Abstract
Evidence for horizontal routes of transmission for Rickettsia felis has come from detection of R. felis infection in vertebrates and multiple blood-feeding arthropods; however, infection of cat fleas, Ctenocephalides felis, during blood feeding has not been demonstrated. In this study, the ability of cat fleas to acquire R. felis through an infectious blood meal with subsequent vertical transmission was examined. Utilizing an artificial feeding system, Rickettsia-naive fleas were exposed to R. felis–infected blood meals and monitored for subsequent infection at weekly intervals for 4 weeks. At 7 days postexposure (dpe) ∼52% of fleas successfully acquired rickettsiae and R. felis DNA; rickettsial transcript and DNA was detected in cat flea feces. Quantitative real-time polymerase chain reaction determined that both the R. felis infection load and R. felis infection density was significantly greater in fleas assessed at later time points. Although a persistent R. felis infection was detected in adult fleas, R. felis infection was not observed in F1 progeny. This study demonstrates that cat fleas are able to acquire R. felis infection from an infectious blood meal and will serve as a model to examine R. felis transmission between arthropod and vertebrate hosts.
Key Words: Cat flea, Flea-borne disease, Rickettsia felis
Introduction
Rickettsia felis, a gram-negative bacterium, has been identified from both vertebrates and arthropods worldwide. In humans, R. felis is considered an emerging pathogen as the etiologic agent of flea-borne rickettsiosis (Perez-Osorio et al. 2008). Clinical characteristics of R. felis rickettsiosis range from a mild to moderate flu-like illness with symptoms similar to other rickettsial infections, including fever, headache, and fatigue to severe disease associated with hepatic, pulmonary, and central nervous system manifestations (Zavala-Velazquez et al. 2000, Zavala-Castro et al. 2009). Molecular and serological surveys have detected R. felis infection of peridomestic mammals that commonly serve as hosts for fleas, including dogs, cats, and opossums (Schriefer et al. 1994, Richter et al. 2002, Labruna et al. 2007); however, a persistently infected vertebrate reservoir has yet to be clearly implicated in the transmission cycle.
Since the first description as the ELB (Elward Lab) agent in a commercial cat flea (Ctenocephalides felis) colony in 1990 (Adams et al. 1990), R. felis has been associated with at least 22 arthropod species worldwide (Berrelha et al. 2009, Blair et al. 2004, Maioli et al. 2009, Reif and Macaluso 2009, Varagnol et al. 2009). Utilization of molecular assays, traditional and quantitative real-time polymerase chain reaction (qPCR), has increased the known flea hosts associated with R. felis; however, only C. felis is currently recognized as a biological vector. Under laboratory conditions, R. felis is predominantly maintained within cat flea cohorts by vertical (transovarial and transstadial) transmission (Azad et al. 1992, Wedincamp and Foil 2002). The capacity to maintain R. felis infection via vertical transmission decreased from ∼65% to <5% when the colony was exposed to rickettsiae-free blood over 12 generations (Wedincamp and Foil 2002). Such decrease in prevalence in the absence of an infectious blood meal, combined with the increasing diversity of infected fleas, suggests that transmission mechanisms outside the vertical route are necessary for maintenance of R. felis in flea populations.
A complex transmission cycle of flea-borne rickettsial diseases has been described previously. Rickettsia typhi, the agent of flea-borne murine typhus, is primarily transmitted to vertebrate hosts via infectious flea feces. However, R. typhi can be ingested by fleas during blood meal acquisition and transmitted both vertically to progeny and horizontally to cofeeding fleas via the vertebrate host blood (Azad 1990). Evidence for horizontal routes of transmission for R. felis has come from detection of R. felis infection in vertebrates and multiple blood-feeding arthropods; however, infection of cat fleas during blood feeding has not been demonstrated. In the current study, the ability of cat fleas to acquire R. felis through an infectious blood meal and subsequently pass the organism to their progeny was examined. Using an artificial flea feeding system, the kinetics of rickettsial infection of Rickettsia-naive cat fleas were examined using a qPCR assay. Additionally, vertical transmission of R. felis to progeny by newly infected cat fleas was assessed. Although acquisition and persistent infection of cat fleas with R. felis was observed, vertical transmission was not evident. Implications of these results in the context of sustained R. felis transmission are discussed.
