Abstract
High-throughput phenotyping approaches (phenomics) are being combined with genome-wide genetic screens to identify alterations in phenotype that result from gene inactivation. Here we highlight promising technologies for 'phenome-scale' analyses in multicellular organisms.
Review
The availability of complete genomic sequences of many model organisms has made it possible to perform highly informative genome-wide functional analyses. For multicellular organisms (including the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, the plants Arabidopsis thaliana and rice, as well as mouse), phenotypic analysis of genetic mutations is still one of the most effective ways to explore the function of a gene. Collections of strains with mutations in nearly every gene are now available, making it possible to analyze the phenotypes of a large number of independent strains. However, conventional analytic approaches, such as high-magnification microscopy at the single-cell level, require manual manipulation of samples and screening by eye, thus limiting throughput and presenting bottlenecks to large-scale genetic studies in multicellular organisms. Therefore, development of high-throughput methods, including automation in phenotyping and screening, is a strategy that is now coming to fruition [1]. Systematic large-scale phenotyping efforts have begun to generate information on a previously unattainable scale. For example, it was recently shown that even a highly dynamic process such as the division of human cells can be studied on a genome-wide scale by live imaging [2].
Cultured cells have also proved amenable to high-throughput phenotyping [2]. Although more challenging, the study of living organisms can provide insights into biological pathways, regulatory networks and/or cellular activity and behavior not obtainable from cultured cells [3-6]. Large-scale acquisition of phenotypic data can then predict important biological outputs, such as the roles of individual genes in development. Thus, high-throughput phenotyping approaches (that is, phenomics) can encompass a broad range of model systems and techniques aimed at understanding the link between genotype and phenotype.
A good example of the evolution of high-throughput phenotyping is provided by RNA interference (RNAi) screens in the worm C. elegans, where recent advances in robotic sample preparation have facilitated high-throughput screens. However, C. elegans is only one of many systems in which innovative technologies for high-throughput studies are being developed. Indeed, the development and use of robotic platforms has also enabled high-throughput phenotypic analysis of plant growth and development at a larger physical scale. Here, we use C. elegans and Arabidopsis as the primary examples of the exciting new wave of approaches to functional genomics [7-10]. We focus on current advances in high-throughput phenotyping (HTP) for the analysis of C. elegans and Arabidopsis, as lessons learned from these organisms can be broadly applied to other animal and plant species.
RNAi and high-throughput phenotyping in C. elegans
Reverse genetic screening has proved a powerful method to identify gene function [11,12]. RNAi is a well-conserved phenomenon observed in many different organisms [13-23]. It was originally discovered in plants, and became one of the first genome-wide techniques used to study loss-of-function phenotypes in several model systems and in mammalian cell culture [24-27]. RNAi screens have become invaluable tools in assessing genotype-phenotype relationships [28,29], and several large-scale RNAi libraries have been generated to identify essential genes and those with novel functions [16,30-32]. For example, an RNAi library of 750 ovary-enriched genes was generated to study the function of genes involved in embryogenesis [31]. RNAi genome-wide screens in Drosophila have been performed using cell culture [12,15]. The genome-wide collection of transgenic constructs that has been prepared for in vivo screening has underpinned a number of studies, including a screen that led to the identification of the sex-peptide receptor of Drosophila [33,34]. Large-scale mutagenesis and phenotyping projects are also under way in mammalian cells, and are likely to yield similarly important results [23,35].
