Abstract
In human corneal epithelium, self-renewal and fate decision of stem cells are highly regulated in a niche microenvironment called palisades of Vogt in the limbus. Herein, we discovered that digestion with dispase, which cleaves off the basement membrane, did not remove the entire basal epithelial progenitor cells. In contrast, digestion with collagenase isolated on cluster consisting of not only entire epithelial progenitor cells but also their closely associated mesenchymal cells because of better preservation of some basement membrane matrix. Collagenase isolated more basal epithelial progenitor cells, which were p63α+ and small in the size (8 μm in diameter), and generated significantly more holoclones and meroclones on 3T3 fibroblast feeder layers than dispase. Further, collagenase isolated more small pan-cytokeratin−/p63α−/vimentin+ cells with the size as small as 5 μm in diameter and heterogeneously expressing vimentin, Oct4, Sox2, Nanog, Rex1, Nestin, N-cadherin, SSEA4, and CD34. Maintenance of close association between them led to clonal growth in a serum-free, low-calcium medium, whereas disruption of such association by trypsin/EDTA resulted in no clonal growth unless cocultured with 3T3 fibroblast feeder layers. Similarly, on epithelially denuded amniotic membrane, maintenance of such association led to consistent and robust epithelial outgrowth, which was also abolished by trypsin/EDTA. Epithelial outgrowth generated by collagenase-isolated clusters was significantly larger in diameter and its single cells yielded more holoclones on 3T3 fibroblast feeder layers than that from dispase-isolated sheets. This new isolation method can be used for exploring how limbal epithelial stem cells are regulated by their native niche cells.
Introduction
Continuous replenishment of healthy corneal epithelial cells relies on stem cells (SCs), which are intermixed with their transient amplifying cells, in the anatomically distinct structure termed the limbal palisades of Vogt, where the stroma is highly vascularized and innervated.1 All limbal basal progenitors are devoid of the expression of cornea-specific differentiation markers such as cytokeratin 3 (CK3),2 CK12,3,4 and gap junction-mediated connexin 43.5 Notably, some limbal basal cells exhibit the slow cycling label-retaining property,6 and have the smallest cell size7,8 and a high proliferative potential in different cultures.9–12 Further, some limbal basal progenitors also preferentially express putative SC markers such as p63,13 especially its ΔNp63α isoform,14 ABCG-2,15–17 integrin α9,18,19 and N-cadherin (N-cad).20
Self-renewal and fate decision of limbal epithelial SCs are highly regulated by a in vivo microenvironment, termed the “niche,” so as to achieve corneal epithelial homeostasis under both normal and injured states.21–23 SCs in their native niche are conceivably mediated by a subset of neighboring cells (including its progeny and subjacent mesenchymal cells), extracellular matrix, and factors sequestered therein. Serial histological sectioning revealed a unique epithelial crypt containing the smallest basal epithelial cells,24–26 suggesting that limbal SCs may lie deeper than expected. The limbal palisades of Vogt have unique ultrastructural features25 and expresses extracellular matrix components such as laminin γ3, SPARC, and tenascin-C.26 Nevertheless, it remains largely unknown whether there exist native niche cells (NCs), and if so whether they may support SC clonal expansion.
Herein, we demonstrate that digestion with dispase, which cleaves the basement membrane,27 fails to isolate all limbal basal epithelial progenitors, whereas additional digestion with trypsin/EDTA (T/E) disrupts the close interaction between limbal basal progenitors and their NCs. In contrast, digestion with collagenase, which removes interstitial collagens, but not basement membrane collagens, effectively isolates the entire limbal basal progenitors together with its closely associated NCs. The validity of such a new isolation method is demonstrated by in vitro clonal assays with or without the use of 3T3 fibroblast feeder layers and by culturing on epithelially denuded human amniotic membrane (dAM) without 3T3 fibroblast feeder layers. The significance of this new isolation method is further discussed regarding how it may be incorporated for ex vivo expansion to engineer a surgical graft containing limbal SCs for treating corneal blindness caused by ocular surface diseases with limbal SC deficiency.
Materials and Methods
Tissue preparation and enzymatic digestion
Human tissue was handled according to the Declaration of Helsinki. In this study, human corneoscleral rims from donors aged 23 to 78 (50.1 ± 22.3) years were provided by the Florida Lions Eye Bank. Immediately after the central corneal button had been used for corneal transplantation, they were transferred in Optisol-GS (Bausch & Lomb; www.bausch.com) and transported at 4°C to the laboratory. The rim was then rinsed three times with Hank's balanced salt solution containing 50 μg/mL gentamicin and 1.25 μg/mL amphotericin B. All materials used for cell culturing are listed as Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/tec). After removal of excessive sclera, conjunctiva, iris, and corneal endothelium, the tissue was cut into 12 one-clock-hour segments, from which a limbal segment was obtained by incisions made at 1 mm within and beyond the anatomic limbus (Fig. 2A). An intact epithelial sheet including basal epithelial cells could be obtained by subjecting each limbal segment to digestion with 10 mg/mL dispase in plastic dishes containing supplemented hormonal epithelial medium (SHEM)27 or defined keratinocyte serum-free medium (D-KSFM)28,29 at 4°C for 16 h under humidified 5% CO2. Alternatively, an intact epithelial sheet consisted of predominant suprabasal epithelial cells was obtained by dispase digestion at 37°C for 2 h and the remaining stroma was then digested with 1 mg/mL collagenase A in SHEM at 37°C for 18 h under humidified 5% CO2 to release cells from the stroma.29 In parallel, each limbal segment, without any further trimming off any stromal tissue, was directly digested with 1 mg/mL collagenase A in SHEM at 37°C for 18 h under humidified 5% CO2 to generate a cell aggregate termed “cluster.” For the above limbal sheets and clusters, single cells were obtained by further digestion with 0.25% trypsin and 1 mM EDTA (T/E) at 37°C for 15 min.
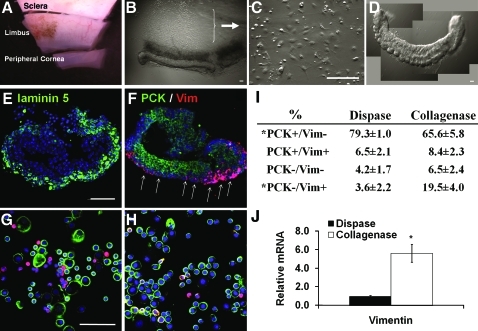
FIG. 2.
Limbal clusters obtained by collagenase digestion. A corneoscleral rim was subdivided into 12 segments, among which each was further trimmed off 1 mm above and below the limbal region (A). Each limbal segment was subjected to collagenase digestion in SHEM at 37°C for 18 h to yield a compact pigmented limbal cluster (B). After additional 24 h culturing on plastic in SHEM, the remaining cells adhered on plastic (C). Each cluster could be manually transferred by a pipette (D). Tangential sectioning of a cluster showed the preservation of laminin 5 (green) on the basal layers (E). Double immunostaining revealed that the cluster contained PCK+ cells (green) and Vim+ (red) cells, which were preferentially distributed on the basement membrane side (F, white arrows). Double immunostaining of cytospin preparations derived from collagenase-isolated clusters revealed more Vim+ cells (G, n = 1200) than those derived from dispase-isolated sheets, which had more PCK+ cells (H, n = 834). Such a difference was confirmed by counting of PCK+ cells and Vim+ cells using Zeiss image analysis (I, from 3 different bright fields and each derived from 3 donors, *p < 0.05). qRT-PCR confirmed a significant sixfold high expression of Vim mRNA in collagenase-isolated clusters than dispase-isolated sheets (J, *p < 0.05, triplicate from 3 donors). Scale bars = 100 μm. SHEM, supplemented hormonal epithelial medium; qRT-PCR, quantitative real-time polymerase chain reaction. Color images available online at www.liebertonline.com/tec
Clonal cultures
The first clonal culture was based on 3T3 fibroblast feeder layers in serum-containing SHEM. The feeder layer was prepared by treating 80% subconfluent 3T3 fibroblasts with 6 μg/mL mitomycin C at 37°C for 2 h in DMEM containing 10% fetal calf serum, and then by seeding mitomycin C-treated 3T3 fibroblasts at a density of 2 × 104/cm2. For initiating clonal growth, a total of 500 single cells derived from limbal sheets or clusters were seeded per 100 mm dish, whereas the same number of cells from the outgrowth derived from limbal sheets or clusters on dAM were seeded in a 35 mm dish. Cultures were maintained in SHEM for 9 days. The second clonal culture was based on collagen type I-precoated six-well plates in D-KSFM, that is, a serum-free low-calcium medium. For initiating clonal growth, 2 × 104 single limbal epithelial cells were seeded with or without the same density of mitomycin C-arrested fibroblast feeder layers as mentioned above. For comparison, 1 × 105 cells dissociated from the remaining stroma were seeded on a noncoated six-well culture plates with or without the same 3T3 fibroblast feeder layer as mentioned above. The culture medium was changed every 3 days, and colony-forming efficiency (CFE) was measured by calculating the percentage of the clone numbers divided by the total number of cells seeded on day 0. Clone morphology was categorized as holoclone, meroclone, and paraclone based on published criteria for skin keratinocytes.30 For analysis, all clones were fixed in cold methanol and subjected to staining with either rhodamine B or crystal violet, or to immunofluorescence staining.
