Abstract
The p53 gene is one of the most frequently mutated genes in human cancer. Some p53 mutations impart additional functions that promote oncogenesis. To investigate how these p53 mutants function, a proteomic analysis was performed. The protein, translocator of the inner mitochondrial membrane 50 (Tim50), was upregulated in a non-small cell lung carcinoma cell line (H1299) that expressed the p53 mutants R175H and R273H compared to cells lacking p53. Tim50 was also elevated in the breast cancer cell lines MDA-MB-468 and SK-BR-3, that endogenously express the p53 mutants R175H and R273H respectively, compared to MCF-10A. The p53 mutants R175H and R273H, but not WT p53, upregulated the expression of a Tim50 promoter construct and chromatin immunoprecipitation (ChIP) analysis indicated increased histone acetylation and increased interaction of the transcription factors Ets-1, CREB and CREB-binding protein (CBP) with the Tim50 promoter in the presence of mutant p53. Finally, reduction of Tim50 expression reduced the growth rate and chemoresistance of cells harboring mutant p53 but had no effect upon cells lacking p53. Taken together, these findings identify the Tim50 gene as a transcriptional target of mutant p53 and suggest a novel mechanism by which p53 mutants enhance cell growth and chemoresistance.
Keywords: p53, Tim50, mutations, mitochondria, cancer, chemoresistance
INTRODUCTION
The tumor suppressor protein p53 is involved in the control of cell cycle progression, DNA integrity, and cell survival. Mutations in the p53 gene are some of the most frequent alterations in human cancers, with most of the mutations resulting in the expression of full-length mutant p53 proteins [1]. Mutant p53 proteins have altered transcriptional activity compared to WT, and often become stable and accumulate to high levels in tumor cells [2–7].
Expression of mutant p53 has been shown to impart increased cell growth [8–10] and chemoresistance to several chemotherapeutic agents in tumor cell lines [9, 11–13]. Mutant p53 expression has also been shown to contribute to metastasis in a mouse model for lung cancer [14, 15] and studies of human cancer indicate that the presence of p53 mutations is associated with poor prognosis in several types of tumors [16–19]. For example, in breast cancer, p53 mutation appears at a very high frequency in HER-2 positive tumors that are prone to metastasize [19]. This follows the ‘gain of function’ (GOF) hypothesis, which predicts that mutations in the p53 gene not only destroy the tumor suppressor function of the WT protein but also impart increased oncogenicity [7, 20, 21]. While significant evidence indicates that mutant p53 contributes to oncogenesis, the exact mechanism of action of mutant p53 is still unclear [12, 22]. One hypothesis is that mutant p53 may act to alter gene expression. Several cell growth and survival related genes whose expression is altered by p53 GOF mutants have been identified by Deb and colleagues [9, 23, 24]. Other laboratories have also reported genes that are influenced by p53 mutants [25–28]. Many of the genes shown are associated with cell proliferation and tumor progression [6–8, 29, 30]. To further explore the mechanism of action of p53 GOF mutants, we performed large-scale mutant p53 immunoprecipitations coupled with mass spectrometry. One of the proteins we identified was Tim50 (Translocase of the inner mitochondrial membrane 50). Tim50 is one component of a large protein complex whose function is to import proteins into the inner mitochondrial matrix [31–33].
In the current work, we show that upregulation of Tim50 can be mediated by mutant p53 and that that loss of Tim50 expression increases the responsiveness of GOF mutant-p53 expressing cancer cells to chemotherapeutic treatment and decreases their rate of cell growth. Our data also identifies the Tim50 gene as direct target of mutant p53 and suggests that increased Tim50 expression may contribute to some GOF phenotypes imparted by mutant p53.
MATERIALS AND METHODS
Cell culture
SK-BR-3, MDA-MB-231, MDA-MB-468, MCF-7, Saos-2 and H1299 cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium) with 10% (v/v) fetal bovine serum. MCF-10A cells were grown in DMEM/F12 supplemented with 5% horse serum, 20 μg/mL EGF, 0.5 μg/ml hydrocortisone, 0.1 μg/ml cholera toxin, and 10 μg/ml insulin. Cells were maintained at 37 °C in an atmosphere of 5 % CO2. Stable H1299 cell lines that expressed vector alone (designated HC5), or p53-R175H or p53-R273H (designated H-p53-R175H or H-p53-R273H respectively) were generated as previously described [23] and maintained in 400 μg/ml gentamicin. The H1299 p53-WT inducible cell line (Hip53) was generated as previously described [34]. p53 in this cell line was induced for 24 hrs with 100 μM Ponasterone A (Invitrogen, Carlsbad, CA)
Large scale mutant p53 immunoprecipitations
One hundred 100-mm dishes of HC5 or H-p53-R175H cells were grown and harvested, (approximately 4 g of cells) and lysed in mammalian lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA and 1X EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN). Cell lysates were clarified by centrifugation at 13,000 × g for 20 min and p53 immunoprecipitated with 250 μg of p53 DO-1 antibody conjugated agarose (#sc-126 AC, Santa Cruz Biotech) overnight at 4 °C. Immunoprecipitants were washed four times with lysis buffer, once with Tris-buffered saline, and incubated for 5 min at 100 °C in 2× SDS sample buffer. Samples were resolved by SDS/PAGE (12% gels), silver-stained, and protein bands excised and in-gel digested with trypsin (Promega, Madison, WI) according to previous procedures [35].
Mass spectrometry
Tryptic peptides were purified with Poros 20 R2 reverse phase packing (Applied Biosystems, Foster City, CA) and subjected to direct infusion nanospray using NanoES spray capillaries (Proxeon, Odense, Denmark) on an Applied Biosystems QSTAR® pulsar XL mass spectrometer. MS spectra were collected both in data dependent acquisition mode and in manual mode using an ion spray voltage of 800 V, a curtain gas of 20, a declustering potential of 75 V and a focusing potential of 280 V. For data-dependent acquisition, MS data were collected for a mass range of 400 m/z to 2000 m/z with a charge state of 2–5 which exceed one count. MS/MS (tandem MS) data were acquired for ions from a mass range of 60–2000 m/z with a dwell time of 15 s per ion. Identification of peptides was achieved by manual interpretation of MS/MS spectra with the aid of Analyst QS 1.1 (MDS Sciex, Concord, ON, Canada) and by searching the non-redundant protein database with the aid of Mascot (Matrix Science, Boston, MA).