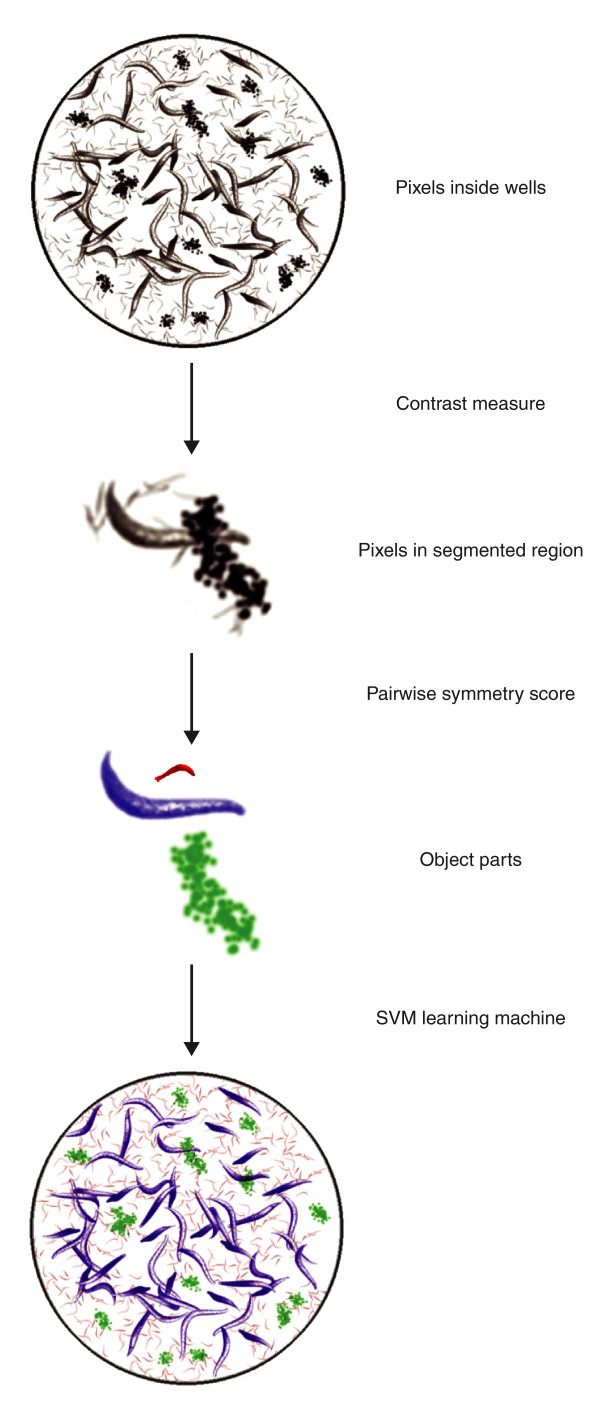
Over the past few years, increasingly sophisticated image-analysis tools have facilitated RNAi screens. Initially, high-throughput RNAi phenotyping focused on endpoint observations, such as worm morphology and viability, and thus were unable to distinguish between the primary and secondary effects of gene silencing. It is now possible to perform rapid and accurate phenotyping of embryonic lethality in different C. elegans developmental stages by analyzing high-throughput image data [36]. The image-analysis system DevStaR uses a hierarchical approach, in which the output of one step is the input for the next, for automatic classification of the developmental stages of worms from a population of mixed stages (including adult, larval and embryonic stages; Figure 1). The system consists of several layers that result in the identification of an area of interest: a segmentation of pixels within this region; a model-based component that breaks the pixel regions into object parts; and finally, a categorization of those objects using a machine-learning approach. This multi-layered object-recognition software offers the computational flexibility for generalized object-recognition problems and, therefore, is not limited to high-throughput worm screens [36].
Figure 1.
Simplified illustration of the DevStaR system. The input images are from 96-well plates containing a population of mixed stages of adult, larva and embryo worms. Each pixel within the wells is first grouped together (contrast measure). Pixels are then grouped into connected components based on a threshold value (pairwise symmetry score). Third, for the object categorization, a support vector machine (SVM) learning method assigned a score to each category. Finally, as a result of the segmentation and labeling, DevStaR distinguishes adult (blue), larva (red) and embryo (green) worms.