Outgrowth on epithelially dAM
Epithelially dAM was prepared by subjecting cryopreserved AM (Bio-Tissue, Inc.) to incubation with 0.02% EDTA in phosphate-buffered saline (PBS) at 37°C for 1 h to loosen amniotic epithelial cells, followed by gentle mechanical scraping with a toothbrush. dAM was then fastened onto a culture insert as previously reported.31 On each dAM, a freshly isolated limbal cluster or sheet was manually transferred through a pipette. For comparison, a limbal cluster was rendered into single cells by T/E before seeded on dAM in SHEM. The culture medium was changed every 2–3 days. Epithelial outgrowth was monitored under phase-contrast microscopy, and terminated on day 7 by crystal violet staining and immunofluorescence staining.
Immunofluorescence staining
Intact limbal segments and dispase-isolated sheet followed by dispase digestion at 4°C for 16 h were cryosectioned to 6 μm thickness. The remaining stroma for clusters obtained by dispase or collagenase digestion were similarly cryosectioned or prepared as a flat mount by air-drying the specimen on a slide before fixation at −20°C in 100% cold methanol, respectively. Single cells from limbal sheets or clusters were also prepared as cytospin onto a slide using Cytofuge® (StatSpin, Inc.) at 1000 rpm for 8 min at the density of 4.0 × 104 cells per chamber. The cytospin preparation was dried at room temperature for 5 min followed by fixation with either 100% cold methanol at −20°C or 4% paraformaldehyde at room temperature for 15 min.
For immunofluorescence staining, some formaldehyde-fixed samples were deparaffinized in xylene, hydrated gradually by graded 100%, 95%, and 80% in ethanol, and incubated in the citrate–EDTA buffer (Sigma-Aldrich; pH 6.2) at 95°C for 20 min to retrieve antigens. Others were permeabilized with 0.2% Triton X-100 in PBS for 15–30 min and blocked with 0.2% bovine serum albumin in PBS for 1 h at room temperature before the addition of primary antibody overnight. Secondary antibodies were then incubated for 1 h before image analysis. Detailed information about primary and secondary antibodies is listed as Supplementary Table S2. Isotype-matched nonspecific IgG antibodies were used as controls. Immunofluorescence micrographs were taken by confocal microscopy (Carl Zeiss, LSM700); cell counting, cell size, and cytolocalization were analyzed using AxioVision software (Zeiss).
Quantitative real-time polymerase chain reaction
Fresh limbal sheets and clusters isolated on day 0 as well as epithelial outgrowth isolated from dAM on day 7 by dispase digestion at 37°C for 2 h were subjected to total RNA extraction by RNeasy Mini RNA isolation kit (Qiagen; www1.qiagen.com), and cDNA was reverse-transcribed from 1 to 2 μg of total RNA by high capacity cDNA transcription kit (Applied Biosystems; www.appliedbiosystems.com). Quantitative polymerase chain reaction (PCR) amplification of different genes was carried out in a 20 μL solution containing cDNA, TaqMan Gene Expression Assay Mix, and universal PCR master Mix (Applied Biosystems). All TagMan Gene Expression Assays with probe sequences used are listed as Supplementary Table S3. All assays were performed in triplicate from each donor; the results were normalized by an internal control, glceraldehyde-3-phosphate dehydrogenase.
Statistical analysis
All assays were performed in triplicate, each with a minimum of three donors. The data are reported as means ± SD. Group means were compared using the appropriate version of Student's unpaired t-test. Test results were reported as two-tailed p-values, where p < 0.05 were considered statistically significant.
Results
Dispase cannot isolate all epithelial cells from the limbal stroma
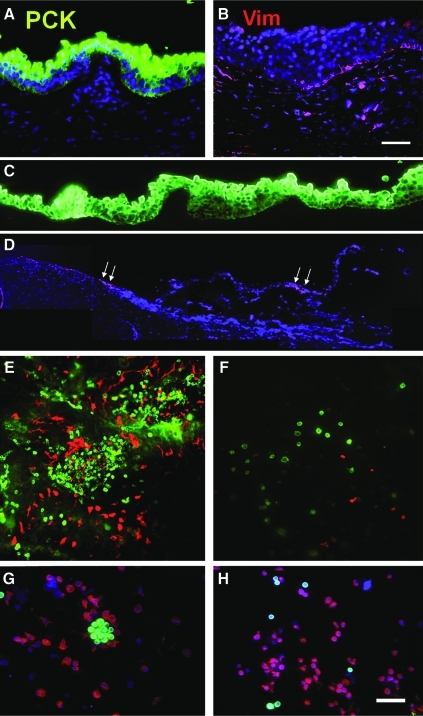
Digestion with dispase, which cleaves off the basement membrane, has been commonly practiced to remove an intact human limbal epithelial sheet27 Given that limbal SCs may lie deep in crypt-like structures in limbal palisades of Vogt,24,32 one may question whether dispase actually removes the entire limbal epithelial progenitors. Even if we assumed that dispase isolated all limbal SCs, we cannot be sure that NCs are also included. To address these questions, we first performed double immunofluorescence staining on cryosectioned human limbal tissues. As expected, the expression of pan-cytokeratin (PCK, green) was found in the full-thickness of limbal epithelium (Fig. 1A), whereas the expression of vimentin (Vim, red) as a mesenchymal cell marker was found close to the basal epithelial cells adjacent to the basement membrane (Fig. 1B). Consistent with what has been reported for the dispase digestion at 4°C/16 h,27 PCK+ staining was distributed to all cell layers, confirming the isolation of an intact limbal epithelial sheet (Fig. 1C). Cryosectioning of the remaining stroma did not reveal any PCK staining, and Vim+ cells were distributed on the stromal surface (Fig. 1D). However, en face optical image of a whole mount preparation the remaining stroma surprisingly showed aggregations of numerous PCK+ and Vim+ cells in the limbus region (Fig. 1E), but only few scattered PCK+ and Vim+ cells in the peripheral cornea (Fig. 1F). Cytospin preparation of the entire cells released from the remaining stroma by collagenase digestion also revealed clusters of small cells in the midst of scattered single cells (not shown). Double immunostaining confirmed that small cells in the cluster were PCK+ and were closely associated with Vim+ cells (Fig. 1G). Additional digestion with T/E rendered them all into either PCK+ or Vim+ single cells (Fig. 1H). Collectively, these results suggested that dispase digestion alone, even using a more extensive regimen of 4°C/16 h, still did not remove all epithelial cells. These deep-seated small epithelial cells could be isolated by further collagenase digestion. When isolated by collagenase, these small epithelial cells aggregated in clusters and were closely associated with Vim+ cells; their close association could be disrupted by additional digestion with T/E.