Plasmid construction and RNA interference
To create a Tim50 promoter construct upstream of luciferase, a 1.97 kb region of the Tim50 gene promoter was amplified by PCR with the following primers: 5′-CCAAGCTTCGAGAGAGACCAAAGGCATC-3′ (with Hind III site underlined) 5′-CCGGGTACCCTCGTTTCTCACTCAAGCCCT-3′ (with Kpn I site underlined). The PCR product was digested with Hind III and Kpn I and ligated into the pGL3 vector (Promega, Madison WI) upstream of the luciferase reporter gene. This construct was designated as pGL3-Tim50. The correct sequence of the Tim50 promoter was verified by sequencing (Molecular Cloning Laboratories, San Francisco, CA). Approximately 3 × 106 cells were transfected for 48 hrs or 96 hrs with either a scrambled siRNA (control), or siRNAs against Tim50 or p53. The siRNA sequences were as follows: Scrambled: 5′-CAUGUCAUGUGUCACAUCACTT-3′; Tim50: 5′-CGAACGGUGCUGGAGCACU-3′ and p53: 5′-GCAUGAACCGGAGGCCCAU-3′. Cells were transfected by electroporation using the Amaxa Nucleofection kit according to the manufacturer’s instructions (Amaxa, Koln, Germany) using appropriate conditions for each cell line. The efficacy of siRNA treatment was confirmed by Western analysis.
Colony survival assays
Approximately 3 × 106 cells were transfected for 48 hrs with scrambled, Tim50 or p53 siRNA as described above. Cells were plated at a density of approximately 1 × 104 cells per 100-mm plate and treated with 25 nM paclitaxel for 48 hrs. After treatment, cells were washed with PBS, and fresh media was replaced. The cells were allowed to form colonies with periodic changes of media for a period of ~2–3 weeks. Colonies were fixed with 100% methanol, stained with 0.02% methylene blue and counted as previously described [9]. Control samples were treated with drug vehicle, DMSO, to measure plating efficiency.
Cell growth assays
Approximately 3 × 106 cells were transfected for 48 hrs with scrambled, Tim50 or p53 siRNA as described above. Cells were plated at a density of 1 × 103 or 1 × 105 cells per 60-mm plate respectively and after 24, 48, 72 and 96 hrs, cells were harvested and counted in a Coulter Counter. All growth assays were performed in triplicate and the experimented was performed twice.
Western blot analysis
Cells were lysed in mammalian cell lysis buffer which contained 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, and protease inhibitor cocktail (Roche, Nutley, NJ). Equal amounts of protein were resolved by SDS/PAGE (8% or 12% gels) and transferred to nitrocellulose membranes. The primary antibodies used for immunoblotting were all used at 1:1000 dilution and included: Tim50 (#IMG-3375, Imgenex, San Diego, CA), Tim23 (#611222, BD Transduction Laboratories, San Jose, CA), NDUFA9 (#MS111, Mitosciences), actin (#1615, Santa Cruz Biotech, Santa Cruz CA), and p53 (#9282, Cell Signaling, Danvers, MA). A monoclonal p53 antibody was also used, prepared as previously described [36]. Primary rabbit polyclonal antibodies were detected using the following secondary antibodies (all at 1:7000 dilution): IRDye800-conjugated affinity-purified rabbit anti-IgG antibody (Rockland Immunochemicals, Gilbertsville, PA), Alexa Fluor® 680-conjugated goat anti-mouse IgG antibody (Molecular Probes, Invitrogen, Carlsbad, PA) and Alexa Fluor® 680-conjugated rabbit anti-goat IgG antibody (Invitrogen, Carlsbad, CA). Proteins were visualized using the Odyssey system (LI-COR) and where indicated, relative amounts of immunoreactive protein in each band were determined by densitometric analysis and normalized to the level of actin.
Quantitative PCR
Q-PCR was performed as described previously [9]. Briefly, total RNA was isolated with Triazol reagent (Invitrogen, Carlsbad, CA). The quality of RNA was checked by 1.2% agarose Tris-borate-EDTA gel electrophoresis and cDNA synthesized using the SuperScript II kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Q-PCR was conducted using a LightCycler system (Roche, Nutley, NJ) as described previously [9]. Primers were designed using OLIGO 5 software (Molecular Biology Insights, Cascade, CO). Reactions were performed in triplicate utilizing SYBR green dye (Invitrogen, Carlsbad, CA) using the following primers: Tim50, 5′-CCGTACTACCAGCCACCCTA-3′ and 5′-TGGGGGTCCACACTATCAAT-3′, Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 5′-GTCAACGGATTTGGTCGTATT-3′ and 5′-GATCTCGCTCCTGGAAGATGG -3′. Tim50 promoter, 5′-GCGTTGGTGGTGGCGAGGTA-3′ and 5′-AGCGGAGGCGGGGAAGG-3′.
Luciferase reporter assays
H1299 were triply transfected with 100 ng of control β-Gal plasmid, 200 ng of the Tim50 promoter-luciferase reporter construct (pGL3-Tim50) and 1500 ng of vector only (pCMVBam) or WT p53, p53-R175H, or -R273H in pCMVBam for 48 hrs [8]. Saos-2 cells were transfected in a similar manner with pGL3-Tim50 and vector only (pCMVBam) or WT p53, p53-R273H, -D281G, or –D281G L22Q/W23S. After transfection, cells were harvested and luciferase activity measured using the Promega luciferase assay kit (#E1500, Promega, Madison, WI) according to the manufacturer’s instructions. Cell extracts were normalized to each other based on total protein concentration and Luciferase activity detected using a Luminometer from Turner Designs. Total luciferase activity was normalized with respect to β-Galactosidase activity measured using the Promega β-Galactosidase Enzyme Assay System (#E2000, Promega, Madison, WI) according to the manufacturer’s instructions.