New computer-aided visualization methods, which automatically distinguish images of worms grown in agar plates, are also available [37]. In addition, automated phenotyping based on machine-learning methods of images obtained from movie frames can also be used to study embryo development [38]. These systems overcome previous bottlenecks in image analysis by scoring image data in a fully automated manner and providing rapid quantitative output that would not be obtainable at high-throughput by manual scoring. Because high-throughput phenotyping generates a large volume of data, which need to be standardized, normalized and analyzed, statistical and bioinformatics approaches are also becoming increasingly available.
Automated screening using worm-sorters
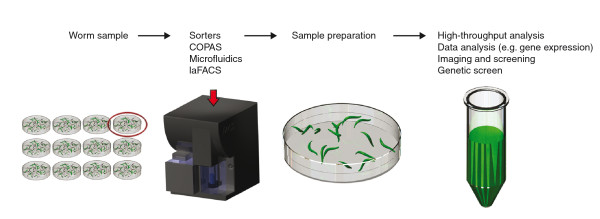
Further advances combining RNAi and sample sorters have enabled rapid selection of organisms with phenotypes of interest for a variety of assays, including genetic screens (Figure 2). Small-animal sorters, such as the BIOSORT/COPAS (complex object parametric analysis and sorter) machine, use a flow-through technique and a profiler system that can analyze up to 100 live animals per second and generate fluorescence emission profiles of the C. elegans body. COPAS has recently been used to analyze the expression pattern of 900 predicted C. elegans genes [39]. By analyzing large numbers of animals from a mixed-stage culture, Dupuy and colleagues [39] generated digitized chronograms of the intensity of gene expression throughout post-embryonic development. This machine allows researchers to study gene expression patterns in a large population of adult animals with a quantitative read-out. However, its sensitivity in sorting non-adult animals, such as embryos and larvae, is limited. Therefore, as a complementary approach, fluorescence-activated cell sorting (FACS) can be employed. By using embryo FACS (eFACS), large numbers of living embryos enriched in any desired embryonic stage can now be selected. Given the availability of different fluorescent marker genes, eFACS enables the assay of embryonic stage-specific gene expression in a high-throughput manner. Moreover, the need for a fast and reliable way of identifying phenotypic alterations in larvae, after modulating or eliminating genes, led researchers to develop a method to sort live C. elegans larvae (laFACS) [40]. Modifying a FACS machine enabled the collection of large quantities of live mutant worms from mixed populations, thereby expanding the arsenal of tools for high-throughput 'sample preparation' for genetic screens. Because these flow-cytometry-based systems sort animals only on one-dimensional intensity profiles, microfluidics chips have been developed to obtain single-cell resolution [41,42]. Microfluidic chips can be designed to function as small-scale sorters with channels and computer-controlled valves that control the environment surrounding the organism and restrict the worms' movements. This technology, when combined with automated image processing, allows high-throughput, non-biased phenotyping, imaging and screening of multicellular organisms [43].
Figure 2.
Outline of general strategies of phenotyping in C. elegans using RNAi and sorters. The sorting techniques COPAS and laFACS can be used to sort live worms. FACS is used to rapidly sort and collect large quantities of live larvae from a mixed population. After laFACs, pure GFP or mutant worms can be used for either genetic or chemical screens, microarray or biochemical assays.
The resolution at which biological samples can be analyzed has greatly increased in recent years as fluorescence microscopy strategies have been developed to characterize gene expression at the single-cell level in C. elegans [1,44]. Methods to quantitatively measure gene-expression dynamics with cellular resolution are anticipated, and will be advantageous to functional genomic studies. However, the challenge of capturing high-resolution images that represent the entire sample remains formidable. Extensive high-throughput time-lapse fluorescent microscopy will only become a reality with improvements to the automation of microscopy imaging and the processing of large datasets.