Materials and Methods
Source of fleas and Rickettsia
Newly emerged, unfed cat fleas (C. felis) were purchased from Elward II (EL). Cat fleas from EL Laboratory have been previously reported to be specifically free of R. felis infection (Pornwiroon et al. 2007). Adult fleas were provided a blood meal via an artificial dog (Wade and Georgi 1988), and eggs, not separated from feces, were reared to adults on sand with artificial diet as previously described (Lawrence and Foil 2002). R. felis (LSU), originally isolated from the Louisiana State University cat flea colony, was propagated in an Ixodes scapularis–derived cell line (ISE6) maintained in modified L15B growth medium, and cellular infection was examined by Diff-Quik (Dade Behring) staining as described previously by Pornwiroon et al. (2006).
Nucleic acid isolation
Individual sampled fleas were assigned sample numbers, placed in 0.5 mL microcentrifuge tubes, and pulverized with sterile plastic pestles in a liquid nitrogen bath. Genomic DNA (gDNA) was extracted using a modified version of the HotSHOT DNA extraction protocol (Truett et al. 2000). Briefly, individual flea lysates were incubated at 95°C for 45 min in 20 μL alkaline lysis reagent (25 mM NaOH and 0.2 mM disodium ethylenediaminetetraacetic acid, pH 12), cooled to 4°C for 5 min, and mixed with 20 μL of neutralizing reagent (40 mM Tris-HCl, pH of 5). All gDNA preparations were stored at −20°C. For flea feces, at 2, 4, 7, 14, 21, and 28 days postexposure (dpe), ∼50 mg of egg-free flea feces were collected and dissolved in 200 μL of 1 × phosphate-buffered saline. Extraction of gDNA from feces was accomplished using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's instructions for extraction of DNA from blood samples and eluted in 40 μL Buffer AE. Total RNA from flea feces was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions for total RNA isolation from cells with the following modifications: ∼50-mg of feces was disrupted and homogenized with two stainless steel 4.77-mm balls in 200 μL of Buffer RLT using a TissueLyser (Qiagen), for 1 min at 30 Hz. Samples were washed as directed with an additional wash in 500 μL Buffer RPE and eluted in 30 μL RNase-free water. RNA samples were DNaseI treated (Promega) according to the manufacturer's instructions. Gene-specific cDNA was synthesized using SuperScript First-Strand Synthesis System (Invitrogen) with R. felis 17-kDa primers (Reif et al. 2008). For all samples no-RT controls were included to verify the absence of DNA contamination.
Rickettsial detection and quantification by PCR
Tissue culture samples or individual fleas were assessed for R. felis infection by qPCR amplification of a 157-bp portion of the R. felis 17-kDa antigen gene as previously described (Reif et al. 2008). To confirm the absence of rickettsial infection before exposure to R. felis, a subset of fleas was additionally screened by qPCR amplification of a 128-bp portion of the rickettsial outer membrane protein B (ompB) gene (Labruna et al. 2007). Briefly, qPCR components and the template that included 2 × iTaq SYBR Green Supermix (BioRad); 100 nM of each primer; DNase/RNase-free water; and 5 μL of gDNA template (samples), water (negative control), or serial 10-fold dilutions (1 × 108 to 10 copies) of pCR4-TOPO-Rf17kDa+Cf18SrDNA were premixed in 35 μL volumes in 96-well plates and aliquoted in triplicate 10 μL reactions on 384-well plates. The qPCR was performed with an ABI 7900HT unit (Applied Biosystems) using conditions previously described (Reif et al. 2008). Results were analyzed with ABI 7900HT sequence detection system (SDS v2.3) software. The specificity of the assay was verified; the expected single peak for the internal control plasmid and positive gDNA samples, but not in the water (negative control) samples, was identified in the dissociation curve. Additionally, representative qPCR products from each trial were verified by gel analysis to confirm the specificity of the reaction; cloning and sequencing confirmed that fleas were infected with R. felis. The sensitivity of this qPCR is 10 copies of plasmid per reaction or ∼267 rickettsiae per 40 μL of flea lysate. For R. felis–positive flea samples, a second assay determined R. felis infection density in individual fleas. Both the R. felis 17-kDa and cat flea 18S rDNA genes were amplified and quantified by extrapolating the individual gene Ct values from serial dilutions of plasmid pCR4-TOPO-Rf17kDa+Cf18SrDNA, which contained single-copy portions of both genes as previously described (Reif et al. 2008).
