Abstract
We amplified 16S rRNA, gltA, and ompA genes from Ixodes pacificus by polymerase chain reaction. Sequencing, BLAST analysis, and phylogenetic constructions indicated that two Rickettsia phylotypes are present in I. pacificus. While phylotype G021 has high homology to Ixodes scapularis endosymbiotic Rickettsia, phylotype G022 is a deeply branched novel spotted fever group Rickettsia.
Key Words: Ixodes pacificus ticks, Rickettsia, Vector-borne
Introduction
The western black-legged tick, Ixodes pacificus, is the most widely distributed tick in the Pacific West Coast region of the United States. It is a primary vector of human diseases, including Lyme borreliosis and anaplasmosis (Burgdorfer et al. 1985). Although Rickettsia rickettsii, the etiological agent of Rocky Mountain spotted fever, has never been detected in I. pacificus, Hughes et al. (1976) identified Tillamook and Grants Pass strains, the spotted fever group agent of rickettsiae from I. pacificus, in western Oregon. Later, Philip et al. (1981) reported that I. pacificus in western California possesses a Rickettsia species that is similar to the Tillamook stain. Alhough several lines of evidence have been collected that support the presence of the Rickettsia species in I. pacificus using serological tests (Hughes et al. 1976, Philip et al. 1981), investigators have not yet used DNA analysis to determine the presence of rickettsiae in the tick. In the present study, we report detection of rickettsial DNA in I. pacificus ticks that were collected in the Napa Valley of California.
Materials and Methods
Tick collection and identification
To detect the presence of rickettsiae in questing I. pacificus, 96 adult I. pacificus Cooley and Kohls ticks were collected in 2003 in the Napa Valley by dragging a white cloth flag along trails with grass vegetation. Genus and species of I. pacificus ticks were confirmed by standard taxonomic characteristics.
DNA extraction and polymerase chain reaction amplification
To isolate DNA, individual ticks were frozen in liquid nitrogen and pulverized using a fitted pestle. Total genomic DNA was extracted from individual ticks using DNeasy Tissue Kit (Qiagen, Valencia, CA), as previously described by Jasinskas et al. (2007). Mock extractions were carried out as a negative control. To detect if rickettsiae were present in the total tick DNA extracts of I. pacificus, aliquots of 96 DNA extracts were mixed; the pool of the total genomic DNA was used as a template for detecting the presence of rickettsiae by polymerase chain reaction (PCR) using 16S rRNA, gltA (encoding citrate synthase protein), and ompA (encoding outer membrane protein A) primer sets. PCR amplification was performed using 50 ng of total genomic DNA of I. pacificus in a 50 μL reaction, containing PCR Master Mix (Promega Corporation, Madison, WI) and 0.1 mM of forward and reverse primers. The primer sequences of the 16S rRNA, gltA, and ompA are 5′-TAAGGAGGTAATCCAGCC-3′ and 5′-CCTGGCTCAGAACGAA-3′, 5′-GGCTAATGAAGCGGTAATAAATATGCTT-3′ and 5′-TTTGCGACGGTATACCCATAGC-3′, and 5′-CACYACCTCAACCGCAGC-3′ and 5′-AAAGTTATATTTCCTAAACCYGTATAAKTATCRGC-3′, respectively. The primers were designed as a result of multiple sequence alignments of each gene of rickettsiae. The PCRs were performed in an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Foster City, CA) with the following conditions: denaturation at 95°C for 5 min, and then 40 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 1 min, and elongation at 72°C for 2 min for 16S rRNA or 30 s for gltA and ompA genes, respectively.
Cloning and sequencing
The PCR products of 16S rRNA, gltA, and ompA genes of rickettsiae were cloned into StrataClone™ PCR cloning vector pSC-A (Agilent Technologies, La Jolla, CA) and sequenced at the MicroChemical Core Facility of San Diego State University using M13 reverse primer (5′-GGAAACAGCTATGACCATG-3′) and custom oligonucleotide primers derived from the 16S rRNA gene (Integrated Device Technology, Coralville, IA). The sequences are listed in GenBank under the accession numbers EU072493 and EU072494 for the 16S gene, GQ375159 and GQ375160 for the gltA gene, GQ375161 and GQ375162 for the ompA gene of the two Rickettsia phylotypes, and GU556973 for the actin gene sequence of I. pacificus.