FIG. 1.
Double immunofluorescence staining of pan-cytokeratins (PCK, green) and Vimentin (Vim, red). PCK+ full-thickness epithelium was present in the cryosectioned human limbal tissue (A), whereas scattered Vim+ cells were subjacent to the epithelium (B). Dispase digestion at 4°C/16 h removed the entire PCK+ epithelial sheet (C). There were no PCK+ cells in the cross section of the remaining stroma, whereas some Vim+ cells were present in the basement membrane area (marked by white arrows) and the stroma (D). Wholemount preparation of the remaining limbal stroma showed in the en face optical image aggregations of numerous PCK+ and Vim+ cells at limbus region (E), but few scattered PCK+ and Vim+ cells in the peripheral cornea (F). Cytospin preparation of cells released from the remaining stroma by collagenase digestion revealed many PCK+ clusters (G) that were associated with Vim+ cells. When these cells were further treated with T/E, PCK+ clusters disappeared and single PCK+ cells were intermixed with Vim+ cells (H). Nuclear counterstaining was performed using Hoechst 33342. Scale bar = 100 μm. T/E, trypsin/EDTA. Color images available online at www.liebertonline.com/tec
Collagenase-isolated clusters contain more Vim+ cells than dispase-isolated sheets
Previously, we noted that collagenase digestion alone differs greatly from dispase or T/E in isolating human corneal endothelial cells by preserving intercellular junctions mediated by N-cad, connexin 43, and ZO-1 as well as cell adhesion to such basement membrane components such as collagen IV, laminin 5, and perlecan.33 Preservation of basement membrane components during isolation is important because we have reported that the success of ex vivo expansion of limbal epithelial progenitor cells is correlated with rapid re-synthesis and deposition of basement membrane components, to which cells maintain their adhesion.33 Because collagenase isolated epithelial cells that are deep-seated in the limbal stroma (Fig. 1), we decided to use it to isolate the limbal epithelium directly from each limbal segment (Fig. 2A). Unlike the sheet configuration generated by dispase digestion,27,29 collagenase digestion yielded a pigmented, compacted cell aggregate, herein termed “cluster” (to distinguish from the “sheet” isolated by dispase) (Fig. 2B). The remaining cells from the limbal stroma mostly adhered on the plastic dish on day 1 in SHEM (Fig. 2C). Thus, collagenase-isolated clusters could easily be transferred to another dish with a pipette (Fig. 2D). Cryosections confirmed the preservation of the basement membrane components, as evidenced by positive staining to laminin 5 (Fig. 2E). Double immunostaining further indicated that the full thickness of epithelial cluster contains PCK+ cells along with Vim+ cells on one side of cluster (Fig. 2F, white arrow). The lack of merge between Vim+ and PCK+ fluorescence in the same cell suggested that Vim+ cells were mostly of a population different from PCK+ cells. To obtain more precise quantitation, we prepared cytospin of single cells obtained by treating dispase-isolated sheets and collagenase-isolated clusters with T/E. Double immunostaining confirmed that collagenase isolated significantly more PCK−/Vim+ cells (Fig. 2G) than dispase (Fig. 2H); PCK−/Vim+ cells comprised 19.5% ± 4.0% of cells derived from collagenase-isolated clusters but only 3.6% ± 2.2% of cells from dispase-isolated sheets (Fig. 2I, p < 0.05, n = 1200 and n = 834 from 3 donors, respectively). Such a difference was further confirmed by a significantly higher expression of Vim transcript in the former using quantitative real-time (qRT)-PCR (Fig. 2J, p < 0.05, n = 3 donors).
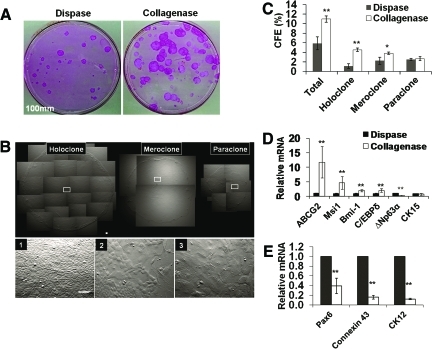
Collagenase isolates significantly more limbal basal progenitors than dispase
The above data also prompted us to examine whether collagenase-isolated clusters contained more limbal basal progenitors, including SCs, than dispase-isolated sheets. To do so, we compared clonal growth by seeding the same density of 500 single cells per 100 mm dish on mitomycin C-arrested 3T3 fibroblast feeder layers. Rhodamine B staining performed on day 9 indeed showed that significantly more epithelial clones were generated by collagenase-isolated clusters compare to dispase-isolated sheets (Fig. 3A). Judged by the criteria provided by Barrandon and Green,30 clones could be classified into three types: holoclone, that is, large round clones with a smooth border and uniformly small and compacted cells in the center and the periphery (Fig. 3B-1); meroclones, that is, smaller round clones with a less smooth border and some large cells in the center (Fig. 3B-2); and paraclones, that is, small irregular clones with an irregular border and large cells in the center (Fig. 3B-3). Collagenase-isolated clusters yielded fivefold more holoclones and twofold more meroclones (4.5% ± 0.4% and 3.9% ± 0.9%, n = 6) than dispase-isolated sheets (0.8% ± 0.2% and 1.7% ± 0.2%, respectively, p < 0.001 and p < 0.05, Fig. 3C). The actual CFE for both holoclones and meroclones should be even higher if we discounted Vim+ cells, which were significantly more in collagenase-isolated clusters than dispase-isolated sheets (Fig. 2). qRT-PCR further illustrated that the mRNA levels of such markers as ABCG2, musashi-1(Msi1), Bmi-1, and C/EBPδ in collagenase-isolated clusters were significantly higher than dispase-isolated sheets (Fig. 3D, n = 3, *p < 0.05 and **p < 0.001). Interestingly, because of the inclusion of more Vim+ cells in collagenase-isolated clusters (Fig. 2), the mRNA levels of ΔNp63α (Fig. 3D, n = 3, **p < 0.01) and those of CK12, connexin 43, and Pax6 were significantly less than dispase-isolated sheets (Fig. 3E, n = 3, **p < 0.001). Collectively, these data suggest that collagenase isolated more limbal basal epithelial progenitors expressing more putative SC markers than dispase.
FIG. 3.