R. felis–infected blood meal preparation
Exposure blood meals were prepared by resuspending a confluent T-75 flask containing ISE6 cells >90% infected with R. felis (passage 4). To prepare the R. felis exposure dose, BacLight viability stain kit (Invitrogen) was used to assess viability and enumerate rickettsiae as previously described (Sunyakumthorn et al. 2008). The concentration of rickettsiae in culture was additionally verified by qPCR amplification of the R. felis 17-kDa gene. The R. felis–infected cell solution was diluted to 5 × 109 rickettsiae, subjected to centrifugation at 13,000 g for 10 min to remove the growth medium, and resuspended in 600 μL of heat-inactivated, defibrinated bovine blood (Rockland Immunochemicals). Intact, R. felis–uninfected ISE6 cells were prepared in an identical manner and served as a negative control for the feeding treatment.
Acquisition of R. felis via infectious blood meal
In two replicate trials, each containing three identical experimental groups and one control group, fleas (125 mixed sex fleas per group) were exposed for 24 h to either a R. felis–infected or –uninfected blood meal. After the exposure meal, cat fleas were maintained on defibrinated bovine blood (not heat-inactivated) that was replaced every 1–3 days for the duration of the experiment. Fleas were examined at weekly intervals for acquisition and persistence of R. felis infection. At each collection point (7, 14, 21, and 28 dpe), gDNA was extracted from 10 individual randomly selected fleas and R. felis infection was determined by qPCR. For each trial the mean (1) incidence of R. felis infection, (2) R. felis infection load per flea lysate, and (3) R. felis infection density were compared at each time point.
Detection of R. felis in cat flea feces
A portion of egg-free flea feces was collected at 2, 4, 7, 14, 21, and 28 dpe from groups of fleas fed either an R. felis–infected or –uninfected blood meal. To determine the presence and quantity of R. felis gDNA, 50-mg samples of feces were examined by qPCR amplification of a portion of the R. felis 17-kDa gene. To assess the viability of R. felis in flea feces, qPCR was used to amplify R. felis 17-kDa-specific cDNA. Due to limited starting material, rickettsial transcript was only assessed in trial 1 on days 9, 21, and 28 dpe.
Vertical transmission of R. felis to flea progeny
In both trials, eggs from fleas exposed to R. felis–infected blood meals were collected, pooled by week postparental blood meal exposure, and reared to adults in an incubator maintained at 27°C and 70% relative humidity. From each group, 10 unfed first-generation (F1) adult cat fleas, collected as eggs 1–4 weeks postparental exposure to the R. felis–infected blood meal, were randomly selected and examined for R. felis infection. Total gDNA was extracted from F1 cat fleas and the presence of R. felis and sample quality were determined by qPCR, as described above.
Statistical analysis
Rickettsial load in fleas and the ratio of Rf17kDa/Cf18S were assessed after the logarithmic transformation of the quantity of the genes of interest (Rf17kDa and Cf18S). Analysis of variance (SAS statistical package, Version 9.1.3, GLM procedure) was performed to examine potential differences between R. felis infection load in fleas and ratio of Rf17kDa/Cf18S copy number over the study period; when overall significance was found, Tukey's honestly significant difference post hoc test was used to examine pairwise differences of means of main effects. Pairwise t-tests of least squares means were performed to determine any interaction effects between trial, group, and day for R. felis infection load and ratio of Rf17kDa/Cf18S. An F-test was used for general comparisons of grouped means. For all comparisons, a p-value of <0.05 was considered significantly different.
Results
Acquisition and growth of R. felis in naive fleas exposed to a R. felis–infected blood meal
To verify the absence of R. felis infection in this colony 20 (10 female and 10 male) newly emerged unfed EL fleas were assessed for R. felis infection by qPCR amplification as previously described (Reif et al. 2008). No rickettsial product was amplified from any flea sample, confirming the absence of R. felis in the EL C. felis colony as documented in previous reports (data not shown) (Pornwiroon et al. 2007). In replicate trials, three groups of fleas were fed R. felis–infected blood meals and a fourth group (control) was fed an uninfected blood meal for 24 h. After the exposure meal 10 fleas per group were assessed for R. felis infection at weekly intervals. The specificity of R. felis infection in fleas was confirmed by sequencing a portion of the rickettsial 17-kDa gene from a representative subset of fleas positive for R. felis infection. All sequenced samples had a 100% nucleotide identity to R. felis URRWXCal2, complete genome (GenBank accession number CP000053).
All groups fed an R. felis–infected blood meal had R. felis–positive individuals for up to 4 weeks postexposure meal, with a 69% and 48% mean incidence of R. felis infection in trials 1 and 2, respectively (Table 1). The incidence of R. felis infection ranged from 30% to 100% and 10% to 90% within trials 1 and 2, respectively.
