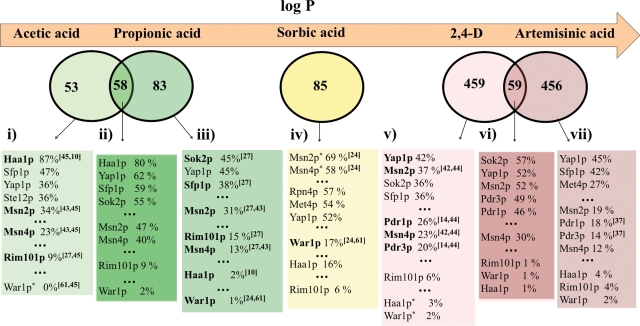
FIG. 4.
Clustering of the genes upregulated in response to acetic, propionic, sorbic, and artemisinic acids (Mira et al., 2009, 2010; Ro et al., 2008; Schuller et al., 2004) or to 2,4-D (Teixeira et al., 2006a) with their documented regulators, according to the information deposited in the YEASTRACT database (March 2010), plus data coming from our recent work (Mira et al., 2010). Based on the level of overlapping, the responsive genes were clustered in three groups: acetic and propionic acid-induced genes, sorbic acid-induced genes and artemisinic acid- and 2,4-D-induced genes. YEASTRACT database was used to cluster, with their documented regulators, the genes that: (1) are activated in cells exposed to acetic acid but not in the presence of propionic acid stress; (2) are upregulated upon exposure to acetic or propionic acid; (3) are activated in cells exposed to propionic acid but not upon acetic acid challenge; (4) are upregulated in yeast cells exposed to sorbic acid; (5) are activated in cells exposed to 2,4D but not in the presence of artemisinic acid stress; (6) are upregulated upon exposure to 2,4-D or artemisinic acid; (7) are activated in cells exposed to artemisinic acid but not upon 2,4-D challenge. The transcription factors were organized by decreasing percentage of documented targets in each dataset. The transcription factors that provide protection toward a given weak acid are distinguished (in bold) from those known to be dispensable for tolerance (indicated with *).