ChIP analysis
Chromatin immunoprecipitations were performed as described [37–39]. Exponentially growing H1299 cells expressing vector (HC5) or mutant p53-R273H (3 × 106) were cross-linked with 1% formaldehyde for 15 min and the reaction stopped by addition of glycine to a final concentration of 0.125M. Cells were collected and washed once with PBS. Pellets were resuspended in RIPA buffer (150 mM NaCl, 50mM Tris pH 8, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40) and then sheared by multiple passages through a 27.5 gauge needle, followed by 25 min of sonication on ice to induce chromatin fragmentation. Following centrifugation, the protein content of the supernatants was determined and equal amounts used for immunoprecipitation overnight at 4°C with gentle tilting with either anti-Acetyl-Histone H3 (#17–615, Millipore, Billerica, MA), anti-Ets-1 (#sc-350, Santa Cruz Biotech, Santa Cruz CA), anti-CREB (#sc-186, Santa Cruz Biotech, Santa Cruz CA) or anti-CBP (#sc-369, Santa Cruz Biotech, Santa Cruz CA), or IgG as a control (Normal rabbit IgG, Millipore, Billerica, MA). Immune complexes were captured the following day with protein A-agarose. The immunoprecipitant was pelleted and washed once with RIPA buffer, once with a high salt wash (500 mm NaCl, 50 mm Tris–HCl, pH 8, 0.1% SDS, 1% NP-40), twice with a LiCl wash (250 mM LiCl, 50 mM Tris–HCl, pH 8, 0.5% sodium deoxycholate, 1% NP-40) and twice with TE buffer. The antibody–DNA complexes were eluted with elution buffer (20% SDS, 10 mM DTT, 100 mM NaHCO3) and the cross linking was reversed by incubation at 65°C overnight. pGEM(3z)f- (3 ng) was added to each sample to act as an internal control and the DNA was then ethanol precipitated. Samples were then dissolved in TE and RNAse (10 mg/ml) and treated with proteinase K (20 mg/ml). Proteins were removed by phenol–chloroform extraction and the DNA isolated by ethanol precipitation. The DNA pellets were dissolved in TE and Q-PCR was carried out as described above.
Statistical analysis
The data from triplicate samples were calculated and expressed as mean ± Standard error of measurement. Differences between groups were determined with Student’s t test, and P < 0.05 was considered significant.
RESULTS
Tim50 expression is upregulated in human lung and breast cells harboring p53 mutants
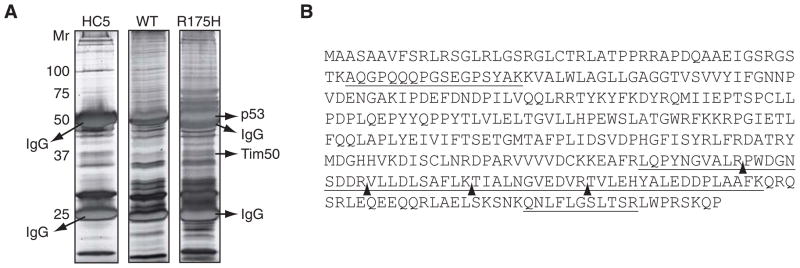
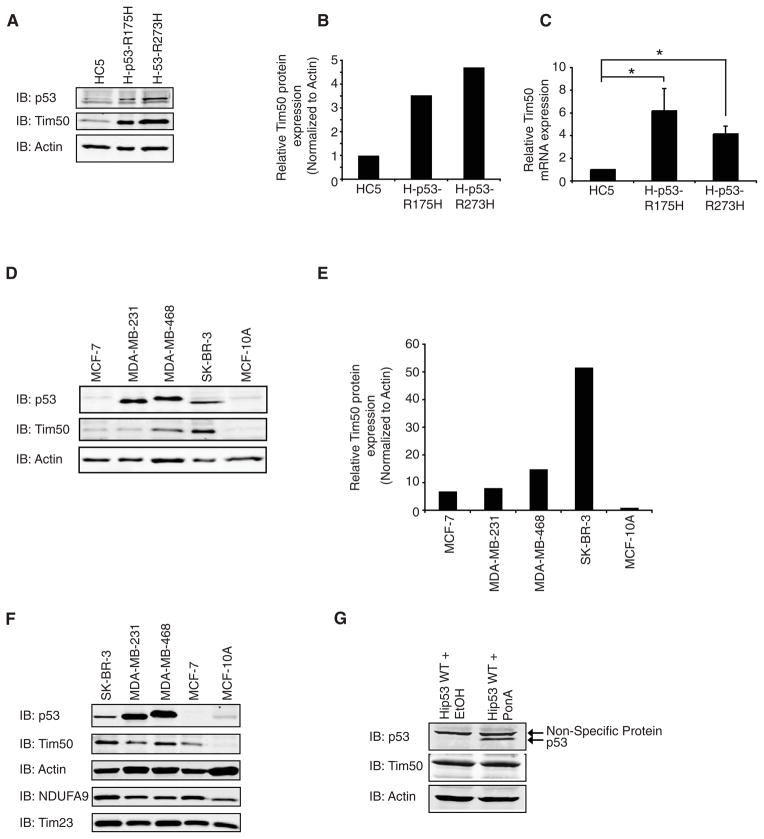
To explore potential mechanisms underlying the GOF phenotype of mutant p53, we conducted a proteomics analysis to identify mutant p53 specific binding proteins. To this end, a non-small lung carcinoma cell line, H1299, that does not express p53, was engineered to stably express the p53 mutants, R175H or R273H (denoted H-p53-R175H and H-p53-R273H) or the control vector only (denoted HC5) [8, 23, 40]. Large scale p53 immunoprecipitations were conducted from HC5, H-p53-R175H or H1299 cells transfected with WT p53 and resolved by SDS-PAGE. A protein of ~37 kDa was observed in p53 immunoprecipitants prepared from H-p53-R175H cells but not from HC5 cells or H1299 cells expressing WT p53 (Fig. 1A). Sequencing of the protein by mass spectrometry identified 7 tryptic peptides: AQGPQQQPGSEGPSYAK (aa 151–167), LQPYNGVALR (aa 359–368), PWDGNSDDR (aa 369–377), VLLDLSAFLK (aa 378–387), TIALNGVEDVR (aa 388–398), TVLEHYALEDDPLAAFK (aa 399–415), and QNLFLGSLTSR (aa 438–448) (Fig 1B). These peptides unequivocally identified the protein as human translocator of the inner mitochondrial membrane (Tim50), a protein required for transport of proteins into the mitochondria (accession number NM_001001563). While the binding of Tim50 to mutant p53 is still under study, it was observed that the amount of endogenous Tim50 protein was elevated approximately 3–4 fold in H-p53-R175H and H-p53-R273H cells compared to HC5 cells using a Tim50 specific antibody (Fig. 2A and B). Consistent with this result, quantitative PCR indicated that Tim50 mRNA was also significantly upregulated in H-p53-R175H and H-p53-R273H cells compared to HC5 (Fig. 2C). To explore the relationship between mutant p53 and Tim50 protein expression further, several breast cancer cell lines were analyzed for p53 and Tim50 expression levels. Tim50 protein levels were significantly elevated in MDA-MB-468 and SK-BR-3 total cell lysates, which harbor the p53 mutants R175H and R273H respectively, compared to MCF-7 and MCF-10A cells that express WT p53 (Fig. 2D and E). The difference observed in the amount of Tim50 protein was not simply due to a difference in the number of mitochondria in the cell lines as the integral mitochondrial protein, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9 (NDUFA9), was similar in the cell lines (Fig. 2F). Moreover, the difference in Tim50 protein levels appeared to be specific as there was no change observed in the amount of Tim23, another protein involved in the transport of proteins into the mitochondria (Fig. 2F) [31]. Since p53 is expressed at relatively low levels in MCF-7 and MCF-10A cells in the absence of cell stress and to determine if WT p53 could alter Tim50 protein expression, we employed H1299 cells engineered to inducibly express WT p53 as previously described [34]. Induction of WT p53 protein expression in this cell line had no discernable affect upon Tim50 protein expression (Fig. 2G).