Table 1.
Incidencea of Rickettsia felis Infection in Cat Fleas Exposed to an Rickettsia felis–Infected Blood meal
| Trial 1 | Trial 2 | |
|---|---|---|
| 7 dpe | 63% ± 13% | 40% ± 15% |
| 14 dpe | 52% ± 19% | 43% ± 24% |
| 21 dpe | 80% ± 12% | 53% ± 13% |
| 28 dpe | 81% ± 12% | 53% ± 15% |
| Mean | 69% | 48% |
Values are means ± standard error of the mean (10 samples per group) of R. felis infection incidence in fleas exposed to an R. felis–infected blood meal.
dpe, days postexposure.
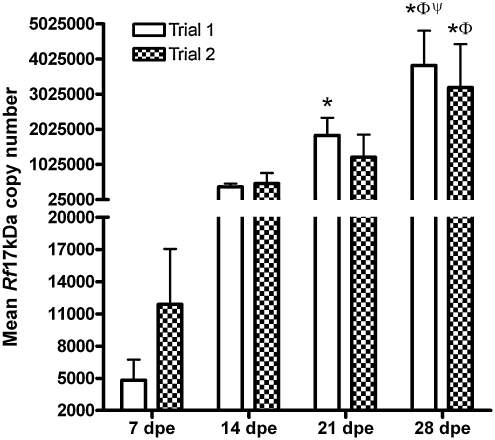
R. felis infection load was determined in R. felis–exposed flea groups by quantifying the copy number of Rf17kDa per individual flea lysate. The mean R. felis infection load per flea lysate was 1.52 × 106 and 1.24 × 106 rickettsiae per flea lysate in trials 1 and 2, respectively. R. felis infection load was significantly greater in fleas assessed at 28 dpe than at 7 dpe (Fig. 1). Specifically, trial 1 fleas had significantly greater R. felis infection loads at 21 and 28 dpe than at 7 dpe. In trial 2 similar trends of increasing R. felis infection load at the later assessment point with a significant increase only observed between 7 and 28 dpe. The range of the R. felis infection load (2.8 × 101 to 1.6 × 107) was expansive. Rickettsial gDNA was never detected in any control fleas fed blood meals with uninfected ISE6 cells.
FIG. 1.
Rickettsial load in fleas exposed to an Rickettsia felis–infected blood meal. In replicate trials, three groups of fleas were fed identical R. felis–infected blood meals for 1 day and 10 fleas per group were individually assessed at weekly intervals for R. felis infection load (Rf17kDa copy number per flea lysate) by quantitative real-time polymerase chain reaction. An overall mean of 1.52 × 106 and 1.24 × 106 rickettsiae per flea lysate was observed in trials 1 and 2, respectively. Bars represent weekly mean Rf17kDa copies per individual flea lysate, ±standard error of the mean. In both trials 1 and 2, a significant increase in the rickettsial load was observed in fleas assessed at the later time points. *Significant difference from 7 days postexposure (dpe), Φsignificant difference from 14 dpe, and Ψsignificant difference from 21 dpe.
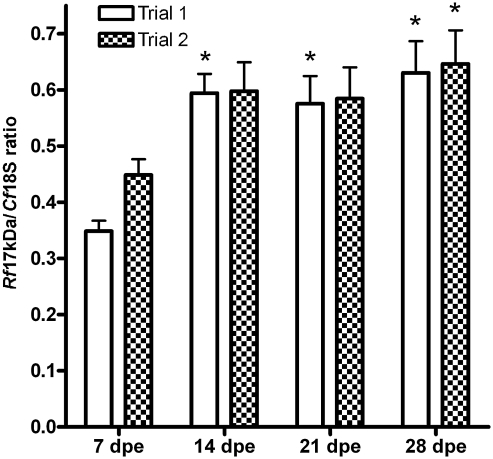
The density of R. felis infection in individual fleas was determined by comparing the mean trial (±standard error of the mean) Rf17kDa/Cf18S ratio for each collection point. The mean Rf17kDa/Cf18S ratio (and range) was 0.54 (0.33–0.80) and 0.57 (0.35–0.71) for trials 1 and 2, respectively. In trial 1, the mean Rf17kDa/Cf18S ratio significantly increased from 7 to 14 dpe and remained significantly greater at all other collection points compared to 7 dpe (Fig. 2). In trial 2, a similar trend of increasing R. felis infection density was observed, but only fleas at 28 dpe had significantly greater rickettsial infection densities compared to fleas at 7 dpe (Fig. 2).