Comparison of clonal growth between dispase-isolated sheets and collagenase-isolated clusters. Significantly more rhodamine B-stained clones were generated by collagenase-isolated clusters than dispase-isolated sheets when 500 single cells were seeded in a 100-mm dish containing mitomycin C-treated 3T3 fibroblast feeder layers and cultured in SHEM for 9 days (A). Three different types of clones, that is, holoclone, meroclone, and paraclone, were identified based on the smoothness of the border and the cell size in the center of the clone with uniformly small cells, mixture of small and large cells, and uniformly large cells, respectively (see 1, 2, and 3 derived from respective insets) (B). Collagenase-isolated clusters had a significantly higher percentage of holoclones and meroclone (C, n = 3, *p < 0.05, and **p < 0.001). qRT-PCR revealed that collagenase-isolated clusters had a significant higher expression of ABCG2, Msi-1, Bmi-1, and C/EBPδ transcripts (D), but a lower transcript expression of ΔNp63α, cytokeratin 12 (CK12), connexin 43, and Pax6 (E) when compared to dispase-isolated sheets (n = 3, **p < 0.001). No significant difference was found in transcript expression of CK15 (D). Scale bar = 100 μm. Color images available online at www.liebertonline.com/tec
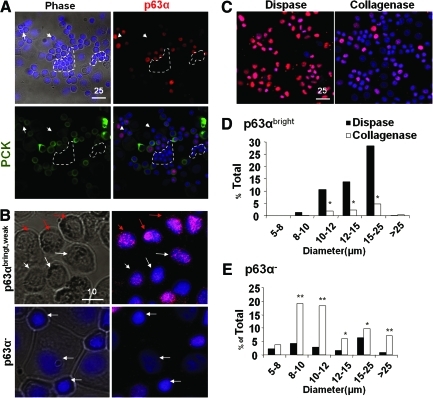
Collagenase isolates more smaller and p63α− NCs than dispase
To further characterize basal epithelial progenitor cells and PCK−/Vim+ cells, we prepared single-cell cytospin preparations of both collagenase-isolated clusters and dispase-isolated sheets. Double immunostaining of PCK and p63α, of which the latter is considered a marker for epithelial progenitor cells,34 confirmed their colocalization as well as the presence of small PCK− and p63α− cells (Fig. 4A). When we subdivided the intensity of nuclear p63α immunostaining into bright, weak, and negative (Fig. 4B), we noted that p63αbright cells were preferentially isolated by dispase, whereas p63αweak cells were preferentially isolated by collagenase (Fig. 4C). When nuclear expression of p63α was correlated with the cell size, we noted that p63αbright cells were significantly more in dispase-isolated sheets in the cell size of 10–12 μm (10.7%), 12–15 μm (13.8%), and 15–25 μm (28.4%) in diameter, which was about five-, six-, and sevenfold higher than collagenase-isolated clusters, respectively (Fig. 4D, p < 0.001). In contrast, p63α− cells tended to be small in the cell size of 8–10 μm (19.2%) and 10–12 μm (18.2%) in diameter, and were significantly more in collagenase-isolated clusters, that is, five- and sixfold, respectively, compared to dispase-isolated sheets (Fig. 4E, p < 0.001). Terminally differentiated large p63α− cells with cell size >25 μm were more in collagenase-isolated clusters than dispase-isolated sheets. No differences were observed in the p63weak population (not shown). These data collectively suggest that collagenase isolated more small p63α− cells than dispase.
FIG. 4.
Correlation between cell size and p63α expression. Cytospin preparations were used to measure the cell size in diameter (μm) and for immunostaining. Double immunostaining confirmed coexpression of p63α (red) and PCK (green) in all small and medium epithelial cells (nuclear counterstaining by Hoechst 33342) and revealed the presence of small PCK−/p63α− cells (circled by white dotted lines) (A). p63α nuclear staining could be subdivided into bright (red arrows), weak (white arrows), and negative, of which the latter tended to be small (B, white arrows in the lower panel). Dispase isolated more p63αbright cells in the size from 10 to 25 μm (C, D), whereas collagenase isolated more p63α− cells in the size spreading from 8 to >25 μm (C, E). Total cells counted in Dispase n = 675 and in Collagenase n = 952, *p < 0.05, **p < 0.001). Scale bar = 25 μm in (A) and (C), and 10 μm in (B). Color images available online at www.liebertonline.com/tec
Phenotypic characterization of small PCK− and p63α− cells in limbal clusters
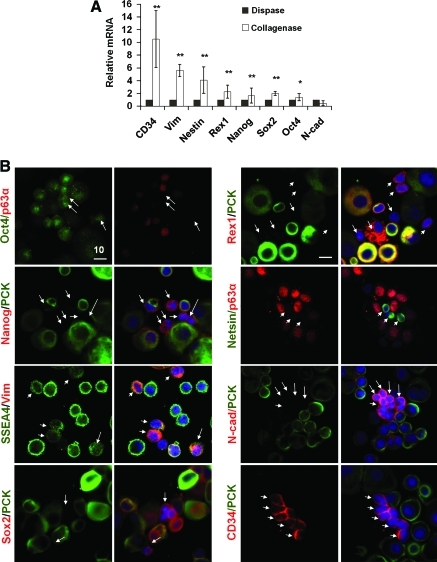
To further characterize the small p63α− cells in collagenase-isolated limbal clusters, qRT-PCR confirmed that mRNA levels of Vim, Nestin, Rex1, Nanog, Sox2, and Oct4 were significantly higher in collagenase-isolated cluster than dispase-isolated sheets (Fig. 5A, n = 3, p < 0.05, 0.001), suggesting that nonepithelial cells expressed SC markers more in collagenase-isolated clusters than dispase-isolated sheets. No difference was noted in the mRNA level of N-cad between the two (Fig. 5A). Double immunostaining between PCK or p63α to identify epithelial cells or Vim to identify mesenchymal cells and the above SC markers showed that these small nonepithelial cells expressed Oct4, Nanog, SSEA4, Sox2, Rex1, Nestin, N-cad, and CD34 (Fig. 5B). These SC markers were not uniformly expressed in these small nonepithelial cells, and were also expressed in some small epithelial cells (not shown). The latter notion could be better appreciated in the staining for Oct4, Nanog, SSEA4, Rex1, and N-cad (Fig. 5B). Cells expressing PCK+ and those expressing CD34+ were mutually exclusive (Fig. 5B); CD34 is expressed in early hematopoietic SC/progenitor in the bone marrow.35 These CD34+ cells were further confirmed to coexpress with Vim+ cells. Some CD34+ cells were also ABCG2+ (not shown). Collectively, these data suggested that small PCK−, p63α−, Vim+, and CD34+ cells in collagenase-isolated clusters heterogeneously expressed embryonic and other SC markers.
FIG. 5.
Phenotypic characterization of putative niche cells from collagenase-isolated clusters. qRT-PCR showed that collagenase-isolated clusters expressed significantly more Vim, Nestin, Rex1, Nanog, and Sox2 transcripts than dispase-isolated sheets (A, n = 3, *p < 0.05, **p < 0.001). Double immunostaining between PCK (green) or p63α (red), of which both were colocalized in epithelial cells (Fig. 4A) or between Vim (red) and other markers revealed that small nonepithelial cells shown in Figure 4 were N-cad+ (red), Sox2+ (red), Nestin+ (green), Oct4+ (green), Rex1+ (red), Nanog+ (red), CD34+ (red), and SSEA4 (green) (B, arrows). Scale bar = 10 μm. Color images available online at www.liebertonline.com/tec
Loss of clonal growth and outgrowth in collagenase-isolated clusters when their close contact with NCs are disrupted by T/E
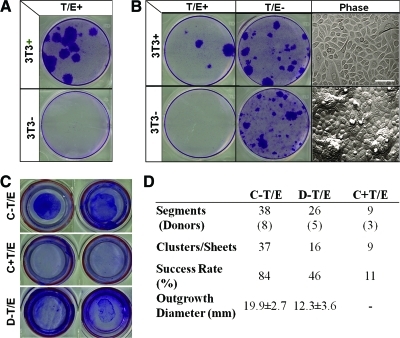
Our recent study showed that the proliferative capacity, clonogenicity, and p63-positive progenitors are better preserved when ex vivo expansion executed by dispase-isolated epithelial sheets were compared to single cells achieved by T/E treatment.36 Therefore, it is conceivable that intercellular junctions disrupted by T/E digestion potentially runs the risk of causing an adverse effect on subsequent expansion of limbal SCs. To examine if disruption of intercellular interaction between limbal epithelial progenitors and surrounding PCK−/Vim+ cells by T/E (Fig. 1) might affect subsequent clonal growth, we first tested it in a serum-free, low-calcium medium, that is, D-KSFM, which we have previously noted to support human and murine corneal/limbal epithelial clonal growth.37,38 Our results showed that clonal growth was absent for dispase-isolated epithelial sheets (without including deep-seated progenitors) if intercellular junction was disrupted by T/E unless 3T3 fibroblast feeder layers were added (Fig. 6A, T/E+). A similar result was noted for cells released from the remaining stroma by collagenase with additional T/E (Fig. 6B, T/E+), which resulted in a mixture of single PCK+ or Vim+ cells (Fig. 1). In contrast, if T/E was not used, cells released from the remaining stroma by collagenase yielded vivid clonal growth (Fig. 6B, T/E−, 3T3−), which contained small PCK+ epithelial clusters closely associated with Vim+ cells (Fig. 1). Phase image illustrated that clones generated by T/E-disrupted single cells cocultured on 3T3 feeder layer (Fig. 6B, T/E+, 3T3+) contained a monolayer of small and large cells, whereas those generated without T/E treatment yielded compact small epithelial cells with scattered elevated mounds (Fig. 6B, Phase).
