Abstract
Comparison of gene expression from transgenes and endogenous genes with or without introns reveals a time-regulating role of introns in natural biological systems.
Biological timing regulated by genetic circuits is essential for many processes in physiology and development. The requirement for transcription of components makes timing in genetic networks potentially sensitive to gene length [1]. It seems likely that introns can affect timing simply by increasing the time necessary to complete the synthesis of a mature mRNA [2].
An attractive way to understand how time is measured and used by organisms is the study of genetic clocks and oscillators. One example is the segmentation clock, which is used by the developing vertebrate embryo to control body segment length and number [3]. Unlike the circadian clock, which has a long period relative to the expected transcription and splicing times of its genes [4], the segmentation clock's relatively short period, in the order of an hour, suggests that gene length and the presence of introns may be significant for timing in this system [5]. Previous work using a simple feedback system has shown that intron length can influence the period of oscillation [6]. Now Takashima et al. [7] have presented work that focuses on the effect of time 'delays' introduced by transcription and splicing of introns in their natural context in the Hes7 gene of the segmentation clock of the mouse embryo [7]. Hes7 is a member of the hairy/enhancer of split (Hes) basic helix-loop-helix family of transcriptional repressor proteins. Their work has two important implications. The first relates to the practical use of transgenes to estimate the dynamics of endogenous genes of interest. The second relates to the mechanisms regulating the segmentation clock.
Presence of introns in reporter genes alters timing of protein expression
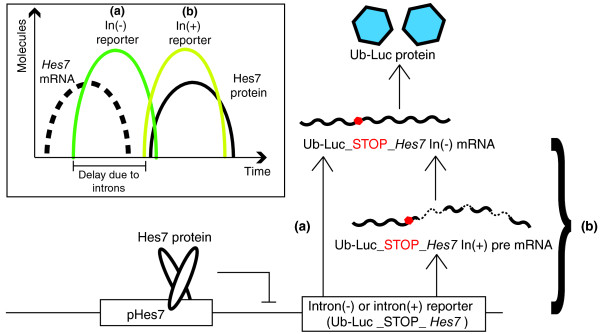
To test whether intron-related time delays could affect gene expression, Takashima et al. [7] generated intron-containing (In(+)) and intron-less (In(-)) reporter constructs (Figure 1). A ubiquitin-destabilized luciferase bioluminescence reporter was placed under the control of the Hes7 promoter region, creating a fusion protein with a half-life comparable to that of Hes7 and an oscillating expression pattern that mimicked endogenous Hes7. Downstream of the luciferase stop codon, they fused either the Hes7 genomic sequence, which contains three introns with total length 1,843 bp, termed In(+), or the corresponding part of the Hes7 cDNA, termed In(-). Thus, translation of either reporter produced ubiquitinated luciferase protein, the difference being that the In(+) construct also required transcribing and splicing of three introns. Following introduction into wild-type mice, the luminescence of each reporter was imaged over time in explanted mouse embryos. Both reporter strains displayed oscillatory expression across the presomitic mesoderm (PSM), with gene expression waves propagating from posterior to anterior. Although these expression patterns mimicked the previously reported patterns of endogenous Hes7, with a period of about 2 h, the onset and therefore the relative timing of luminescence differed between the In(-) and In(+) reporters (Figure 1, inset).
Figure 1.
Design and results of Hes7 In(-) and In(+) reporters studied by Takashima et al. [7]. The ubiquitinated luciferase (Ub-Luc) sequence tagged with either intronless In(-) or intron-containing In(+) Hes7 transcript sequence is placed under the control of the Hes7 promoter region (pHes7). From the mRNA, only the Ub-Luc sequence before the stop codon (red dot) is translated, and this is detected by luminescence imaging in vivo. In contrast to the In(-) reporter (a), the synthesis and processing of introns (dotted RNA segments) from the In(+) transcript introduce an additional delay into the expression timing of the In(+) reporter (b). Inset, the timing of Hes7 mRNA (black dashed line) and Hes7 protein (continuous black line) is compared with In(-) and In(+) luminescence reporter signals (green and yellow lines, respectively).