FIG. 2.
Rickettsial infection density in fleas fed an R. felis–infected blood meal 7–28 dpe. The R. felis infection density was determined by quantitative real-time polymerase chain reaction of individual cat flea samples. Bars represent the mean (±standard error of the mean) Rf17kDa/Cf18S ratio of three experimental groups exposed to identical R. felis exposure meals for 1 day and assessed at weekly intervals in two replicate trials. Mean R. felis infection density was 0.54 and 0.57 in trials 1 and 2, respectively, and significantly greater density was observed in fleas sampled at later time points. *Significant difference from 7 dpe.
Detection of R. felis in flea feces
Total gDNA was extracted from samples of egg-free flea feces from each group and assessed at 2, 4, 7, 14, 21, and 28 dpe for fecal shedding of rickettsiae. Rickettsial gDNA was detectable in flea feces up to 28 dpe and was observed in most of the R. felis–exposed groups at each collection point. Significant variations in R. felis 17-kDa copy number in feces across collection time points were assessed by comparing the logarithmically transformed mean R. felis 17-kDa copy number per 50-mg flea feces at individual collection points. An initial significant decline in R. felis 17-kDa copies in flea feces was observed between 2 dpe (1.57 × 107 rickettsiae) and 4 dpe (1.7 rickettsiae). At 4 dpe rickettsial gDNA could only be detected in one group in trial 1 and no groups in trial 2. For both trials, mean rickettsial load detected in feces at 7, 14, 21, and 28 dpe was 4.37 × 105 rickettsiae per 50-mg feces. At these weekly collection points, rickettsial load did not vary significantly among groups within trials. The shedding pattern of individual R. felis–infected adult fleas was not assessed in this study. Feces from all control flea groups and environmental controls were consistently negative for R. felis DNA. At 9, 21, and 28 dpe in trial 1, the viability of R. felis in flea feces was assessed by amplification of R. felis 17-kDa cDNA synthesized from flea feces total RNA extracts. Rickettsial transcription of the 17-kDA antigen gene was detected in flea feces only at 21 dpe.
Vertical transmission of R. felis
F1 progeny were collected as eggs or larvae, reared to adults, and assessed for R. felis infection. From batches of eggs (pooled by week postparental R. felis–infected blood meal) 10 unfed F1 adults from every group were individually examined for vertically transmitted R. felis infection by qPCR amplification of a portion of the rickettsial 17-kDa antigen gene. Despite R. felis infection in adults, R. felis infection was not detected in any F1 progeny.
Discussion
The present study demonstrates horizontal acquisition (with a resulting persistent infection) of R. felis by an arthropod feeding on an infectious blood meal. The present study demonstrates horizontal acquisition (with a resulting persistent infection) of R. felis by an arthropod feeding on an infectious blood meal. The absence of documented horizontal transmission of R. felis between arthropod and vertebrate hosts via blood feeding suggests an inability of arthropods to acquire R. felis infection from an infected vertebrate host and that vertical transmission between fleas is the only transmission route involved in the maintenance of R. felis in nature (Weinert et al. 2009). While the ability of R. felis to be transmitted between two hosts has yet to be unequivocally demonstrated, oral acquisition of a new infection by hematophagous arthropods is a perquisite to successful horizontal transmission.
Potential R. felis horizontal transmission mechanisms and routes have been examined previously, but were unsuccessful in infecting fleas. Cat fleas fed an R. felis–infected blood using an artificial host for 2–3 days and their progeny were PCR negative for R. felis infection (Wedincamp and Foil 2002). In another experiment, uninfected fleas were fed on R. felis seropositive cats; however, all fleas and their progeny were negative for R. felis infection (Wedincamp and Foil 2002). The contrast of successful R. felis acquisition between earlier infection attempts and the current study may be due to the infectious meal preparation. Previous studies utilized undefined amounts of rickettsiae delivered in ground R. felis–infected flea lysates in nonheat-inactivated blood (Wedincamp and Foil 2002), compared to quantified cultures of R. felis–infected intact ISE6 cells fed to fleas in heat-inactivated blood in the current study. Heat inactivation of the delivery medium would limit the potential for complement-mediated lysis of rickettsiae in the preparation. In experiments using seropositive animals, lack of R. felis acquisition by fleas may be the result of minimal or already resolved infection in the animals (Wedincamp and Foil 2002), as a seropositive animal does not imply an active infection.