High-throughput phenotyping for plant biotechnology
The identification of genes that underlie phenotypic variation for complex agronomic traits such as biomass and drought tolerance will be key to biotechnology-aided crop improvement. Because such traits are often controlled by many genes that are also heavily influenced by the environment, the discovery of their genetic basis often requires large-scale phenotyping strategies. Mutational methods such as chemical or fast neutron mutagenesis can be used in forward genetic screens, whereas insertional mutagenesis via T-DNA lines or transposons is used to generate libraries of loss-of-function mutants for reverse genetic screens. Arabidopsis has led the way in plant phenotypic profiling because insertional mutations of most genes are available [45-51]. Rice, as a leading experimental model for monocotyledonous crops, also has a panel of insertional mutant lines [52]. Insertional mutagenesis has also been applied to other crops, including maize and Medicago truncatula [53,54]. However, advances in phenomics will be essential to fully realize the potential of these powerful genetic resources.
The investigation of complex traits such as root morphology, leaf size, plant height, flower shape or seed weight requires analyzing hundreds to thousands of plants, which poses a major challenge. Furthermore, gene response as a function of the environment must be accounted for. For this reason, tools specific for digital phenotyping together with automation of this process in controlled environments are necessary for high-throughput screening of plant phenotypes. Digital phenotyping offers the major advantage that data can be reanalyzed when new traits of interest or new types of measurements emerge. As the demand for digital image-acquisition technologies increases, several efforts have been made to generate software tools capable of producing objective and quantitative analyses of large image sets. Automated platforms have been developed for Arabidopsis and for crop plants to allow different aspects of automated visualization and image quantification. For example, the PHENOPSIS platform was used to dissect plant responses to soil water deficit in a collection of natural accessions of Arabidopsis [55]. The PHENODYN platform imposes drought scenarios and has been used to image maize and rice plants [56]. In addition, several efforts to improve aspects of automated visualization and image quantification for high-throughput phenotype scoring (for example, seed germination, hypocotyl growth, leaf-area development and root growth dynamics) have been made for Arabidopsis. Specifically, the high-throughput seed-germination analysis platform GERMINATOR was used to screen for natural variation in a population of 165 recombinant inbred lines, which revealed several quantitative trait loci (QTLs) for salt tolerance [57]. High-resolution measurements of hypocotyl growth and shape have been obtained by automated quantification of time series of electronic images using HYPOTrace [58]. Other examples of fully or partially automated imaging platforms for non-destructive image-based phenotyping are LeafAnalyser, LAMINA and GROWSCREEN 3 D [59-61]. These computer-based tools provide quantitative descriptors for leaf shape and size. A shortcoming of most of these tools is that they are designed to address very specific questions. Moreover, most traditional phenotype-scoring systems are based on endpoint analysis, and therefore do not easily capture the dynamic aspects of complex traits.
Recent approaches to capture these aspects have incorporated time-course data acquisition so that transient events and subtle temporal changes can be observed. However, the challenge of observing dynamic growth processes and responses to environmental stimuli, through the combination of automated time-lapse imaging with automated image analysis, remains [62]. Many image-analysis-based software tools have focused on quantifying root growth rates and root structure. Advances in machine vision and computation of automatic trait evaluation have facilitated digital reconstruction of root systems and have potentially increased the levels of throughput for phenotyping in plants. Examples of software that allow higher-throughput phenotyping are RootTrace [63], KineRoot [64], SmartRoot [65], RootLM [66], Phytomorph [67,68], RootFlow [69] and WinRhizo [70].