FIG. 6.
Loss of clonal growth and outgrowth by disrupting close contact with niche cells by T/E. In defined keratinocyte serum-free medium, single cells derived from dispase-isolated sheets (37°C/2 h) by T/E did not generate any clonal growth as judged by crystal violet staining on day 10 in six-well plates, unless 3T3 fibroblast feeder layers were added (A). Similarly, single cells derived from the remaining stroma released by collagenase and treated with T/E (Fig. 1) also did not generate any clonal growth unless 3T3 fibroblast feeder layers were added (B, T/E+). In contrast, cells released from the remaining stroma by collagenase without T/E, which showed epithelial clusters associated with nonepithelial cells (Fig. 1), could generate vivid clonal growth without 3T3 fibroblast feeder layers (B, T/E−). Clones generated on 3T3 fibroblast feeder layers consisted of a monolayer of epithelial cells, whereas those without 3T3 feeder layers exhibited compacted small cells with scattered mounts (B, Phase). In SHEM, an outgrowth on dAM inserts in six-well plates, judged by crystal violet staining on day 7, was consistently generated by collagenase-isolated clusters without T/E (C-T/E) but abolished with T/E (C+T/E), and to a lesser extent by dispase-isolated sheets without T/E (D-T/E) (C, two representative segments from the same donor). Using more limbal segments from different donors, C-T/E gave an overall success rate of 84% and achieved a round shape outgrowth within the diameter of 19.9 ± 2.7 mm in 7 days. In contrast, C+T/E gave a success rate of 11% and no measurable diameter, whereas D-T/E yielded a 46% success rate and generated an uneven round shape with a smaller diameter of 12.3 ± 3.6 mm (D, p < 0.05). Scale bar = 100 μm in (B). dAM, denuded amniotic membrane. Color images available online at www.liebertonline.com/tec
Consistent successful outgrowth was generated by collagenase-isolated clusters on dAM (Fig. 6C, C-T/E). In contrast, no outgrowth was noted if collagenase-isolated clusters were treated with T/E before seeding on dAM (Fig. 6C, C+T/E). As a comparison, outgrowth generated by dispase-isolated sheets was smaller (Fig. 6C, D-T/E). Collagenase digestion yielded a higher success rate of 84% in generating an outgrowth with a significantly larger diameter of 19.9 ± 2.7 mm. In contrast, dispase digestion achieved a success rate of 46% and outgrowth diameter of 12.3 ± 3.6 mm from 26 limbal segments obtained from 5 donors (Fig. 6D, p < 0.05). When collagease-isolated clusters were treated with T/E, the success rate was reduced to 11% with negligible growth (p < 0.001).
Outgrowth of collagenase-isolated clusters on dAM generates higher clonal growth than that of dispase-isolated sheets
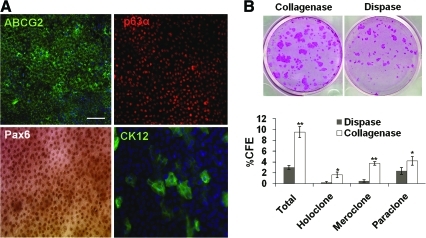
The overall growth curve of collagenase-isolated clusters was slow between day 1 and 3 after seeding on dAM, but faster from day 5 to 7, reaching a significantly larger outgrowth size (Fig. 6D). On day 7, epithelial cells of the outgrowth of collagenase-isolated clusters were smaller than those of dispase-isolated sheets (not shown). Immunostaining confirmed universal expression of ABCG2 in cell membranes and p63α in nuclei of these small epithelial cells, whereas Pax6 was expressed in nuclei and CK12 on some scattered suprabasal cells (Fig. 7A). These results confirmed the expansion of progenitor cells and adoption of a corneal fate for some differentiated cells. When 500 single cells obtained from such outgrowth removed from dAM by dispase followed by T/E were seeded on 3T3 fibroblast feeder layers in a 35 mm dish, we noted that clonal growth on day 9 from collagenase-isolated clusters was significantly more than that from dispase-isolated sheets (Fig. 7B). Outgrowth generated by collagenase-isolated clusters yielded a significant higher CFE of holoclones (1.6% ± 0.4% vs. 0.3% ± 0.1%, p < 0.001), meroclone (3.7% ± 0.3% vs. 0.5% ± 0.2%, p < 0.001), and paraclone (4.2% ± 0.8% vs. 2.3% ± 0.6%, p < 0.05) than that generated by dispase-isolated sheets.
FIG. 7.
Comparison of outgrowth on dAM generated by collagenase-isolated clusters and dispase-isolated sheets. Immunostaining confirmed universal expression of ABCG2 in cell membranes and p63α and Pax6 expression in nuclei of small basal epithelial cells, whereas CK12 was expressed by few large suprabasal cells (A). Single cells derived from the outgrowth isolated from dAM by dispase followed by T/E gave significantly more clones comprising all three classes of holoclones, meroclones, and paraclones from collagenase-isolated clusters when compared to dispase-isolated sheets (B, rhodamin B staining on day 9, *p < 0.05, **p < 0.001). Scale bar = 100 μm. Color images available online at www.liebertonline.com/tec
Discussion
In this study, we have identified a potential pitfall of using dispase digestion for isolating human limbal epithelial progenitor cells. Although dispase can successfully isolate intact human limbal27 and murine corneolimbal epithelial sheets,28,39 it failed to remove all limbal basal epithelial progenitors from the human limbus (Fig. 1). Given that dispase primarily cleaves basement membrane components,27,40 we speculate that the aforementioned failure might be contributed by the unique features noted in the SC-harboring limbal palisades of Vogt such as undulated and fenestrated basement membrane,24,41 unique ultrastructural features,25 and enrichment of laminin γ3, SPARC, and tenascin-C.26 Further, we cannot ignore the likelihood that limbal SCs might actually reside in unique epithelial crypt structures on a plane deeper than the general basement membrane.24,32,42 Nevertheless, digestion with collagenase, which removes interstitial collagens, but not basement membrane collagens,43,44 presents with several advantages. Besides the fact that the underlying stroma did not need to be trimmed before digestion, the entire limbal epithelium was isolated as a cluster from most unwanted stromal cells left on the dish. In addition, the collagenase-isolated clusters retained adhesion to the basement membrane (Fig. 2E), hence not only retaining all basal progenitors, including SCs, but also subjacent Vim+ mesenchymal cells (Fig. 2F).
The evidence that collagenase-isolated clusters included more epithelial progenitors, including SCs, than dispase-isolated sheets was supported by higher clonal growth with significantly higher numbers of holoclones and meroclones on 3T3 fibroblast feeder layers (Fig. 3). Such a difference was supported by the findings that collagenase isolated more small epithelial progenitor cells with a cell size in the range from 8 to12 μm (not shown), p63αweak nuclear staining (Fig. 4C), and significantly higher transcript levels of SC markers such as ABCG2, Bmi-1, Musashi-1, and C/EBPδ (Fig. 3D). Previously, several studies have demonstrated that p63 is a transcription factor heralding the onset of stratified epithelial morphogenesis,45,46 and that its ΔNp63α isoform is a putative marker for human limbal SCs.14,47 Our finding of p63αweak nuclear staining in the smallest epithelial progenitors in the cytospin preparation of collagenase-isolated clusters was new, highlighting the likelihood that such cells might escape the detection in cryosectioned specimens. C/EBPδ has been noted to help induce G0/G1 cell cycle arrest and promote self-renewal of SCs in the mammary gland48,49 and the human limbus.47 Therefore, a notably higher ratio between the transcript level of C/EBPδ and that of ΔNp63α (Fig. 3D) might suggest the presence of more quiescent SCs in collagenase-isolated clusters than dispase-isolated sheets, which contained epithelial cells with p63αbright nuclear staining (Fig. 4C). Hence, collagenase-isolated clusters are a better source for future investigation of SC subpopulations and the function of the above SC markers to resolve whether there exists a bona fide marker for labeling human limbal epithelial SCs.