Interestingly, in an experiment on R. felis vertical transmission, R. felis prevalence waned (63%–2.5% over 12 generations) in fleas artificially fed uninfected blood meals, versus fleas fed on a cat host that retained a 65% R. felis infection prevalence (Wedincamp and Foil 2002), supporting the likelihood of flea acquisition of R. felis via a blood meal. Transmission of R. felis could require a minimum threshold infection in the vertebrate (e.g., during periods of bacteremia) for a feeding arthropod to ingest a sufficient amount of bacteria to become infected. The quantity of ingested bacteria necessary to result in arthropod infection can vary among bacterial species and strains. For example, R. typhi is acquired by feeding fleas during periods of rickettsemia (days 3–20 postinoculation) (Farhang-Azad et al. 1983), but requires ingestion of only a few rickettsiae for rat fleas to become infected and subsequently able to transmit R. typhi to another vertebrate (Vaughan and Azad 1990). Compared to R. typhi, acquisition and infection of fleas with R. felis likely requires a greater infection dose or host bacteremia. In the current study, fleas were exposed to a ∼8 × 106 R. felis per microliter of blood and were able to acquire a persistent R. felis infection. As fleas were allowed to feed freely on the R. felis–infected blood meal the specific infectious dose was not determined. Future studies are needed to define additional vertebrate-to-arthropod R. felis transmission parameters, including the minimum infectious dose.
In a previous study, R. felis infection load and density were examined over time in fleas naturally infected with R. felis, feeding on a cat host (Reif et al. 2008). Although the R. felis incidence was observed to decrease in the population over time during individual experiments, the R. felis infection load and infection density remained relatively stable with no association between increased rickettsial replication and flea reproduction observed. Despite a reduced mean infection density compared to vertically R. felis–infected fleas, fleas in the current study were able to acquire and develop a persistent R. felis infection for several weeks (similar to other pathogenic rickettsial species) after feeding on an infectious blood meal (Azad and Beard 1998).
The predominant route of transmission for most insect-transmitted pathogenic Rickettsia (e.g., R. typhi and R. prowazekii) is via fecal transmission (Azad 1990). As the cat flea is the only currently known biological vector of R. felis, the potential of horizontal transmission via fecal transmission must be considered. Previously, R. felis DNA was detected in the feces of cat fleas that fed on bovine blood containing R. felis–infected flea homogenates, but not human blood containing R. felis from culture 6 days post–R. felis meal (Wedincamp and Foil 2002). Acquisition of R. felis infection by flea larvae feeding on feces, flea eggs, and younger instar larvae positive for R. felis gDNA was also examined, but all resulting adults were negative for R. felis infection (Wedincamp and Foil 2002). In the current study, the potential for fecal transmission was also considered, as R. felis gDNA was detected in feces, not only directly after an R. felis–infected blood meal, but also up to 28 dpe. The high Rf17kDa copy number in the feces at 2 dpe reflects the large exposure dose of R. felis in the infected blood meals, followed by the significant decrease observed at 4 dpe as fleas finished digesting and excreting the exposure meal and continued to feed on uninfected blood. R. felis–positive feces at time points after 4 dpe were the result of rickettsial shedding by fleas, not contamination from a digested R. felis–infected blood meal as fleas were transferred into clean feeding cage several times during the study. The continued presence of R. felis gDNA in flea feces at later time points suggests active rickettsial infection. Differences in flea number and R. felis infection incidence in a group at any one time likely contributed to the variable presence of R. felis DNA observed in groups at each collection time point.
To address the viability of R. felis in flea feces, rickettsial RNA was amplified from flea feces. Due to limited sample, RNA was extracted from fecal samples only in trial 1 at 9, 21, and 28 dpe. Despite R. felis RNA being detected at only 21 dpe, the presence of viable R. felis in flea feces may indicate an additional transmission route. Further, the sensitivity of R. felis transcript detection in flea could be improved by targeting constitutively expressed transcripts (e.g., 16S rRNA). Fecal-borne transmission of other rickettsial species traditionally transmitted by saliva during arthropod feeding has also been studied. Under experimental conditions, Rickettsia rickettsii and Rickettsia conorii (both tick-transmitted rickettsial species) have been identified in louse feces by Immunofluorescence and PCR amplification of the rickettsial ompA gene (Houhamdi and Raoult 2006). As with R. rickettsii and R. conorii in lice feces, the viability and infectiousness of R. felis in flea feces is unknown and additional studies will be needed to determine if any of these rickettsial species, similar to Bartonella species, in insect feces are infectious to either arthropods or vertebrates.