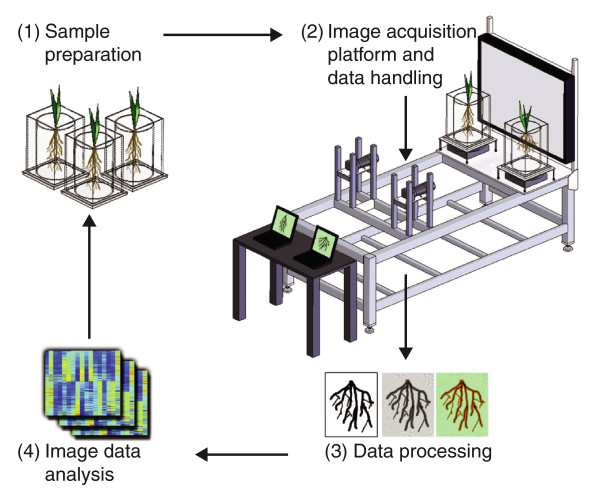
Many high-throughput methods have been developed for Arabidopsis, aided by its small size. For crop plants, an automatic imaging system has been applied to monitoring rice growth [71]. Moreover, a foundation for high-throughput automatic phenotyping for QTL analysis of root system architecture (RSA) traits of crop plants has been laid recently. To capture the root-system topologies of diverse rice cultivars, inbred lines were grown in a transparent gel substrate and imaged at high resolution. The resulting images were combined in an analysis pipeline that automatically extracted RSA measurements. Using a machine-learning approach, these measurements were able to distinguish between closely related genotypes [72] (Figure 3). Alternative methods exist for the non-destructive capture of images of crop root systems grown in solid substrates, such as X-ray tomography and positron emission tomography (PET), but these are limited by throughput, resolution or cost [73,74].
Figure 3.
The general strategies of phenotyping in plants. Illustration of the root-imaging platform. (1) Rice plants are grown in cylinders in gel-based media (sample preparation). (2) The cylinders are placed in a box containing water on the imaging turntable with backlighting. Computers control cameras attached to a four-post support system, which permits adjustments vertically and horizontally. Images are acquired through 360° (image-acquisition platform and data handling). (3) Cropped images from multiple angles are used for analysis (data processing). (4) Feature maps of root architecture that record values for a variety of root features, such as perimeter, depth, bushiness and volume, of each image (image/data analysis).
Low-cost packages for high-throughput phenotyping allow the handling of large-scale experiments, and downstream software pipelines offer flexibility for analysis of numerous lines and treatments. The improved efficiency and absence of subjectivity are great advantages of computer-aided assessment. In the past few years, the generation of phenotypic databases for large numbers of mutants has become a collaborative effort. For example, large-scale phenotypic analysis has been reported in rice using several mutant resources and several phenotype databases are now available (Table 1) [75-79]. Web-accessible collections of visible phenotypes observed for other crop plants, such as barley, maize, tomato and soybean, are also available (Table 1) [80-82].
Table 1.
Phenotypic databases for crop plants
| Plant species | Database name | Website | Reference |
|---|---|---|---|
| Rice | Oryza Tag Line (OTL) | http://urgi.versailles.inra.fr/OryzaTagLine | [76] |
| Rice Mutant Database (RMD) | http://rmd.ncpgr.cn | [78] | |
| Tos17 | http://pc7080.abr.affrc.go.jp/phenotype | [79] | |
| OryGenesDB | http://orygenesdb.cirad.fr/index.html | [77] | |
| Barley | SCRI Barley Mutants | http://bioinf.scri.ac.uk/barley/ | [80] |
| Maize | maizeGDB | http://www.maizegdb.org/rescuemu-phenotype.php | [81] |
| Tomato | Tomato Mutant Database | http://zamir.sgn.cornell.edu/mutants/ | [82] |
| LycoTILL | http://www.agrobios.it/tilling/index.html | [85] | |
| Soybean | Soybean Mutation Database | http://www.soybeantilling.org/psearch.jsp | [86] |
Conclusions
Mutational analysis remains the gold standard for identifying and characterizing gene function and this is being facilitated by high-throughput phenotyping. Given the demand for high-throughput phenotypic analysis in many organisms, we can expect the further development of large-scale phenotyping to unravel complex genotype-phenotype relationships. As an example, automated microscopy provides the opportunity to collect vast amounts of data that need to be standardized, normalized and analyzed. This increases the need for community access to store and search these large datasets. It would be of great benefit if large-scale phenotypic data could be easily compared and shared between labs. However, current limitations to the reuse and sharing of such data include the lack of standardized vocabulary terms, experimental parameters and quantitative benchmarks. Therefore, there is a pressing need for clearly defined standards and terms agreed upon by a given community. To achieve this goal, databases that contain phenotypic information and, especially, integration of phenomic and other genome-wide data are required. Multi-organism phenotype-genotype databases that facilitate cross-species identification of genes associated with orthologous phenotypes are now becoming available (for example, PhenomicDB) [83,84]. In the next few years, the ability to harvest the full benefit of such large datasets can only be obtained by combining the genomic, epigenomic, transcriptomic, proteomic, metabolomic and phenomic data into shared databases. This resource will be invaluable for the investigation and eventual elucidation of molecular mechanisms regulating the biology of multicellular organisms, and will form a comprehensive description of the whole organism, opening new paths into systems biology.