Compared to dispase, collagenase also enriched a population of PCK−/Vim+ nonepithelial cells subjacent to the basement membrane (Figs. 1 and 2F). Based on a strong correlation in the coexpression of PCK and p63α in epithelial cells (Fig. 4A), cytospin preparations further revealed that these PCK−/p63α− nonepithelial cells comprised around 20% of collagenase-isolated clusters, significantly >3.6% found in dispase-isolated sheets (Fig. 2). We were surprised to note that some PCK−/p63α− cells had a cell size ranging from 5 to 10 μm in diameter (Fig. 4E), which was even smaller than the smallest p63αweak cells with a cell size of 8 to 12 μm in diameter previously reported for limbal SCs.7,8 Using qRT-PCR and double immunostaining, we confirmed that these small nonepithelial cells indeed expressed Oct4, Nanog, SSEA4, Sox2, Rex1, Nestin, N-cad, and CD34 (Fig. 5), markers expressed by embryonic SCs as well as other types of SCs. They were heterogeneously expressed in the entire population of PCK−/p63α− cells, suggesting that future characterization of the differentiation status among them might help understand their inter-relationship. In addition, we were surprised by the finding that some of the aforementioned markers were also expressed by PCK+ or p63α+ epithelial cells (Fig. 5B). Whether such expression in epithelial progenitors represents genuine SCs needs to be further investigated. The expression of embryonic and other SCs markers in cells isolated from the limbal regions with the potential of adopting neuronal and other fates have been reported by others.50–54 Herein, for the first time, our data point out that one likely source is the small nonepithelial cells subjacent to the limbal basement membrane. As a result, one should be cautious in using nestin, N-cad, and ABCG2 as a marker to define limbal SCs.
We speculate that the aforementioned nonepithelial cells contain native NCs that may support limbal epithelial SCs. This speculation was supported by their close association (Figs. 1 and 2) and expression of a set of SC markers (Fig. 5). More importantly, maintenance of such close association by collagenase digestion (Fig. 1) was sufficient to support clonal growth in a serum-free, low-calcium D-KSFM medium without 3T3 fibroblasts (Fig. 6). In this system, dispase-isolated sheets failed to yield any clonal growth unless 3T3 fibroblast feeder layers were added (Fig. 6). However, when the association of NCs and SCs was disrupted by T/E (Fig. 1), clonal growth depended on 3T3 fibroblast feeder layers (Fig. 6). Hence, these nonepithelial cells, mimicking NCs, function like 3T3 fibroblast feeder layers in promoting clonal expansion of epithelial progenitors. Further investigation to identify factors involved in the interaction between SCs and NCs can be pursued using the ground work established in our study. Thus, our finding that SC expansion in vitro relies on close interaction with its adjacent mesenchymal NCs bear resemblance to SKP cells in the epidermal dermal papilla,55 myofibroblsts in the intestinal crypt,56 and vascular endothelial cells in the bone marrow.57
Several ex vivo expansion protocols have been employed to engineer a surgical graft containing limbal epithelial progenitors for human transplantation in treating corneal blindness caused by limbal SC deficiency.58–64 In contrast to the protocols that used limbal explants as the source,60–63 we used collagenase-isolated clusters without including those fibroblast-like cells adherent onto the plastic dish. At this moment, it remains unclear whether there is any benefit or harm of including these cells during ex vivo expansion as well as in clinical transplants that are used directly without ex vivo expansion. Based on the line of reasoning provided above, one may understand why 3T3 fibroblast feeder layers are routinely used to achieve expansion for those protocols using T/E alone or dispase followed by T/E to dissociate the human limbal epithelium into single cells.58,59,62 Besides one group, which uses fibrin gel as the substrate,65,66 all others use either intact AM60 or dAM.59–63 We67,68 and others69,70 have reported that the resultant epithelial phenotype expanded on intact AM resembles the limbal epithelium, whereas that on dAM turns into the corneal epithelium. That is why 3T3 fibroblasts are also used in a coculturing system when human limbal epithelial progenitors are expanded on dAM.59,62,64 The inclusion of 3T3 fibroblasts casts a regulatory concern of transmitting murine viral diseases. Although less and slower, dispase-isolated sheets also generated outgrowth on dAM without 3T3 fibroblast feeder layers (Fig. 6C). However, outgrowth derived from collagenase-isolated clusters yielded higher and faster (Fig. 6C), expressed more presumed SC markers (Fig. 6D), and reached a transplantable size within 1 week larger than dispase-isolated sheets (Fig. 7). Compared to CFE on 3T3 feeder layers derived from collagenase-isolated cluster on day 0, CFE for three types of clones from the outgrowth on dAM was significantly lower, suggesting that further improvement of this new method for better ex vivo expansion of limbal SCs is needed for treating corneal blindness caused by limbal SC deficiency.
Supplementary Material
Acknowledgments
Authors thank Ek Kia Tan, Shunsuke Sakurai, and Lorraine Chua for technical assistant on article preparation. This work was supported by Grant RO1 EY06819 from National Eye Institute, National Institutes of Health, Bethesda, Maryland. In part supported by Grant NHRI-99A1-PDCO-0108117 from the National Health Research Institutes, Taiwan.
Disclosure Statement
S.C.G.T. and his family are >5% share holders of TissueTech, Inc., which owns U.S. Patent nos. 6,152,142 and 6,326,019 on the method of preparation and clinical uses of human amniotic membrane distributed by Bio-Tissue, Inc.