Arthropods are the primary host of all rickettsial species and many rickettsial species are inherited and maintained via transovarial and transstadial transmission within arthropods (Sakurai et al. 2005). Vertical transmission of R. felis has been described in detail and is the primary transmission strategy in the LSU cat flea colony (Wedincamp and Foil 2002). Although R. felis acquisition and sustained infection in fleas was demonstrated in the current study, all F1 progeny examined for vertically transmitted R. felis infection were negative. Potential factors for the lack of vertical transmission detection include the nature of the delivery preparation (Rickettsia-infected tick cells), the constitutive microbial profile of the cat fleas (Pornwiroon et al. 2007), or the rarity of vertical transmission events as an artifact of the in vitro model utilized.
The ability of cat fleas to acquire R. felis infection with subsequent development of a persistent infection after a single R. felis–infected blood meal is demonstrated. Acquisition of R. felis by this route results in mean infection densities less than that observed for vertically infected fleas. Further studies are needed to examine the dissemination of horizontally acquired R. felis in cat fleas and other factors that could influence vertical transmission to progeny. Also, additional studies examining the viability and infectiousness of R. felis in flea feces are needed, as R. felis gDNA can be detected in flea feces almost 1 month after infection. These experiments confirm that cat fleas are able to acquire R. felis infection from an R. felis–infected blood meal and should serve as a platform to develop in vivo models of R. felis transmission between arthropod and vertebrate hosts.
Acknowledgments
The authors would like to thank J.A. Macaluso for help with article preparation and Mark Guillotte for technical assistance. This research was conducted as a partial component of KER dissertation research. This work was supported by the National Institutes of Health, National Institutes of Allergy and Infectious Disease (AI069248).
Disclosure Statement
No competing financial interest exists.
References
- Adams JR. Schmidtmann ET. Azad AF. Infection of colonized cat fleas, Ctenocephalides felis (Bouche), with a rickettsia-like microorganism. Am J Trop Med Hyg. 1990;43:400–409. doi: 10.4269/ajtmh.1990.43.400. [DOI] [PubMed] [Google Scholar]
- Azad AF. Epidemiology of murine typhus. Annu Rev Entomol. 1990;35:553–569. doi: 10.1146/annurev.en.35.010190.003005. [DOI] [PubMed] [Google Scholar]
- Azad AF. Beard CB. Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1998;4:179–186. doi: 10.3201/eid0402.980205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AF. Sacci JB., Jr. Nelson WM. Dasch GA, et al. Genetic characterization and transovarial transmission of a typhus-like rickettsia found in cat fleas. Proc Natl Acad Sci U S A. 1992;89:43–46. doi: 10.1073/pnas.89.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrelha J. Briolant S. Muller F. Rolain JM, et al. Rickettsia felis and Rickettsia massiliae in Ivory Coast, Africa. Clin Microbiol Infect. 2009;15:251–252. doi: 10.1111/j.1469-0691.2008.02273.x. [DOI] [PubMed] [Google Scholar]
- Blair PJ. Jiang J. Schoeler GB. Moron C, et al. Characterization of spotted fever group rickettsiae in flea and tick specimens from northern Peru. J Clin Microbiol. 2004;42:4961–4967. doi: 10.1128/JCM.42.11.4961-4967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhang-Azad A. Traub R. Wisseman CL., Jr. Rickettsia mooseri infection in the fleas Leptopsylla segnis and Xenopsylla cheopis. Am J Trop Med Hyg. 1983;32:1392–1400. doi: 10.4269/ajtmh.1983.32.1392. [DOI] [PubMed] [Google Scholar]
- Houhamdi L. Raoult D. Experimentally infected human body lice (Pediculus humanus humanus) as vectors of Rickettsia rickettsii and Rickettsia conorii in a rabbit model. Am J Trop Med Hyg. 2006;74:521–525. [PubMed] [Google Scholar]