Fig. 1.
Identification of Tim50 as a putative p53 interacting protein. (A) Large scale p53 immunoprecipitations were performed from HC5, H-p53-R175H, or H1299 cells transfected with WT-p53 and proteins resolved on 12% polyacrylamide gels. A silver stained gel is shown. Arrows indicate the identity of some proteins determined by mass spectrometry. (B) The protein sequence of human Tim50 and all peptides identified by mass spectrometry. Underlined sequences represent peptides identified, and separate peptides are denoted by arrows.
Fig. 2.
Expression of mutant p53 correlates with elevated levels of Tim50 mRNA and protein expression. (A) Total cell lysates from HC5, H-p53-R175H or H-p53-R273H cells were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. A representative image is shown and the experiment was performed twice with similar results. (B). Quantitation of Tim50 protein levels was performed and normalized to actin levels. The amount of Tim50 protein expression in HC5 cells was set to 1 and the relative amount of Tim50 expression in H-p53-R175H or H-p53-R273H cells is shown. (C) Total RNA was extracted from HC5, H-p53-R175H or H-p53-R273H cells and quantitative real-time PCR was performed to determine Tim50 mRNA levels. The amount of Tim50 expression was determined using a relative standard curve after normalization to the internal standard, GAPDH mRNA. The normalized Tim50 mRNA level in the control cell line, HC5, was set to 1 and the relative level of Tim50 mRNA in H-p53-R175H or H-p53-R273H cells is shown. Results are mean (+/−) S.E.M (n=6). * = significance p< 0.05. (D) Equal protein amounts of total cell lysates from the indicated cell lines were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (E) Tim50 and actin protein levels were quantitated and the ratio of Tim50/actin protein levels are shown. (F) Equal protein amounts of total cell lysates from the indicated cell lines were immunoblotted with the indicated antibodies. (G) H1299 cells stably transfected with ecdysone-inducible WT p53 (Hip53 WT cells) were incubated in the presence of ethanol vehicle (EtOH) or 100 μM Ponasterone A (PonA) for 24 hrs. Total cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Tim50 promoter activity is upregulated in human lung cells harboring p53 mutants
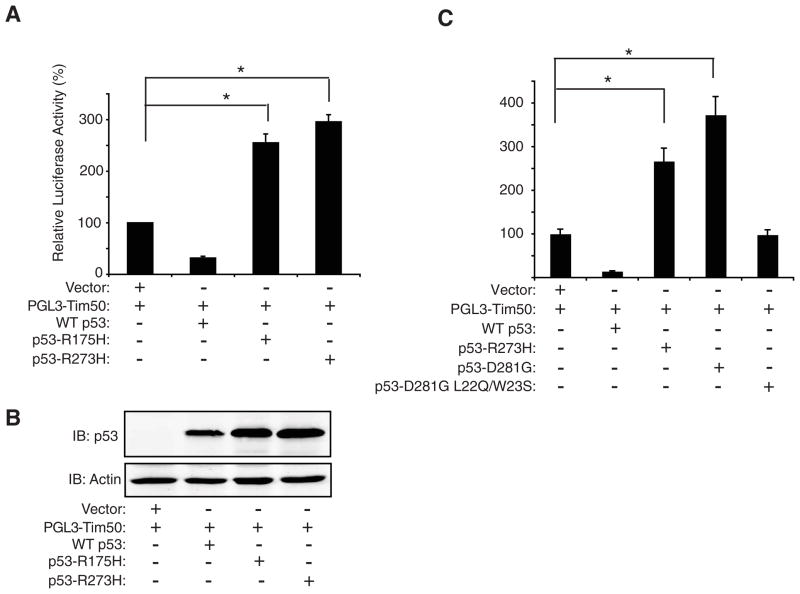
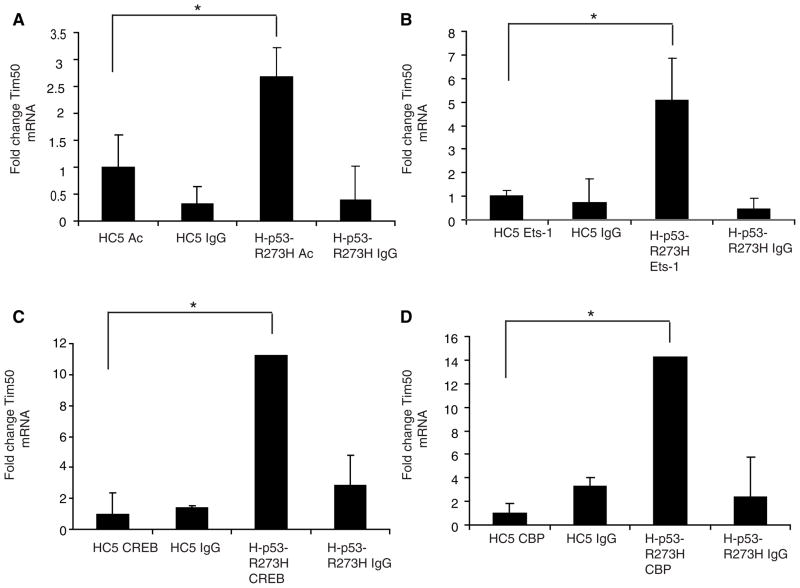
To determine if mutant p53 could affect Tim50 expression through upregulation of the Tim50 promoter, we constructed a Tim50 promoter construct consisting of 1.97 Kb of the Tim50 promoter region cloned from genomic DNA (using information obtained in the NCBI database) and inserted into the pGL3 luciferase reporter plasmid (materials and methods). The Tim50 reporter construct, termed pGL3-Tim50, was then co-transfected along with WT p53 or the p53 mutants, R175H and R273H in H1299 cells. The p53 mutants, R175H and R273H, upregulated luciferase activity approximately 2.5 and 3 fold respectively, but WT p53, in contrast, inhibited Tim50 promoter activity (Fig. 3A). Immunoblotting indicated that all p53 proteins were expressed in this assay (Fig. 3B). Similar results were observed in Saos-2 cells, a p53-null, human osteosarcoma cell line (Fig. 3C). To further examine a role for p53 transactivation in the upregulation of Tim50, we transfected Saos-2 cells with the p53 mutant, D281G or its corresponding transactivation-deficient mutant, D281G, L22Q/W23S [9]. The addition of the L22Q and W23S mutations to the D281G mutant was shown to abrogate the transactivation function of this p53 mutant [9]. Transfection with p53-D281G upregulated Tim50 luciferase activity, but no Tim50 luciferase activity was observed with p53-D281G L22Q/W23S (Fig. 3C). This result suggested that