To estimate the difference in the timing of expression, Takashima et al. [7] compared the expression patterns of the luciferase reporters with the pattern of the Hes7 transcript and the endogenous Hes7 protein in the PSM. This comparison was achieved by first imaging the PSM for luminescence, then fixing immediately and staining with either an RNA intron probe or an antibody for Hes7 protein, and measuring the distance between neighboring endogenous and luminescence expression stripes. This spatial measurement was then converted into an estimate of time difference using the propagation speed of the expression wave from the luminescence reporter movies. The time estimate relies on the assumption that the endogenous mRNA and protein are produced during the same cycle of the clock as the reporter, and that the timing between different steps in gene expression is constant across the PSM. The In(-) reporter was visible an average of 10 min after onset of Hes7 transcription, and 21 min before the Hes7 protein. In contrast, the In(+) reporter was visible only 29 min after the appearance of Hes7 mRNA, and almost simultaneously with Hes7 protein. Thus the presence of Hes7 introns seems to delay translation of the reporter by approximately 19 min. As further confirmation of this intron-related delay, luminescence was detected earlier from In(-) than In(+) reporter constructs under the control of the Hes1 promoter region in mouse fibroblasts [7].
These experiments [7] provide an important measurement of expression delays in vivo and are a landmark for groups wishing to infer timing information from reporter genes. Estimates in the literature of RNA polymerase II transcription speed and intron splicing duration vary, but recent genomic-scale studies indicate a surprisingly high net transcription rate of 3.8 kb/min and splicing times of 5 to 10 min [8]. Given these values, it is possible that the observed reporter gene expression delay [7] arises largely from the splicing of introns and not from the extra time required to transcribe them. Beyond the key observation of the delaying effects of introns, another lesson is that the inclusion of an early short intron in a reporter gene should create a useful marker for the timing of transcriptional initiation.
Intron-based delays affect the somitogenesis clock
In the second part of their paper, Takashima et al. [7] tackle the question of whether an intron-based transcriptional delay affects the function of the somitogenesis clock. Although a consensus has not been reached about the genetic circuit responsible for the oscillatory activity of the segmentation clock, there is evidence from several vertebrate species that places the Hes proteins at the center of the mechanism [3,5,9]. One influential class of models uses explicit delays to account for the time taken to synthesize or transport molecules in the network, and the Hes7 negative-feedback oscillator model of Takashima et al. [7] uses such explicit delays to represent the time required to transcribe and splice Hes7. In these models the delays have a critical role in determining whether the network will oscillate and, if so, what the period will be [5]. Of course, delays do not actually exist in biochemistry, rather they are convenient simplifications to combine many steps into one parameter, but their use is well-justified in many circumstances [10]. In fact, other models of the segmentation clock introduce additional molecular species whose dynamics fill in the gaps represented by explicit delays.
The model used by Takashima et al. [7] predicts that oscillations of the clock will be eliminated when the delay for the production of mature mRNA is adjusted to 19 min faster than the wild-type, the value estimated from the reporter experiments [7]. To test this prediction the authors generated 'intronless' mice, in which the coding region of the endogenous Hes7 locus was replaced with Hes7 cDNA. Strikingly, these mice have a morphological phenotype comparable to Hes7 null mice, with badly segmented axial skeletons, strongly reduced body length and defects in somite polarity markers. The production pattern of Hes7 during embryogenesis differed from the wild-type, as no dynamic spatiotemporal pattern of Hes7 mRNA or protein could be detected in the intronless embryos. Furthermore, imaging the In(+) reporter in the intronless mouse showed that dynamic regulation of the Hes7 promoter was lost: no oscillations, no propagating waves, only an even luminescence signal across the PSM. Thus, the prediction of the model about the effect of intronic delays seems to be confirmed.