Acknowledgements
We are grateful to Tim Horn and Chris Topp for providing the 'Root Imaging Platform' design. We thank Ryan Baugh and Brad Martsberger, the members of the Benfey lab and the reviewers for their helpful comments. Our work in this area is funded by the NIH as well as the NSF Arabidopsis 2010 program and NSF Plant Genome Research Program.
References
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Hériché JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wünsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P. et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahringer J. Turn to the worm!. Curr Opin Genet Dev. 1997;7:410–415. doi: 10.1016/S0959-437X(97)80157-8. [DOI] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:662–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sundaresan V. Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr Opin Biotechnol. 2000;11:157–161. doi: 10.1016/S0958-1669(00)00075-6. [DOI] [PubMed] [Google Scholar]
- Jorgensen EM, Mango SE. The art and design of genetic screens Caenorhabditis elegans. Nat Rev Genet. 2002;3:356–369. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Somma MP, Fasulo B, Cenci G, Cundari E, Gatti M. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol Biol Cell. 2002;13:2448–2460. doi: 10.1091/mbc.01-12-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/S0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA. Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet. 2003;4:29–38. doi: 10.1038/nrg982. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–344. doi: 10.1016/0167-7799(90)90220-R. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlü N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 2000;10:1619–1622. doi: 10.1016/S0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, Kim SK, Kemphues KJ. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol. 2002;12:1959–1964. doi: 10.1016/S0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Yapici N, Kim YJ, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451:33–37. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- Tsujii H, Eguchi Y, Chenchik A, Mizutani T, Yamada K, Tsujimoto Y. Screening of cell death genes with a mammalian genome-wide RNAi library. J Biochem. 2010;148:157–170. doi: 10.1093/jb/mvq042. [DOI] [PubMed] [Google Scholar]
- White AG, Cipriani PG, Kao HL, Lees B, Geiger D, Sontag E, Gunsalus KG, Piano F. Rapid and accurate developmental stage recognition of C. elegans from highthroughput image data. IEEE. 2010. 978-1-4244-6985-7/10. [DOI] [PMC free article] [PubMed]
- Towards automated high-throughput screening of C. elegans on agar. http://arxiv.org/abs/1003.4287
- Ning F, Delhomme D, LeCun Y, Piano F, Bottou L, Barbano PE. Toward automatic phenotyping of developing embryos from videos. IEEE Trans Image Process. 2005;14:1360–1371. doi: 10.1109/TIP.2005.852470. [DOI] [PubMed] [Google Scholar]
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis AR, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabási AL, Baillie D, Vidal M. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- Fernandez AG, Mis EK, Bargmann BO, Birnbaum KD, Piano F. Automated sorting of live C. elegans using laFACS. Nat Methods. 2010;7:417–418. doi: 10.1038/nmeth.f.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Microfluidic system for on-chip high-throughput whole-animal sorting and screening at subcellular resolution. Proc Natl Acad Sci USA. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- Stirman JN, Brauner M, Gottschalk A, Lu H. High-throughput study of synaptic transmission at the neuromuscular junction enabled by optogenetics and microfluidics. J Neurosci Methods. 2010;191:90–93. doi: 10.1016/j.jneumeth.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJ, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Bevan M, Barnes WM, Chilton MD. Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 1983;11:369–385. doi: 10.1093/nar/11.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella L, Block MD, Messens E, Hernalsteens JP, Montagu MV, Schell J. Chimeric genes as dominant selectable markers in plant cells. EMBO J. 1983;2:987–995. doi: 10.1002/j.1460-2075.1983.tb01532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, Clarke JD, Cotton D, Bullis D, Snell J, Miguel T, Hutchison D, Kimmerly B, Mitzel T, Katagiri F, Glazebrook J, Law M, Goff SA. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res. 2007;120:157–165. doi: 10.1007/s10265-006-0048-x. [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Jeong DH, Jung KH, Lee S. Reverse genetic approaches for functional genomics of rice. Plant Mol Biol. 2005;59:111–123. doi: 10.1007/s11103-004-4037-y. [DOI] [PubMed] [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, Li L, O'Brien BA, Fajardo DS, Wroclawska E, Tseung CW, Lai J, Hunter CT, Avigne WT, Baier J, Messing J, Hannah LC, Koch KE, Becraft PW, Larkins BA, McCarty DR. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics. 2007;8:116. doi: 10.1186/1471-2164-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, Ratet P, Mysore KS. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, Muller B, Simonneau T, Tardieu F. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006;169:623–635. doi: 10.1111/j.1469-8137.2005.01609.x. [DOI] [PubMed] [Google Scholar]
- Sadok W, Naudin P, Boussuge B, Muller B, Welcker C, Tardieu F. Leaf growth rate per unit thermal time follows QTL-dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant Cell Environ. 2007;30:135–146. doi: 10.1111/j.1365-3040.2006.01611.x. [DOI] [PubMed] [Google Scholar]
- Joosen RV, Ligterink W, Hilhorst HW, Keurentjes JJ. Advances in genetical genomics of plants. Curr Genomics. 2009;10:540–549. doi: 10.2174/138920209789503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Uilecan IV, Assadi AH, Kozmik CA, Spalding EP. HYPOTrace: image analysis software for measuring hypocotyl growth and shape demonstrated on Arabidopsis seedlings undergoing photomorphogenesis. Plant Physiol. 2009;149:1632–1637. doi: 10.1104/pp.108.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weight C, Parnham D, Waites R. LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J. 2008;53:578–586. doi: 10.1111/j.1365-313X.2007.03330.x. [DOI] [PubMed] [Google Scholar]
- Bylesjo M, Segura V, Soolanayakanahally RY, Rae AM, Trygg J, Gustafsson P, Jansson S, Street NR. LAMINA: a tool for rapid quantification of leaf size and shape parameters. BMC Plant Biol. 2008;8:82. doi: 10.1186/1471-2229-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup B, Scharr H, Fischbach A, Wiese-Klinkenberg A, Schurr U, Walter A. Diel growth cycle of isolated leaf discs analyzed with a novel, high-throughput three-dimensional imaging method is identical to that of intact leaves. Plant Physiol. 2009;149:1452–1461. doi: 10.1104/pp.108.134486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Fisahn J. Analysis of Arabidopsis thaliana root growth kinetics with high temporal and spatial resolution. Ann Bot. 2010;105:783–791. doi: 10.1093/aob/mcq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A, Ubeda-Tomas S, Holman TJ, Bennett MJ, Pridmore T. High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol. 2009;150:1784–1795. doi: 10.1104/pp.109.140558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu P, Pal A, Lynch JP, Brown KM. A novel image-analysis technique for kinematic study of growth and curvature. Plant Physiol. 2007;145:305–316. doi: 10.1104/pp.107.103226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Draye X, Bennett MJ. Modelling root growth and development. SEB Exp Biol Ser. 2008;61:195–211. [PubMed] [Google Scholar]