the p53 mutants may operate directly at the promoter level of Tim50 to upregulate its expression. To explore a potential role for mutant p53 at the Tim50 promoter region, we measured the level of histone acetylation, as one indicator of chromatin structure, by Chromatin Immunoprecipitation (ChIP) assays in the absence and presence of mutant p53. ChIP assays were performed using anti-acetylated histone specific antibodies in combination with quantitative PCR directed against a region of the Tim50 promoter (bp 756–890). Using this approach, it was found that the Tim50 promoter region was quantitatively enhanced in the anti-acetylated histone immunoprecipitants from H-p53-R273H cells but not from HC5 cells (Fig. 4A).
Fig. 3.
Mutant p53 up-regulates Tim50 promoter activity. (A) H1299 cells were transfected with a plasmid containing the Tim50 promoter region upstream of the luciferase reporter gene (pGL3-Tim50), the β-galactosidase control plasmid, and pCMVBam control plasmid (vector), or the indicated p53 plasmid for 48 hrs. After transfection, luciferase activity was detected using a luciferase reporter assay and values normalized to β-galactosidase values to control for transfection efficiency. The value of normalized luciferase activity in the vector control was set to 100 and all other samples compared to it. (B) Cell lysates from the transfections were blotted with the indicated antibodies. Similar results were obtained in two additional experiments. (C) Saos-2 cells were transfected with the indicated plasmids and luciferase activity was measured as previously described. The value of normalized luciferase activity in the vector control was set to 100 and all other samples compared to it. Similar results were obtained in two additional experiments. * = significance p< 0.05.
Fig. 4.
Mutant p53 expression enhances histone acetylation and transcription factor recruitment at the Tim50 promoter. Chromatin immunoprecipitation (ChIP) assays were performed from HC5 or H-p53-R273H cells using antibodies specific for: (A) acetylated Histone H3 (Ac); (B) Ets-1; (C) CREB; or (D) CBP using equal amounts of protein. ChIP assays were also performed with non-specific IgG as a control in each case. The immunoprecipitants were assayed for the presence of the Tim50 promoter by quantitative PCR and Tim50 mRNA levels normalized to the internal standard, GAPDH mRNA. The normalized Tim50 mRNA level in the control cell line, HC5, was set to 1 and compared to the Tim50 mRNA levels in H-p53-R273H cells. * = significance p< 0.05.
Similar results were observed using a portion of the Tim50 coding region (bp 733–922) in the PCR reaction (data not shown). No significant enrichment of the Tim50 promoter region was observed using control antibodies directed against human IgG. We also analyzed the Tim50 promoter for potential transcription factor binding sites. Potential transcription factor binding sites on the Tim50 promoter were identified TFSEARCH software, version 1.3 (available on the World Wide Web at www.cbrc.jp/htbin/nph-tfsearch) with the cut-off threshold of 85%. Six putative CREB sites (bp 271–339), four putative CBP sites (bp 298–302), and four putative Ets-1 sites (bp 949–1710) were identified. To determine if mutant p53 expression enhanced the presence of these transcription factors at the Tim50 promoter, we performed ChIP assays using antibodies directed against Ets-1, CREB and CBP. The Tim50 promoter region was greatly enhanced in anti-Ets-1, -CREB and -CBP immunoprecipitants from H-p53-R273H cells but not from HC5 cells (Fig. 4B, C and D). Together these results indicate alteration of chromosome structure and an enrichment of several known transcription factors at the Tim50 promoter in the presence of mutant p53.