Phylogenetic analysis
To study evolutionary relatedness of identified rickettsiae, phylogenetic trees of 16S rRNA, gltA, and ompA genes were constructed as described by Jasinskas et al. (2007). Briefly, multisequence alignments were performed using ClustalX version 1.83.1 (University College Dublin); phylogeny of each gene was constructed by PHYLO_WIN (version 2). Galtier and Gouy's method was used to determine evolutionary distance values; the values were then used to construct phylograms by a neighbor-joining method.
Transmission electron microscopy
To observe bacteria in I. pacificus ticks, midgut from an adult male I. pacificus was imaged using a transmission electron microscope. The dissected tick midgut was fixed in 3% glutaraldehyde in 50 mM cacodylate buffer (pH 7.0). Samples were chilled to 10°C, and microwaved for 40 s at a power setting that produced a temperature increase of 10°C–15°C using a PELCO™ Microwave System (Ted Pella, Redding, CA). Specimens were again chilled, and microwaved for 40 s. Samples were washed 3× with 50 mM cacodylate buffer (pH 7.0), and then postfixed with 1.5% aqueous osmium tetroxide in the microwave (40 s, 2×). After osmication, tissues were rinsed three times with distilled water, and then dehydrated in a graded ethanol series, 40 s of microwave energy per grade, and temperature restriction of 37°C. Next, samples were infiltrated with a 1:1 mixture of ethanol ERL4206 (ERL)-Quetol (3,4-epoxy cyclohexyl methyl 3,4-epoxy cyclohexyl carboxylale) resin (Ted Pella) for 15 min using the microwave set at a temperature restriction of 45°C. Infiltration was completed by replacing the 1:1 mixture of ethanol and resin with 100% ERL-Quetol for 15 min in the microwave with the same temperature restriction, and then repeating this step a second time. Samples were embedded in 100% ERL-Quetol and polymerized overnight at 60°C in a vacuum oven. Specimens were sectioned on a Reichert ultramicrotome using a Diatome diamond knife, stained with aqueous uranyl acetate followed by Reynold's lead citrate. The sections were examined using a transmission electron microscope (Phillips EM 208S; FEI, Hillsboro, OR).
Results
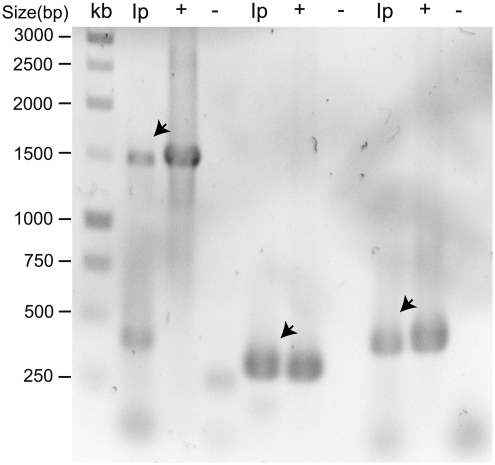
16S rRNA, gltA, and ompA genes were detected by PCR in pooled I. pacificus tick DNA (Fig. 1). “Classifier” analysis at the Ribosomal Database Project II Web site (http://rdp.cme.msu.edu/index.jsp) placed the amplified 16S rRNA sequences in this study in the genus Rickettsia. Sequencing of 10 clones for each gene determined that the size of the gltA clone is 341 base pairs (bp). However, the sizes of 16S rRNA and ompA clones are 1482 and 1483 bp, and 444 and 438 bp, respectively. The sequences of the two 16S rRNA amplicons shared 98% of their nucleotide-sequence identity. Similar nucleotide sequence identity (97%) was observed in the gltA sequences. When the ompA nucleotide sequences were compared with each other, a significantly low nucleotide identity of 81% was observed. Comparison at the amino acid level revealed 72% identity between the two translated ompA nucleotide sequences. BLAST analysis of the sequences of the 16S rRNA, gltA, and ompA genes against the NCBI DNA database showed nucleotide sequence similarities with spotted fever group Rickettsia. The closest matches of the 16S rRNA nucleotide sequences exhibited 98% and 99% nucleotide sequence identities to 16S rRNA of Rickettsia massiliae MTU5 (accession number CP000683.1). One of the gltA sequences exhibited 100% nucleotide sequence identities with the gltA gene of Rickettsia raoultii strain Khabarovsk (accession number DQ365804.1), whereas the other gltA sequence was most identical (99% nucleotide sequence identities) to the gltA gene sequence of Rickettsia monacensis strain CN45Kr (accession number FJ009429.1). The ompA sequences were also compared with sequences of the ompA gene of Rickettsia species in GenBank. One of the ompA sequences had 99% nucleotide sequence identity to the ompA gene of Rickettsia sp. CAMWD07-2 from I. pacificus (accession number EU544298.1) and 98% nucleotide sequence identity to the gltA gene of Ixodes scapularis endosymbiont TX125 (accession number EF689735.1), whereas the other ompA sequence had 88% nucleotide sequence identity to the gltA gene of Rickettsia amblyommii species TNLBL06-3 (accession number EU544295.1).
