Effects of ethanol, octanoic and decanoic acids on fermentation and the passive influx of protons through the plasma membrane of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1993;38:656–663. [Google Scholar]
- Stratford M. Anslow P.A. Comparison of the inhibitory action on Saccharomyces cerevisiae of weak-acid preservatives, uncouplers, and medium-chain fatty acids. FEMS Microbiol Lett. 1996;142:53–58. doi: 10.1111/j.1574-6968.1996.tb08407.x. [DOI] [PubMed] [Google Scholar]
- Stratford M. Plumridge A. Archer D.B. Decarboxylation of sorbic acid by spoilage yeasts is associated with the PAD1 gene. Appl Environ Microbiol. 2007;73:6534–6542. doi: 10.1128/AEM.01246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.C. Sá-Correia I. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem Biophys Res Commun. 2002;292:530–537. doi: 10.1006/bbrc.2002.6691. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C. Telo J.P. Duarte N.F. Sá-Correia I. The herbicide 2,4-dichlorophenoxyacetic acid induces the generation of free-radicals and associated oxidative stress responses in yeast. Biochem Biophys Res Commun. 2004;324:1101–1107. doi: 10.1016/j.bbrc.2004.09.158. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C. Santos P.M. Fernandes A.R. Sá-Correia I. A proteome analysis of the yeast response to the herbicide 2,4-dichlorophenoxyacetic acid. Proteomics. 2005;5:1889–1901. doi: 10.1002/pmic.200401085. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C. Fernandes A.R. Mira N.P. Becker J.D. Sá-Correia I. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 2006a;6:230–248. doi: 10.1111/j.1567-1364.2006.00041.x. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C. Monteiro P. Jain P. Tenreiro S. Fernandes A.R. Mira N.P., et al. The YEASTRACT database: a tool for the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006b;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.C. Duque P. Sá-Correia I. Environmental genomics: mechanistic insights into toxicity of and resistance to the herbicide 2,4-D. Trends Biotechnol. 2007;25:363–370. doi: 10.1016/j.tibtech.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Tenreiro S. Rosa P.C. Viegas C.A. Sá-Correia I. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast. 2000;16:1469–1481. doi: 10.1002/1097-0061(200012)16:16<1469::AID-YEA640>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Tenreiro S. Nunes P.A. Viegas C.A. Neves M.S. Teixeira M.C. Cabral M.G., et al. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2002;292:741–748. doi: 10.1006/bbrc.2002.6703. [DOI] [PubMed] [Google Scholar]
- Thakur J.K. Arthanari H. Yang F. Pan S.J. Fan X. Breger J, et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- Viegas C.A. Sá-Correia I. Activation of plasma membrane ATPase of Saccharomyces cerevisiae by octanoic acid. J Gen Microbiol. 1991;137:645–651. doi: 10.1099/00221287-137-3-645. [DOI] [PubMed] [Google Scholar]
- Viegas C.A. Almeida P.F. Cavaco M. Sá-Correia I. The H(+)-ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl Environ Microbiol. 1998;64:779–783. doi: 10.1128/aem.64.2.779-783.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas C.A. Cabral M.G. Teixeira M.C. Neumann G. Heipieper H.J. Sá-Correia I. Yeast adaptation to 2,4-dichlorophenoxyacetic acid involves increased membrane fatty acid saturation degree and decreased OLE1 transcription. Biochem Biophys Res Commun. 2005;330:271–278. doi: 10.1016/j.bbrc.2005.02.158. [DOI] [PubMed] [Google Scholar]
- Warth A.D. Effect of benzoic acid on glycolytic metabolite levels and intracellular pH in Saccharomyces cerevisiae. Appl Environ Microbiol. 1991;57:3415–3417. doi: 10.1128/aem.57.12.3415-3417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]