Reduction of Tim50 protein expression inhibits the growth of human lung and breast cancer cells expressing p53 mutants
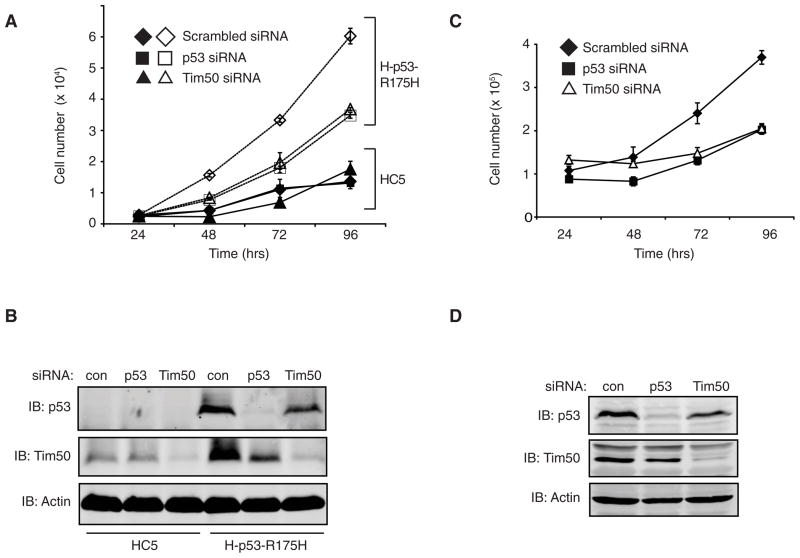
Previously it was shown that the p53 mutant, R175H imparts a growth rate advantage when expressed in breast cancer cells [9]. To determine whether elevated Tim50 expression contributes to a growth rate advantage imparted by p53 GOF function mutants, Tim50 expression was reduced using RNAi in H-p53-R175H and HC5 cells. The role of mutant p53 in growth rate enhancement in this cell line was similarly analyzed by p53 siRNA. Reduction of Tim50 protein expression significantly reduced the growth rate of H-p53-R175H cells but had no effect on HC5 cells that lack p53 (Fig. 5A). Reduction of p53 protein expression, as expected, also reduced the growth rate of H-p53-R175H cells but had no effect upon HC5 cells. The efficacy of RNAi in reducing Tim50 or p53 expression was confirmed by Western analysis (Fig. 5B). To determine if the loss of Tim50 protein expression impacts a cancer cell line that endogenously expresses a p53 mutant, we examined the effect of Tim50 RNAi upon the growth rate of SK-BR-3 cells, a breast cancer cell that harbors the p53 mutant, R175H. Consistent with the results obtained in H-p53-R175H cells, reduction of p53 or Tim50 protein expression by siRNA (Fig. 5D) significantly reduced the growth rate of SK-BR-3 cells (Fig. 5C).
Fig. 5.
Loss of Tim50 expression impairs the growth rate of mutant p53 expressing cells. (A) HC5 cells (closed symbols) or H-p53-R175H cells (open symbols) were transfected with scrambled siRNA (diamonds) or siRNA directed against p53 (squares) or Tim50 (triangles). At the indicated time points, cell numbers were determined. (B) Western analysis of HC5 or H-p53-R175H cell lysates treated with siRNA. Total cell lysates were harvested 96 hrs post-transfection and immunoblotted with the indicated antibodies. (C) SK-BR-3 cells were transfected with scrambled siRNA (diamonds) or siRNA directed against p53 (squares) or Tim50 (triangles) and at the indicated time points, cell numbers were determined. * = significance p< 0.05 (D) Western analysis of SK-BR-3 cell lysates treated with siRNA. Cell lysates were harvested 96 hrs post-transfection and immunoblotted with the indicated antibodies. All assays were performed in triplicate and similar results were obtained in two additional experiments.
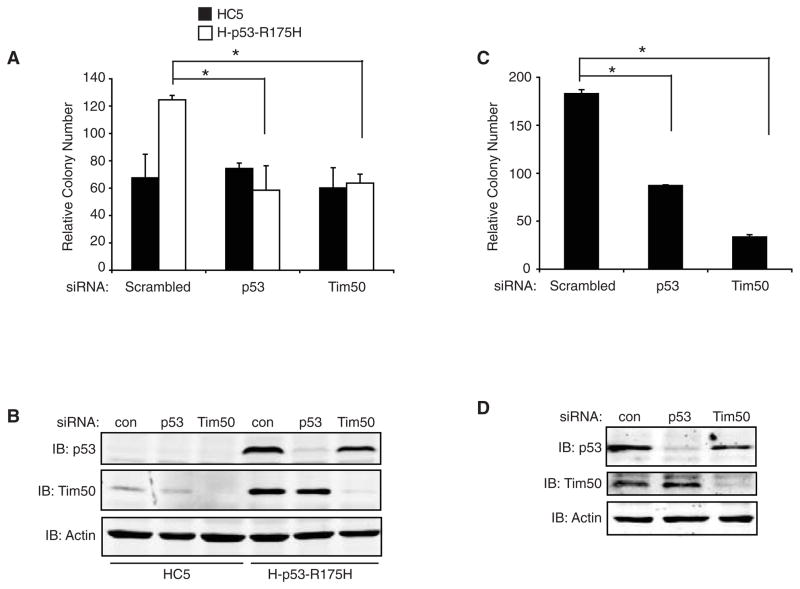
Reduction of Tim50 protein expression inhibits the chemoresistance of human lung and breast cancer cells expressing p53 mutants
The presence of mutant p53 expression in cancer cells has been shown to increase chemoresistance from treatment with different therapeutic agents [9, 11–13] and reduction of mutant p53 levels results in increased chemosensitivity (unpublished results, Dr. Sumitra Deb). To determine if Tim50 protein expression contributes to chemoresistance imparted by p53 mutants, we reduced Tim50 expression and analyzed cells for their survival from treatment with Paclitaxel, a drug presently in clinical use. Reduction of Tim50 protein expression resulted in a significant decrease in the survival of H-p53-R175H cells treated with paclitaxel but had no effect upon HC5 cells as measured by colony formation assays (Fig. 6A and B). Reduction of p53 protein expression, as expected, also reduced the survival of p53-R175H cells but not HC5 cells after treatment with paclitaxel. To determine if Tim50 protein expression contributed to the chemoresistance of cancer cells expressing endogenous mutant p53, we performed Tim50 and p53 siRNA in SK-BR-3 cells and measured cell survival from paclitaxel treatment. Consistent with the results obtained in H-p53-R175H cells, reduction of Tim50 or p53 protein expression in SK-BR-3 cells resulted in a significant decrease in cell survival from paclitaxel treatment (Fig. 6C and D).
Fig. 6.