FIG. 1.
Detection of 16S rRNA, gltA, and ompA genes by PCR amplification of Ixodes pacificus tick extracts. Ninety-six ticks were collected in the Napa Valley, California. Tick DNA was extracted, pooled, and used as template for PCR amplification. The PCR products were electrophoresed and observed by staining with ethidium bromide. kb, 1 kb DNA ladder (Promega). Ip, I. pacificus tick extract; (+), Rickettsia conorii DNA positive control; (−), no template negative control; arrow, PCR amplicons. PCR, polymerase chain reaction.
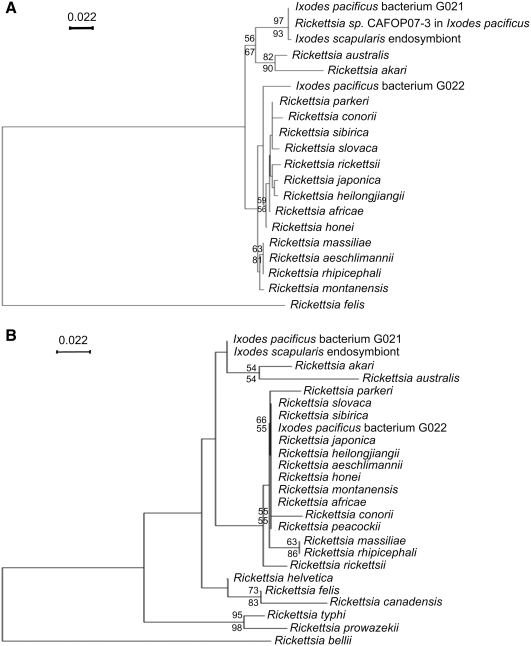
The neighbor-joining tree shown in Figure 2 was based on the results of a distance matrix analysis of all available gltA and ompA sequences of rickettsiae in GenBank. The phylogenetic trees revealed that two PCR sequences of each gene, which were amplified from I. pacificus in this study, clustered with species of the spotted fever group of the genus Rickettsia. Since the two PCR sequences of each gene belong to different lineages, as shown in Figure 2, we have named the rickettsiae in I. pacificus as G021 and G022 phylotypes. The neighbor-joining tree indicated that the ompA gene sequence of the phylotype G021 was related to known Rickettsia australis and Rickettsia akari. However, the sequence was more closely related to the ompA gene sequence of Rickettsia sp. CAFOP07-3 in I. pacificus, and I. scapularis Rickettsia endosymbiont. All of the above Rickettsia species, including the phylotype G021, formed a monophyletic group that is distinct from the rest of spotted fever group rickettsiae. On the other hand, the ompA gene of the phylotype G022 constituted a distinct and deeply branched phylogenetic lineage within the spotted fever group of Rickettsia. The phylogeny placed the phylotype G022 as a sister lineage to the clade of members of rickettsiae in the spotted fever group, including Rickettsia slovaca, Rickettsia heilongjiangii, Rickettsia japonica, Rickettsia rickettsii, Rickettsia parkeri, Rickettsia sibirica, Rickettsia africae, Rickettsia honei, and Rickettsia conorii (Fig. 2A). Although the branching order of the two Rickettsia phylotypes failed to be resolved with other Rickettsia species based on the 16S rRNA sequences (data not shown), results of the phylogeny of the gltA gene sequences supported the phylogeny of the ompA gene sequences; the phylotype G021 formed a monophyletic group with R. australis, R. akari, and the endosymbiotic Rickettsia species in I. scapularis, whereas the phylotype G022 had close relatives with members of the spotted fever group of rickettsiae (Fig. 2B). Similar tree topologies were obtained when either maximum-likelihood or maximum-parsimony was used for tree constructions.
FIG. 2.