References
- 1.Lavker R.M. Tseng S.C. Sun T.T. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res. 2004;78:433. doi: 10.1016/j.exer.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Schermer A. Galvin S. Sun T.-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W.Y. Mui M.M. Kao W.W. Liu C.Y. Tseng S.C. Conjunctival epithelial cells do not transdifferentiate in organotypic cultures: expression of K12 keratin is restricted to corneal epithelium. Curr Eye Res. 1994;13:765. doi: 10.3109/02713689409047012. [DOI] [PubMed] [Google Scholar]
- 4.Liu C.-Y. Zhu G. Converse R. Kao C.W.-C. Nakamura H. Tseng S.C.G. Mui M.-M. Seyer J. Justice M.J. Stech M.E., et al. Characterization and chromosomal localization of the cornea-specific murine keratin gene Krt1.12. J Biol Chem. 1994;269:24627. [PubMed] [Google Scholar]
- 5.Matic M. Petrov I.N. Rosenfeld T. Wolosin J.M. Alterations in connexin expression and cell communication in healing corneal epithelium. Invest Ophthalmol Vis Sci. 1997;38:600. [PubMed] [Google Scholar]
- 6.Cotsarelis G. Cheng S.Z. Dong G. Sun T.-T. Lavker R.M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 7.Romano A.C. Espana E.M. Yoo S.H. Budak M.T. Wolosin J.M. Tseng S.C. Different cell sizes in human limbal and central corneal basal epithelia measured by confocal microscopy and flow cytometry. Invest Ophthalmol Vis Sci. 2003;44:5125. doi: 10.1167/iovs.03-0628. [DOI] [PubMed] [Google Scholar]
- 8.De Paiva C.S. Pflugfelder S.C. Li D.Q. Cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Stem Cells. 2006;24:368. doi: 10.1634/stemcells.2005-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebato B. Friend J. Thoft R.A. Comparison of central and peripheral human corneal epithelium in tissue culture. Invest Ophthalmol Vis Sci. 1987;28:1450. [PubMed] [Google Scholar]
- 10.Kruse F.E. Tseng S.C.G. A tumor promoter-resistant subpopulation of progenitor cells is present in limbal epithelium more than corneal epithelium. Invest Ophthalmol Vis Sci. 1993;34:2501. [PubMed] [Google Scholar]
- 11.Lindberg K. Brown M.E. Chaves H.V. Kenyon K.R. Rheinwald J.G. In vitro preparation of human ocular surface epithelial cells for transplantation. Invest Ophthalmol Vis Sci. 1993;34:2672. [PubMed] [Google Scholar]
- 12.Pellegrini G. Golisano O. Paterna P. Lambiase A. Bonini S. Rama P. De Luca M. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J Cell Biol. 1999;145:769. doi: 10.1083/jcb.145.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellegrini G. Dellambra E. Golisano O. Martinelli E. Fantozzi I. Bondanza S. Ponzin D. McKeon F. De Luca M. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci USA. 2001;98:3156. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Iorio E. Barbaro V. Ruzza A. Ponzin D. Pellegrini G. De L.M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc Natl Acad Sci USA. 2005;102:9523. doi: 10.1073/pnas.0503437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe K. Nishida K. Yamato M. Umemoto T. Sumide T. Yamamoto K. Maeda N. Watanabe H. Okano T. Tano Y. Human limbal epithelium contains side population cells expressing the ATP-binding cassette transporter ABCG2. FEBS Lett. 2004;565:6. doi: 10.1016/j.febslet.2004.03.064. [DOI] [PubMed] [Google Scholar]
- 16.De Paiva C.S. Chen Z. Corrales R.M. Pflugfelder S. Li D.-Q. ABCG2 transporter identifies a population of clonogenic human limbal epithelial cells. Stem Cells. 2005;23:63. doi: 10.1634/stemcells.2004-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budak M.T. Alpdogan O.S. Zhou M. Lavker R.M. Akinci M.A. Wolosin J.M. Ocular surface epithelia contain ABCG2-dependent side population cells exhibiting features associated with stem cells. J Cell Sci. 2005;118:1715. doi: 10.1242/jcs.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stepp M.A. Zhu L. Sheppard D. Cranfill R. Localized distribution of alpha 9 integrin in the cornea and changes in expression during corneal epithelial cell differentiation. J Histochem Cytochem. 1995;43:353. doi: 10.1177/43.4.7534781. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z. De Paiva C.S. Luo L. Kretzer F.L. Pflugfelder S.C. Li D.Q. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22:355. doi: 10.1634/stemcells.22-3-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi R. Yamato M. Sugiyama H. Sumide T. Yang J. Okano T. Yano Y. Nishida K. N-cadherin is expressed by putative stem/progenitor cells and melanocytes in the human limbal epithelial stem cell niche. Stem Cells. 2006;25:289. doi: 10.1634/stemcells.2006-0167. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs E. Tumbar T. Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 22.Xie T. Li L. Stem cells and their niche: an inseparable relationship. Development. 2007;134:2001. doi: 10.1242/dev.002022. [DOI] [PubMed] [Google Scholar]
- 23.Li L. Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 24.Dua H.S. Shanmuganathan V.A. Powell-Richards A. Tiqhe P.J. Joseph A. Limbal epithelial crypts: a novel anatomical structure and a putative limbal stem cell niche. Br J Ophthalmol. 2005;89:529. doi: 10.1136/bjo.2004.049742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shortt A.J. Secker G.A. Munro P.M. Khaw P.T. Tuft S.J. Daniels J.T. Characterization of the limbal epithelial stem cell niche: novel imaging techniques permit in vivo observation and targeted biopsy of limbal epithelial stem cells. Stem Cells. 2007;25:1402. doi: 10.1634/stemcells.2006-0580. [DOI] [PubMed] [Google Scholar]
- 26.Schlotzer-Schrehardt U. Dietrich T. Saito K. Sorokin L. Sasaki T. Paulsson M. Kruse F.E. Characterization of extracellular matrix components in the limbal epithelial stem cell compartment. Exp Eye Res. 2007;85:845. doi: 10.1016/j.exer.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Espana E.M. Romano A.C. Kawakita T. Di Pascuale M. Smiddy R. Tseng S.C. Novel enzymatic isolation of an entire viable human limbal epithelial sheet. Invest Ophthalmol Vis Sci. 2003;44:4275. doi: 10.1167/iovs.03-0089. [DOI] [PubMed] [Google Scholar]
- 28.Kawakita T. Espana E.M. He H. Yeh L.K. Liu C.Y. Tseng S.C. Calcium-induced abnormal epidermal-like differentiation in cultures of mouse corneal-limbal epithelial cells. Invest Ophthalmol Vis Sci. 2004;45:3507. doi: 10.1167/iovs.04-0266. [DOI] [PubMed] [Google Scholar]
- 29.Hayashida Y. Li W. Chen Y.T. He H. Chen C.-Y. Kheirkah A. Zhu Y.T. Tseng S.C.G. Heterogeneity of limbal basal epithelial progenitor cells. Cornea. 2010;29:11. doi: 10.1097/ICO.0b013e3181ea4b13. [DOI] [PubMed] [Google Scholar]
- 30.Barrandon Y. Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meller D. Tseng S.C.G. Conjunctival epithelial cell differentiation on amniotic membrane. Invest Ophthalmol Vis Sci. 1999;40:878. [PubMed] [Google Scholar]
- 32.Shanmuganathan V.A. Foster T. Kulkarni B.B. Hopkinson A. Gray T. Powe D.G. Lowe J. Dua H.S. Morphological characteristics of the limbal epithelial crypt. Br J Ophthalmol. 2007;91:514. doi: 10.1136/bjo.2006.102640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W. Sabater A.L. Chen Y.T. Hayashida Y. Chen S.Y. He H. Tseng S.C. A novel method of isolation, preservation, and expansion of human corneal endothelial cells. Invest Ophthalmol Vis Sci. 2007;48:614. doi: 10.1167/iovs.06-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeon F. p63 and the epithelial stem cell: more than status quo? Genes Dev. 2004;18:465. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- 35.Krause D.S. Fackler M.J. Civin C.I. May W.S. CD34: structure, biology, and clinical utility. Blood. 1996;87:1. [PubMed] [Google Scholar]
- 36.Kawakita T. Shimmura S. Higa K. Espana E.M. He H. Shimazaki J. Tsubota K. Tseng S.C. Greater growth potential of p63-positive epithelial cell clusters maintained in human limbal epithelial sheets. Invest Ophthalmol Vis Sci. 2009;50:4611. doi: 10.1167/iovs.08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawakita T. Espana E.M. He H. Yeh L.K. Liu C.Y. Tseng S.C. Calcium-induced abnormal epidermal-like differentiation in cultures of mouse corneal-limbal epithelial cells. Invest Ophthalmol Vis Sci. 2004;45:3507. doi: 10.1167/iovs.04-0266. [DOI] [PubMed] [Google Scholar]