Loss of Tim50 expression reduces survival of mutant p53 expressing cell lines to paclitaxel. (A) HC5 cells (filled bars) or H-p53-R175H cells (open bars) were transfected with scrambled siRNA or siRNA directed against p53 or Tim50, treated with 25 nM paclitaxel for 48 hrs and colony survival assays performed. (B) Western analysis of HC5 and H-p53-R175H cell lysates treated with siRNA. Cell lysates were harvested 48 hrs post-transfection and immunoblotted with the indicated antibodies. (C) SK-BR-3 cells were transfected with scrambled siRNA or siRNA directed against p53 or Tim50, treated with 25 nM paclitaxel for 48 hrs and colony survival assays performed. (D) Western analysis of SK-BR-3 cell lysates treated with siRNA. Cell lysates were harvested 48 hrs post-transfection and immunoblotted with the indicated antibodies. All assays were performed in triplicate and similar results were obtained in two additional experiments. Colony numbers were adjusted to account for plating differences based on control plates treated with vehicle (DMSO). Relative colony numbers are shown. * = significance p< 0.05
DISCUSSION
Some mutations in the tumor suppressor protein, p53, have been termed ‘gain of function’ mutations because in addition to disabling the functions of WT p53, these mutations appear to impart additional functions to the protein that contribute to an increased oncogenic phenotype in cells in which they are expressed [8–13]. However, it remains unclear how p53 GOF mutants act to promote a GOF phenotype. To explore the mechanism of p53 GOF mutants, we conducted a proteomics study to identify mutant p53-specific interacting proteins. Using large scale immunoprecipitation of mutant p53 from H1299 cells expressing the p53 mutant, R175H, we detected a co-precipitating protein that was not present in controls. Sequencing by mass spectrometry identified the protein as translocator of the inner mitochondrial membrane (Tim50). Although additional experiments are underway to study the binding of Tim50 to mutant p53, it was observed that endogenous Tim50 protein levels were greatly upregulated in cells expressing mutant p53 compared to cells that lacked p53 or expressed WT p53. Elevated levels of Tim protein expression were also observed in several breast cancer cell lines that expressed endogenous levels of mutant p53, compared to cells expressing WT p53, indicating that alteration of Tim50 protein levels was not simply due to mutant p53 overexpression.
Our studies also suggest that the upregulation of Tim50 protein expression observed may be a result of mutant p53 acting either directly or indirectly at the Tim50 promoter. This conclusion is supported by the result that Tim50 transcriptional activity was upregulated by mutant p53 but not by WT p53 using a construct consisting of the Tim50 promoter. Moreover, ChIP assays also indicated that the chromatin structure of the Tim50 promoter was altered (as judged by histone acetylation) in the presence of mutant p53. The interaction of several transcription factors with the Tim50 promoter was also elevated in the presence of a p53 GOF mutant, R273H. Precedent for the interaction of p53 mutants with transcription factors was shown by the interaction of the p53 GOF mutant, D281G, with Ets-1 and the selective up-regulation of endogenous human MDR1 expression [41]. Taking together, these findings strongly suggest that the Tim50 gene is a transcriptional target of p53 GOF mutants. While the p53 utilized in these studies, R273H, was found to behave qualitatively in a similar manner to p53-R175H with regard to Tim50 activation and expression, it should be noted that p53-R175H and p53-R273H represent different classes of p53 mutants: p53R-175H is a conformational mutant, whereas p53-R273H is a DNA contact site mutant [42]. Our conclusions may therefore be relevant to a broad range of p53 mutants implicated in human cancer. Further studies in human cancers will be necessary to determine if there is a correlation between mutant p53 status, Tim50 expression, and tumorigenicity.
What might be the role of increased Tim50 protein expression in the action of p53 GOF mutants? Tim50 forms part of a large protein complex that functions to import proteins with mitochondrial presequences into the mitochondrial matrix [33, 43]. This includes all mitochondrial matrix and a number of inner mitochondrial membrane proteins required for normal mitochondrial function. Specifically, Tim50 is thought to facilitate transfer of proteins from the outer membrane of the inner mitochondrial space across the inner mitochondrial membrane. In addition to participating in transport of proteins, it is thought that Tim50 maintains the permeability barrier of mitochondria by closing the translocation pore [33]. Indeed loss of Tim50 results in the loss of mitochondrial membrane potential (MMP) in yeast [33] and flies [44]. Perhaps as a result of the defect in mitochondrial function, Tim50 null flies were also found to be reduced in size and cells derived from the flies showed a proliferation defect [44]. Thus, Tim50, through its essential role in the transport or proteins into the mitochondrial matrix, has been shown to play an active role in the modulation of growth and development [44].
How might upregulation of the Tim50 protein contribute to the increased survival from paclitaxel treatment observed? Paclitaxel treatment has been shown to induce apoptosis in lung cancers [45] whereas reduction of Tim50 expression was shown to increase the sensitivity of human cell lines to death stimuli by increasing the rate of cytochrome C release from mitochondria [46]. For example, it was shown that loss of Tim50 expression by siRNA enhanced cytochrome C release and cell death after treatment of HEK293T cells with UV or staurosporine [46]. Therefore, it is conceivable that increasing the level of Tim50 protein may confer a greater resistance to cytotoxic agents such as paclitaxel.
In summary, we have shown for the first time that a protein required for the import of proteins into the mitochondrial matrix, Tim50, is upregulated in cells that harbor p53 GOF mutants. Our data also suggests that upregulation of protein import into the mitochondria or perhaps maintenance of mitochondrial membrane potential may confer a selective growth or chemoresistance advantage to cancer cells. In this context, these findings suggest that the upregulation of the Tim50 protein by mutant p53 may be a survival strategy for tumors that harbor p53 mutations and it may allow for the development of unique strategies centered around protein import into the mitochondria that could allow for the selective targeting of cancer cells. Alternatively, if Tim50 upregulation is a widespread mechanism for cancer cell survival, measurement of Tim50 expression in cancer cells may be a valuable biomarker for oncogenic potential and tumor development.
Highlights.
The p53 gene is one of the most frequently mutated genes in human cancer and the resultant proteins often impart oncogenic properties to cells in which they are expressed.
Cells expressing mutant p53 have higher levels of Tim50, a protein required for maintenance of mitochondrial membrane potential and for transport of proteins into the mitochondria compared to cells that express no p53 or wild-type p53.
These studies identify the Tim50 gene as a transcriptional target of mutant p53 and suggest that overexpression of Tim50 may contribute to the oncogenic phenotypes observed in cells that harbor mutant p53 including increased growth and chemoresistance.
Acknowledgments
This work was supported by a Ruth L. Kirschstein National Research Service Award T32-CA085159-7 and a Department of Defense Postdoctoral Training Fellowship to Heidi Sankala (W81XW11-09-1-0390) and by grants from NIH to Sumitra Deb (CA70712 and CA121144). Paul Graves is supported by a grant from the American Heart Association (0835471N). We thank Lathika Mohanraj and Jennifer Harris for their technical assistance.