Phylogenetic trees of ompA (A) and gltA (B) genes of the two Rickettsia phylotypes in I. pacificus. Other selected sequences on the phylogenetic tree of the ompA gene are Rickettsia sp. CAFOP07-3 (EU544297.1), Ixodes scapularis endosymbiont (AB002268.1), Rickettsia aeschlimannii (DQ379981.1), Rickettsia massiliae (DQ212707.1), Rickettsia rhipicephali (EU109177.1), Rickettsia australis (AF149108.1), Rickettsia montanensis (AF045223.1), Rickettsia sibirica (AABW01000001.1), Rickettsia rickettsii (AY319293.1), Rickettsia slovaca (EU622810.1), Rickettsia honei (AF018075.1), Rickettsia africae (EU622980.1), Rickettsia parkeri (EU715288.1), Rickettsia heilongjiangii (AF179362.2), Rickettsia conorii (U43794.1), Rickettsia felis (AY727036.1), Rickettsia peacockii (AY319292.1), Rickettsia japonica (U83442.1), and Rickettsia akari (L01461.1). Other selected sequences on the phylogenetic tree of the gltA gene are I. scapularis endosymbiont (EF662058.1), Rickettsia felis (CP000053.1), Rickettsia helvetica (DQ821857.1), Rickettsia akari (U59717.1), Rickettsia canadensis (BAF79997.1), Rickettsia australis (U59718.1), Rickettsia africae (CP001612.1), Rickettsia japonica (AY743327.1), Rickettsia parkeri (EF102236.1), Rickettsia slovaca (AY129301.1), Rickettsia montanensis (U74756.1), Rickettsia honei (U59726.1), Rickettsia sibirica (DQ124930.1), Rickettsia typhi (U59714.1), Rickettsia heilongjiangensis (AY285776.1), Rickettsia bellii (NC_007940.1), Rickettsia prowazekii (NC_000963.1), Rickettsia rickettsii (DQ150689.1), Rickettsia conorii (NC_003103.1), Rickettsia aeschlimannii (DQ235776.1), Rickettsia massiliae (U59719.1), Rickettsia rhipicephali (U59721.1), and Rickettsia peacockii (DQ100162.1). The tree is inferred from the comparison of either ompA or gltA sequences by the neighbor-joining method using PHYLO_WIN. The numbers at nodes are the bootstrap values obtained from 1000 replicates. Bootstrap values >50 are shown at the nodes. Bootstrap values determined by the neighbor-joining method and maximum-parsimony are shown above the line and below the line, respectively. Bar, nucleotide distance.
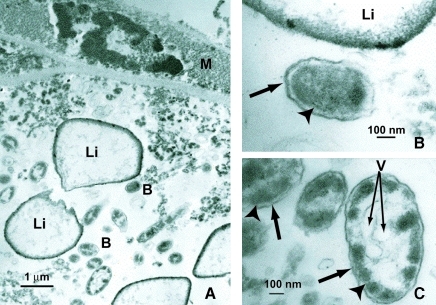
Transmission electron microscope observations of the midgut of I. pacificus revealed intracellular microorganisms in the cytoplasm of the cells in the midgut. The microorganisms appeared to be rod-shaped bacteria, with distinct gram-negative cell walls that included an outer membrane and a cytoplasmic membrane. The bacteria varied in shape from straight to curved and the size ranged from 0.3 to 0.6 μm wide by 0.5 to 0.9 μm long. The bacterial cells also possessed intracellular inclusions, and some of them appeared to be electron dense, whereas other intracellular inclusions were electron transparent. The number of the rod-shaped bacteria varied among cells, ranging from none to hundreds per cell (Fig. 3). No ultrastructural evidence of the presence of more than one type of bacteria was found in the midgut of I. pacificus.
FIG. 3.
Transmission electron micrographs of the midgut tissue from an Ixodes pacificus tick. (A) Longitudinal view of the midgut tissue: B, bacteria; Li, lipid inclusion. (B, C) Higher magnification of bacteria: thick arrows point to the outer membrane; arrowheads point to the inner membrane. Notice the electron-lucent vacuole-like structures (V) within the bacteria.
Discussion
In this study, we detected and identified two Rickettsia phylotypes in I. pacificus ticks from Napa Valley, California. The two phylotypes belong to different lineages: while the phylotype G021 is a close relative of a Rickettsia symbiont in I. scapularis, the phylotype G022 is a deeply branched novel spotted fever group Rickettsia. Although there were a couple of studies identifying Rickettsia species in I. pacificus using less-sensitive serological tests (Hughes et al., 1976, Philip et al. 1981), our investigation documents the presence of Rickettsia species using molecular biology tools.