- Xiaojun Q, Ji Q, Wu Y. RootLM: a simple color image analysis program for length measurement of primary roots in Arabidopsis. Plant Root. 2007;1:10–16. doi: 10.3117/plantroot.1.10. [DOI] [Google Scholar]
- Miller ND, Parks BM, Spalding EP. Computer-vision analysis of seedling responses to light and gravity. Plant J. 2007;52:374–381. doi: 10.1111/j.1365-313X.2007.03237.x. [DOI] [PubMed] [Google Scholar]
- Brooks TL, Miller ND, Spalding EP. Plasticity of Arabidopsis root gravitropism throughout a multidimensional condition space quantified by automated image analysis. Plant Physiol. 2010;152:206–216. doi: 10.1104/pp.109.145292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weele CM, Jiang HS, Palaniappan KK, Ivanov VB, Palaniappan K, Baskin TI. A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth. Roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol. 2003;132:1138–1148. doi: 10.1104/pp.103.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenault J-L, Pouleur S, Messier C, Guay R. WinRHIZO™, a root-measuring system with a unique overlap correction method. HortScience. 1995;30:906. [Google Scholar]
- Ishizuka T, Tanabata T, Takano M, Shinomura T. Kinetic measuring method of rice growth in tillering stage using automatic digital imaging system. Environ Control Biol. 2005;43:83–96. [Google Scholar]
- Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, Weitz JS, Benfey PN. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 2010;152:1148–1157. doi: 10.1104/pp.109.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy SR, Roberts JA, Black CR, McNeill A, Davidson R, Mooney SJ. The X-factor: visualizing undisturbed root architecture in soils using X-ray computed tomography. J Exp Bot. 2010;61:311–313. doi: 10.1093/jxb/erp386. [DOI] [PubMed] [Google Scholar]
- Kiser MR, Reid CD, Crowell AS, Phillips RP, Howell CR. Exploring the transport of plant metabolites using positron emitting radiotracers. HFSP J. 2008;2:189–204. doi: 10.2976/1.2921207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata N, Miyoshi K, Nonomura K, Yamazaki Y, Ito Y. Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol. 2005;46:48–62. doi: 10.1093/pcp/pci506. [DOI] [PubMed] [Google Scholar]
- Larmande P, Gay C, Lorieux M, Périn C, Bouniol M, Droc G, Sallaud C, Perez P, Barnola I, Biderre-Petit C, Martin J, Morel JB, Johnson AA, Bourgis F, Ghesquière A, Ruiz M, Courtois B, Guiderdoni E. Oryza Tag Line, a phenotypic mutant database for the Genoplante rice insertion line library. Nucleic Acids Res. 2008. pp. D1022–D1027. [DOI] [PMC free article] [PubMed]
- Droc G, Perin C, Fromentin S, Larmande P. OryGenesDB 2008 update: database interoperability for functional genomics of rice. Nucleic Acids Res. 2009. pp. D992–D995. [DOI] [PMC free article] [PubMed]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006. pp. D745–D748. [DOI] [PMC free article] [PubMed]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.). Plant J. 2004;40:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC. MaizeGDB's new data types, resources and activities. Nucleic Acids Res. 2007. pp. D895–D900. [DOI] [PMC free article] [PubMed]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. In silico screening of a saturated mutation library of tomato. Plant J. 2004;38:861–872. doi: 10.1111/j.1365-313X.2004.02088.x. [DOI] [PubMed] [Google Scholar]
- Kahraman A, Avramov A, Nashev LG, Popov D, Ternes R, Pohlenz HD, Weiss B. PhenomicDB: a multi-species genotype/phenotype database for comparative phenomics. Bioinformatics. 2005;21:418–420. doi: 10.1093/bioinformatics/bti010. [DOI] [PubMed] [Google Scholar]
- PhenomicDB. http://www.phenomicdb.de
- LycoTILL. http://www.agrobios.it/tilling/index.html
- Soybean Mutation Database. http://www.soybeantilling.org/psearch.jsp