Abbreviations
- CREB
cyclic AMP response element binding protein
- CBP
CREB-binding protein
- ChIP
Chromatin Immunoprecipitation
- MMP
mitochondrial membrane potential
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- Tim50
Translocase of the inner mitochondrial membrane 50
- NDUFA9
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9
- GOF
gain of function
- siRNA
small interfering RNA
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hussain SP, Harris CC. Recent Results Cancer Res. 1998;154:22–36. doi: 10.1007/978-3-642-46870-4_2. [DOI] [PubMed] [Google Scholar]
- 2.Crook T, Vousden KH. Embo J. 1992;11:3935–3940. doi: 10.1002/j.1460-2075.1992.tb05487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crook T, Wrede D, Tidy JA, Mason WP, Evans DJ, Vousden KH. Lancet. 1992;339:1070–1073. doi: 10.1016/0140-6736(92)90662-m. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell C, Zambetti GP. Gene. 2001;277:15–30. doi: 10.1016/s0378-1119(01)00696-5. [DOI] [PubMed] [Google Scholar]
- 5.Sigal A, Rotter V. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- 6.Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, Brown DR, Deb SP, Deb S. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deb S, Jackson CT, Subler MA, Martin DW. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deb D, Scian M, Roth KE, Li W, Keiger J, Chakraborti AS, Deb SP, Deb S. Oncogene. 2002;21:176–189. doi: 10.1038/sj.onc.1205035. [DOI] [PubMed] [Google Scholar]
- 9.Scian MJ, Stagliano KE, Anderson MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. Mol Cell Biol. 2005;25:10097–10110. doi: 10.1128/MCB.25.22.10097-10110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan W, Gao L, Jin D, Otterson GA, Villalona-Calero MA. Transgenic Res. 2008;17:355–366. doi: 10.1007/s11248-007-9154-3. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Sutphin PD, Schwartz D, Matas D, Almog N, Wolkowicz R, Goldfinger N, Pei H, Prokocimer M, Rotter V. Oncogene. 1998;16:3269–3277. doi: 10.1038/sj.onc.1201867. [DOI] [PubMed] [Google Scholar]
- 12.Blandino G, Levine AJ, Oren M. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 13.Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. Nucleic Acids Res. 2007;35:2093–2104. doi: 10.1093/nar/gkm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR. J Clin Invest. 2007;117:1893–1901. doi: 10.1172/JCI31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Levesque MA, Katsaros D, Yu H, Zola P, Sismondi P, Giardina G, Diamandis EP. Cancer. 1995;75:1327–1338. doi: 10.1002/1097-0142(19950315)75:6<1327::aid-cncr2820750615>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Gemba K, Ueoka H, Kiura K, Tabata M, Harada M. Lung Cancer. 2000;29:23–31. doi: 10.1016/s0169-5002(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 18.Turner BC, Gumbs AA, Carbone CJ, Carter D, Glazer PM, Haffty BG. Cancer. 2000;88:1091–1098. [PubMed] [Google Scholar]
- 19.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 21.Oren M, Rotter V. Cold Spring Harb Perspect Biol. 2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Oncogene. 2006;25:304–309. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 23.Scian MJ, Stagliano KE, Deb D, Ellis MA, Carchman EH, Das A, Valerie K, Deb SP, Deb S. Oncogene. 2004;23:4430–4443. doi: 10.1038/sj.onc.1207553. [DOI] [PubMed] [Google Scholar]
- 24.Scian MJ, Stagliano KE, Ellis MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. Cancer Res. 2004;64:7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 25.Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K. Hepatology. 2007;46:759–768. doi: 10.1002/hep.21736. [DOI] [PubMed] [Google Scholar]
- 26.Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. Oncogene. 2007;26:2212–2219. doi: 10.1038/sj.onc.1210296. [DOI] [PubMed] [Google Scholar]
- 27.Weisz L, Damalas A, Liontos M, Karakaidos P, Fontemaggi G, Maor-Aloni R, Kalis M, Levrero M, Strano S, Gorgoulis VG, Rotter V, Blandino G, Oren M. Cancer Res. 2007;67:2396–2401. doi: 10.1158/0008-5472.CAN-06-2425. [DOI] [PubMed] [Google Scholar]
- 28.Zalcenstein A, Weisz L, Stambolsky P, Bar J, Rotter V, Oren M. Oncogene. 2006;25:359–369. doi: 10.1038/sj.onc.1209061. [DOI] [PubMed] [Google Scholar]
- 29.Chin KV, Ueda K, Pastan I, Gottesman MM. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 30.Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. J Biol Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 31.Endo T, Yamamoto H, Esaki M. J Cell Sci. 2003;116:3259–3267. doi: 10.1242/jcs.00667. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]
- 33.Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, Rehling P. Science. 2006;312:1523–1526. doi: 10.1126/science.1127628. [DOI] [PubMed] [Google Scholar]
- 34.Flatt PM, Tang LJ, Scatena CD, Szak ST, Pietenpol JA. Mol Cell Biol. 2000;20:4210–4223. doi: 10.1128/mcb.20.12.4210-4223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 36.Banks L, Matlashewski G, Crawford L. Eur J Biochem. 1986;159:529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- 37.Smith ER, Allis CD, Lucchesi JC. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 38.Bronder JL, Moran RG. J Biol Chem. 2003;278:48861–48871. doi: 10.1074/jbc.M304844200. [DOI] [PubMed] [Google Scholar]
- 39.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 40.Lanyi A, Deb D, Seymour RC, Ludes-Meyers JH, Subler MA, Deb S. Oncogene. 1998;16:3169–3176. doi: 10.1038/sj.onc.1201857. [DOI] [PubMed] [Google Scholar]
- 41.Sampath J, Sun D, Kidd VJ, Grenet J, Gandhi A, Shapiro LH, Wang Q, Zambetti GP, Schuetz JD. J Biol Chem. 2001;276:39359–39367. doi: 10.1074/jbc.M103429200. [DOI] [PubMed] [Google Scholar]
- 42.Bullock AN, Fersht AR. Nat Rev Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 43.Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K. Embo J. 2003;22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugiyama S, Moritoh S, Furukawa Y, Mizuno T, Lim YM, Tsuda L, Nishida Y. Genetics. 2007;176:927–936. doi: 10.1534/genetics.107.072074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Kim JE, Reed E, Li QQ. Int J Oncol. 2005;27:247–256. [PubMed] [Google Scholar]
- 46.Guo Y, Cheong N, Zhang Z, De Rose R, Deng Y, Farber SA, Fernandes-Alnemri T, Alnemri ES. J Biol Chem. 2004;279:24813–24825. doi: 10.1074/jbc.M402049200. [DOI] [PubMed] [Google Scholar]