One potential caveat to this interpretation is the observation that in intronless mice Hes7 protein was reduced to 34% of wild-type levels. To rule out the possibility that low protein levels caused the phenotype, a series of experiments compared the ability of In(+) and In(-) Hes7 transgenes with multiple copy number to rescue the Hes7 null mutant. In(-) Hes7 transgenes producing Hes7 protein levels equivalent to and higher than the wild-type failed to rescue, whereas even low levels of Hes7 protein from In(+) Hes7 transgenes rescued the phenotype. These controls offer compelling evidence that the absence of introns, and not the level of protein, is responsible for the morphological phenotype and stopped clock of the Hes7 intronless mutant embryos.
From these data [7], it is tempting to conclude that 'delays' introduced by the transcription and splicing of Hes7 introns are a critical mechanism of the mouse segmentation clock, and indeed these elegant experiments are entirely consistent with this possibility. However, to play the devil's advocate, there are many ways to break an oscillator, and a broken oscillator may not easily reveal the internal dynamics that led to its failure. One way around this is to predict and experimentally confirm the quantitative change to a property, such as the period, of an oscillator that is still running. This was possible in the synthetic oscillator network mentioned earlier, in which the period was found to depend on intron length [6], and a similar strategy in the segmentation clock using multiple Hes7 alleles with varying intron length and/or number might yield a series of informative period changes before breaking the clock. Another option might be to predict how the combination of two individually non-functional but compensatory modifications could restore the function of the clock. If such a truly diagnostic and quantitative comparison of an altered phenotype and the model's prediction could be made, it would significantly bolster interpretation, and thereby also further support a central role for the Hes7 feedback loop in the mouse segmentation clock. Nevertheless, along with previous work from the same group on the stability of Hes7 protein [9], this study [7] supports the basic mechanisms of the auto-inhibition with transcriptional delay model for the vertebrate segmentation clock [5]. This work [7] also convincingly demonstrates that introns can have a significant role in the timing of gene expression during development in vivo, with implications both for future experiments designed to measure rapid changes in gene expression and for our understanding of the regulation of timing in biological systems.
Abbreviations
bp: base pair; kb: kilobase; PSM: presomitic mesoderm.
Acknowledgements
AO is a recipient of a Dresden International Graduate School (DIGS-BB) postgraduate fellowship. ACO is supported by the Max Planck Society and by the European Research Council under the European Communities Seventh Framework Programme (FP7/2007-2013)/ERC Grant no. 207634. We thank Laurel Rohde and Cristian Schröter for comments.
References
- Thummel CS. Mechanisms of transcriptional timing in Drosophila. Science. 1992;255:39–40. doi: 10.1126/science.1553530. [DOI] [PubMed] [Google Scholar]
- Swinburne IA, Silver PA. Intron delays and transcriptional timing during development. Dev Cell. 2008;14:324–330. doi: 10.1016/j.devcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter C, Oates AC. Segment number and axial identity in a segmentation clock period mutant. Curr Biol. 2010;20:1254–1258. doi: 10.1016/j.cub.2010.05.071. [DOI] [PubMed] [Google Scholar]
- Cibois M, Gautier-Courteille C, Legagneux V, Paillard L. Post-transcriptional controls - adding a new layer of regulation to clock gene expression. Trends Cell Biol. 2010;20:533–541. doi: 10.1016/j.tcb.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/S0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Swinburne IA, Miguez DG, Landgraf D, Silver PA. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 2008;22:2342–2346. doi: 10.1101/gad.1696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Ohtsuka T, Gonzalez A, Miyachi H, Kageyama R. Intronic delay is essential for oscillatory expression in the segmentation clock. Proc Natl Acad Sci USA. 2011;108:3300–3305. doi: 10.1073/pnas.1014418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- MacDonald N. Biological Delay Systems: Linear Stability Theory. Vol. 8. Cambridge: Cambridge University Press; 1989. [Google Scholar]