- Labruna MB. Ogrzewalska M. Moraes-Filho J. Lepe P, et al. Rickettsia felis in Chile. Emerg Infect Dis. 2007;13:1794–1795. doi: 10.3201/eid1311.070782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence W. Foil LD. The effects of diet upon pupal development and cocoon formation by the cat flea (Siphonaptera: Pulicidae) J Vector Ecol. 2002;27:39–43. [PubMed] [Google Scholar]
- Maioli G. Horta MC. Ogrzewalska M. Capelli G, et al. First detection of Rickettsia felis in Ctenocephalides felis fleas from Italy. Clin Microbiol Infect. 2009;15:222–223. doi: 10.1111/j.1469-0691.2008.02146.x. [DOI] [PubMed] [Google Scholar]
- Perez-Osorio CE. Zavala-Velazquez JE. Arias Leon JJ. Zavala-Castro JE. Rickettsia felis as emergent global threat for humans. Emerg Infect Dis. 2008;14:1019–1023. doi: 10.3201/eid1407.071656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornwiroon W. Kearney MT. Husseneder C. Foil LD, et al. Comparative microbiota of Rickettsia felis-uninfected and -infected colonized cat fleas, Ctenocephalides felis. ISME J. 2007;1:394–402. doi: 10.1038/ismej.2007.38. [DOI] [PubMed] [Google Scholar]
- Pornwiroon W. Pourciau SS. Foil LD. Macaluso KR. Rickettsia felis from cat fleas: isolation and culture in a tick-derived cell line. Appl Environ Microbiol. 2006;72:5589–5595. doi: 10.1128/AEM.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif KE. Macaluso KR. Ecology of Rickettsia felis: a review. J Med Entomol. 2009;46:723–736. doi: 10.1603/033.046.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif KE. Stout RW. Henry GC. Foil LD, et al. Prevalence and infection load dynamics of Rickettsia felis in actively feeding cat fleas. PLoS ONE. 2008;3:e2805. doi: 10.1371/journal.pone.0002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. Fournier PE. Petridou J. Haussinger D, et al. Rickettsia felis infection acquired in Europe and documented by polymerase chain reaction. Emerg Infect Dis. 2002;8:207–208. doi: 10.3201/eid0802.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M. Koga R. Tsuchida T. Meng XY, et al. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl Environ Microbiol. 2005;71:4069–4075. doi: 10.1128/AEM.71.7.4069-4075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriefer ME. Sacci JB., Jr. Taylor JP. Higgins JA, et al. Murine typhus: updated roles of multiple urban components and a second typhuslike rickettsia. J Med Entomol. 1994;31:681–685. doi: 10.1093/jmedent/31.5.681. [DOI] [PubMed] [Google Scholar]
- Sunyakumthorn P. Bourchookarn A. Pornwiroon W. David C, et al. Characterization and growth of polymorphic Rickettsia felis in a tick cell line. Appl Environ Microbiol. 2008;74:3151–3158. doi: 10.1128/AEM.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE. Heeger P. Mynatt RL. Truett AA, et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Varagnol M. Parola P. Jouan R. Beaucournu JC, et al. First detection of Rickettsia felis and Bartonella clarridgeiae in fleas from Laos. Clin Microbiol Infect. 2009;15:334–335. doi: 10.1111/j.1469-0691.2008.02272.x. [DOI] [PubMed] [Google Scholar]
- Vaughan JA. Azad AF. Acquisition of murine typhus rickettsiae by fleas. Ann N Y Acad Sci. 1990;590:70–75. doi: 10.1111/j.1749-6632.1990.tb42209.x. [DOI] [PubMed] [Google Scholar]
- Wade SE. Georgi JR. Survival and reproduction of artificially fed cat fleas, Ctenocephalides felis Bouche (Siphonaptera: Pulicidae) J Med Entomol. 1988;25:186–190. doi: 10.1093/jmedent/25.3.186. [DOI] [PubMed] [Google Scholar]
- Wedincamp J., Jr. Foil LD. Vertical transmission of Rickettsia felis in the cat flea (Ctenocephalides felis Bouche) J Vector Ecol. 2002;27:96–101. [PubMed] [Google Scholar]
- Weinert LA. Werren JH. Aebi A. Stone GN, et al. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009;7:6. doi: 10.1186/1741-7007-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala-Castro J. Zavala-Velazquez J. Walker D. Perez-Osorio J, et al. Severe human infection with Rickettsia felis associated with hepatitis in Yucatan, Mexico. Int J Med Microbiol. 2009;299:529–533. doi: 10.1016/j.ijmm.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala-Velazquez JE. Ruiz-Sosa JA. Sanchez-Elias RA. Becerra-Carmona G, et al. Rickettsia felis rickettsiosis in Yucatan. Lancet. 2000;356:1079–1080. doi: 10.1016/S0140-6736(00)02735-5. [DOI] [PubMed] [Google Scholar]