Rickettsia species have been repeatedly isolated from Ixodes ticks in the United States; more than 20 novel species of spotted fever group rickettsiae have been reported in Ixodes tick species other than I. pacificus (Fournier et al. 2003). Using a hemolymph test or direct immunofluorescence, spotted fever group rickettsiae have been detected in I. scapularis (Magnarelli et al. 1979) and Ixodes cookei nymphs (Magnarelli et al. 1985). Later, PCR detected that a rickettsial endosymbiont is present in I. scapularis (Moreno et al. 2006) and that R. rickettsii is present in Ixodes texanus (Anderson et al. 1986). Recently, a bacterial symbiont of the genus Rickettsia was identified in I. scapularis in the tick genome project (Nene 2009). All of the previous descriptions of spotted fever group rickettsiae in association with Ixodes ticks represent the great genetic diversity within the spotted fever group rickettsiae.
The pathogenicity of Rickettsia phylotypes G021 and G022 remains to be determined. The discovery of these phylotypes also prompts future research on their transmission and prevalence in tick populations.
Acknowledgments
We thank Dr. Alan G. Barbour at the University of California, Irvine for providing I. pacificus tick samples. We thank Dr. David H. Walker at the University of Texas Medical Branch for providing genomic DNA of R. conorii. We also thank Dr. Patricia L. Siering for providing comments on the manuscript. This work was supported by Dr. Jianmin Zhong's NIH R15 grant 1 R15 AI 82515-01 and Humboldt State University New Faculty Seed Grant and by Dr. Jacob P. Varkey's Howard Hughes Medical Institute grant 52005127.
Disclosure Statement
No competing financial interests exist
References
- Anderson JF. Magnarelli LA. Philip RN. Burgdorfer W. Rickettsia rickettsii and Rickettsia montana from Ixodid ticks in Connecticut. Am J Trop Med Hyg. 1986;35:187–191. doi: 10.4269/ajtmh.1986.35.187. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W. Lane RS. Barbour AG. Gresbrink RA, et al. The western black-legged tick, Ixodes pacificus: a vector of Borrelia burgdorferi. Am J Trop Med Hyg. 1985;34:925–930. doi: 10.4269/ajtmh.1985.34.925. [DOI] [PubMed] [Google Scholar]
- Fournier PE. Dumler JS. Greub G. Zhang J, et al. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–5465. doi: 10.1128/JCM.41.12.5456-5465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes LE. Clifford CM. Gresbrink R. Thomas LA, et al. Isolation of a spotted fever group rickettsia from the Pacific Coast tick, Ixodes pacificus, in Oregon. Am J Trop Med Hyg. 1976;25:513–516. doi: 10.4269/ajtmh.1976.25.513. [DOI] [PubMed] [Google Scholar]
- Jasinskas A. Zhong J. Barbour AG. Highly prevalent Coxiella sp. bacterium in the tick vector Amblyomma americanum. Appl Environ Microbiol. 2007;73:334–336. doi: 10.1128/AEM.02009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnarelli LA. Anderson JF. Burgdorfer W. Rocky mountain spotted fever in Connecticut: human cases, spotted-fever group rickettsiae in ticks, and antibodies in mammals. Am J Epidemiol. 1979;110:148–155. doi: 10.1093/oxfordjournals.aje.a112799. [DOI] [PubMed] [Google Scholar]
- Magnarelli LA. Anderson JF. Burgdorfer W. Philip RN, et al. Spotted fever group rickettsiae in immature and adult ticks (Acari: Ixodidae) from a focus of Rocky Mountain spotted fever in Connecticut. Can J Microbiol. 1985;31:1131–1135. doi: 10.1139/m85-213. [DOI] [PubMed] [Google Scholar]
- Moreno CX. Moy F. Daniels TJ. Godfrey HP, et al. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol. 2006;8:761–772. doi: 10.1111/j.1462-2920.2005.00955.x. [DOI] [PubMed] [Google Scholar]
- Nene V. Tick genomics—coming of age. Front Biosci. 2009;14:2666–2673. doi: 10.2741/3404. [DOI] [PubMed] [Google Scholar]
- Philip RN. Lane RS. Casper EA. Serotypes of tick-borne spotted fever group rickettsiae from western California. Am J Trop Med Hyg. 1981;30:722–727. doi: 10.4269/ajtmh.1981.30.722. [DOI] [PubMed] [Google Scholar]