- 38.Kawakita T. Shimmura S. Hornia A. Higa K. Tseng S.C. Stratified epithelial sheets engineered from a single adult murine corneal/limbal progenitor cell. J Cell Mol Med. 2008;12:1303. doi: 10.1111/j.1582-4934.2008.00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawakita T. Espana E.M. He H. Yeh L.-K. Liu C.-Y. Tseng S.C.G. Epidermal differentiation promoted by high calcium serum free condition in mouse corneal/limbal epithelial culture. Invest Ophthalmol Vis Sci. 2005;45:3507. doi: 10.1167/iovs.04-0266. [DOI] [PubMed] [Google Scholar]
- 40.Spurr S.J. Gipson I.K. Isolation of corneal epithelium with Dispase II or EDTA. Effects on the basement membrane zone. Invest Ophthalmol Vis Sci. 1985;26:818. [PubMed] [Google Scholar]
- 41.Gipson I.K. The epithelial basement membrane zone of the limbus. Eye. 1989;3(Pt 2):132. doi: 10.1038/eye.1989.21. [DOI] [PubMed] [Google Scholar]
- 42.Yeung A.M. Schlotzer-Schrehardt U. Kulkarni B. Tint N.L. Hopkinson A. Dua H.S. Limbal epithelial crypt: a model for corneal epithelial maintenance and novel limbal regional variations. Arch Ophthalmol. 2008;126:665. doi: 10.1001/archopht.126.5.665. [DOI] [PubMed] [Google Scholar]
- 43.Tuori A. Uusitalo H. Burgeson R.E. Terttunen J. Virtanen I. The immunohistochemical composition of the human corneal basement membrane. Cornea. 1996;15:286. doi: 10.1097/00003226-199605000-00010. [DOI] [PubMed] [Google Scholar]
- 44.Li W. He H. Kuo C.L. Gao Y. Kawakita T. Tseng S.C. Basement membrane dissolution and reassembly by limbal corneal epithelial cells expanded on amniotic membrane. Invest Ophthalmol Vis Sci. 2006;47:2381. doi: 10.1167/iovs.05-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koster M.I. Kim S. Mills A.A. DeMayo F.J. Roop D.R. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senoo M. Pinto F. Crum C.P. McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 47.Barbaro V. Testa A. Di I.E. Mavilio F. Pellegrini G. De L.M. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J Cell Biol. 2007;177:1037. doi: 10.1083/jcb.200703003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Rourke J. Yuan R. DeWille J. CCAAT/enhancer-binding protein-delta (C/EBP-delta) is induced in growth-arrested mouse mammary epithelial cells. J Biol Chem. 1997;272:6291. doi: 10.1074/jbc.272.10.6291. [DOI] [PubMed] [Google Scholar]
- 49.Hutt J.A. O'Rourke J.P. DeWille J. Signal transducer and activator of transcription 3 activates CCAAT enhancer-binding protein delta gene transcription in G0 growth-arrested mouse mammary epithelial cells and in involuting mouse mammary gland. J Biol Chem. 2000;275:29123. doi: 10.1074/jbc.M004476200. [DOI] [PubMed] [Google Scholar]
- 50.Zhao X. Das A.V. Thoreson W.B. James J. Wattnem T.E. Rodriguez-Sierra J. Ahmad I. Adult corneal limbal epithelium: a model for studying neural potential of non-neural stem cells/progenitors. Dev Biol. 2002;250:317. [PubMed] [Google Scholar]
- 51.Seigel G.M. Sun W. Salvi R. Campbell L.M. Sullivan S. Reidy J.J. Human corneal stem cells display functional neuronal properties. Mol Vis. 2003;9:159. [PubMed] [Google Scholar]
- 52.Dravida S. Pal R. Khanna A. Tipnis S.P. Ravindran G. Khan F. The transdifferentiation potential of limbal fibroblast-like cells. Brain Res Dev Brain Res. 2005;160:239. doi: 10.1016/j.devbrainres.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Umemoto T. Yamato M. Nishida K. Yang J. Tano Y. Okano T. Limbal epithelial side-population cells have stem cell-like properties, including quiescent state. Stem Cells. 2006;24:86. doi: 10.1634/stemcells.2005-0064. [DOI] [PubMed] [Google Scholar]
- 54.Polisetty N. Fatima A. Madhira S.L. Sangwan V.S. Vemuganti G.K. Mesenchymal cells from limbal stroma of human eye. Mol Vis. 2008;14:431. [PMC free article] [PubMed] [Google Scholar]
- 55.Kellner J.C. Coulombe P.A. Preview. SKPing a hurdle: Sox2 and adult dermal stem cells. Cell Stem Cell. 2009;5:569. doi: 10.1016/j.stem.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Samuel S. Walsh R. Webb J. Robins A. Potten C. Mahida Y.R. Characterization of putative stem cells in isolated human colonic crypt epithelial cells and their interactions with myofibroblasts. Am J Physiol Cell Physiol. 2009;296:C296. doi: 10.1152/ajpcell.00383.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler J.M. Nolan D.J. Vertes E.L. Varnum-Finney B. Kobayashi H. Hooper A.T. Seandel M. Shido K. White I.A. Kobayashi M., et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pellegrini G. Traverso C.E. Franzi A.T. Zingirian M. Cancedda R. De Luca M. Long-term restoration of damaged corneal surface with autologous cultivated corneal epithelium. Lancet. 1997;349:990. doi: 10.1016/S0140-6736(96)11188-0. [DOI] [PubMed] [Google Scholar]
- 59.Schwab I.R. Reyes M. Isseroff R.R. Successful transplantation of bioengineered tissue replacements in patients with ocular surface disease. Cornea. 2000;19:421. doi: 10.1097/00003226-200007000-00003. [DOI] [PubMed] [Google Scholar]
- 60.Tsai R.J.F. Li L.-M. Chen J.-K. Reconstruction of damaged corneas by transplantation of autologous limbal epithelial cells. N Engl J Med. 2000;343:86. doi: 10.1056/NEJM200007133430202. [DOI] [PubMed] [Google Scholar]
- 61.Sangwan V.S. Vemuganti G.K. Iftekhar G. Bansal A.K. Rao G.N. Use of autologous cultured limbal and conjunctival epithelium in a patient with severe bilateral ocular surface disease induced by acid injury: a case report of unique application. Cornea. 2003;22:478. doi: 10.1097/00003226-200307000-00016. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura T. Inatomi T. Sotozono C. Koizumi N. Kinoshita S. Successful primary culture and autologous transplantation of corneal limbal epithelial cells from minimal biopsy for unilateral severe ocular surface disease. Acta Ophthalmol Scand. 2004;82:468. doi: 10.1111/j.1395-3907.2004.00285.x. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura T. Inatomi T. Sotozono C. Ang L.P. Koizumi N. Yokoi N. Kinoshita S. Transplantation of autologous serum-derived cultivated corneal epithelial equivalents for the treatment of severe ocular surface disease. Ophthalmology. 2006;113:1765. doi: 10.1016/j.ophtha.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 64.Shimazaki J. Aiba M. Goto E. Kato N. Shimmura S. Tsubota K. Transplantation of human limbal epithelium cultivated on amniotic membrane for the treatment of severe ocular surface disorders. Ophthalmology. 2002;109:1285. doi: 10.1016/s0161-6420(02)01089-8. [DOI] [PubMed] [Google Scholar]
- 65.De Luca M. Pellegrini G. Green H. Regeneration of squamous epithelia from stem cells of cultured grafts. Regen Med. 2006;1:45. doi: 10.2217/17460751.1.1.45. [DOI] [PubMed] [Google Scholar]
- 66.Rama P. Matuska S. Paganoni G. Spinelli A. De L.M. Pellegrini G. Limbal stem-cell therapy and long-term corneal regeneration. N Engl J Med. 2010;363:147. doi: 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- 67.Li W. Hayashida Y. He H. Kuo C.L. Tseng S.C. The fate of limbal epithelial progenitor cells during explant culture on intact amniotic membrane. Invest Ophthalmol Vis Sci. 2007;48:605. doi: 10.1167/iovs.06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grueterich M. Espana E. Tseng S.C. Connexin 43 expression and proliferation of human limbal epithelium on intact and denuded amniotic membrane. Invest Ophthalmol Vis Sci. 2002;43:63. [PubMed] [Google Scholar]
- 69.Hernandez Galindo E.E. Theiss C. Steuhl K.P. Meller D. Gap junctional communication in microinjected human limbal and peripheral corneal epithelial cells cultured on intact amniotic membrane. Exp Eye Res. 2003;76:303. doi: 10.1016/s0014-4835(02)00314-7. [DOI] [PubMed] [Google Scholar]
- 70.Hernandez Galindo E.E. Theiss C. Steuhl K.P. Meller D. Expression of Delta Np63 in response to phorbol ester in human limbal epithelial cells expanded on intact human amniotic membrane. Invest Ophthalmol Vis Sci. 2003;44:2959. doi: 10.1167/iovs.02-0776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.