Abstract
The signal transducer and activator of transcription-3 (Stat3) is a member of the STAT family of cytoplasmic transcription factors. Overactivation of Stat3 is detected with high frequency in human cancer and is considered a molecular abnormality that supports the tumor phenotype. Despite concerted investigative efforts, the molecular mechanisms leading to the aberrant Stat3 activation and Stat3-mediated transformation and tumorigenesis are still not clearly defined. Recent evidence reveals a crosstalk close relationship between Stat3 signaling and members of the Rho family of small GTPases, including Rac1, Cdc42 and RhoA. Specifically, Rac1, acting in a complex with the MgcRacGAP (male germ cell RacGAP), promotes tyrosine phosphorylation of Stat3 by the IL6-receptor family/Jak kinase complex, as well as its translocation to the nucleus. Studies have further revealed that the mutational activation of Rac1 and Cdc42 results in Stat3 activation, which occurs in part through the upregulation of IL6 family cytokines that in turn stimulates Stat3 through the Jak kinases. Interestingly, evidence also shows that the engagement of cadherins, cell to cell adhesion molecules, specifically induces a striking increase in Rac1 and Cdc42 protein levels and activity, which in turn results in Stat3 activation. In this review we integrate recent findings clarifying the role of the Rho family GTPases in Stat3 activation in the context of malignant progression.
Keywords: Rho GTPases, MgcRacGAP, Stat3
Introduction
Normal or tumor tissues consist of cells which are in constant contact with their neighbors in a three-dimensional structure, and recent findings revealed that cell to cell adhesion may influence fundamental cellular processes such as cell division, differentiation and apoptosis [6,47]. In this context, recent studies have shown that the engagement of cadherins, calcium-dependent cell to cell adhesion molecules, causes a dramatic increase in the levels and activity of the signal transducer and activator of transcription-3 (Stat3), a member of the STAT family of cytoplasmic transcription factors, known to play a key role in a variety of cancers. Despite extensive efforts, the molecular mechanisms of Stat3 activation that lead to tumorigenesis are poorly understood. A family of molecules that are dramatically affected by the engagement of cadherins is the Rho, small GTPases (Rho). It has been demonstrated that mutationally activated forms of the Rac1, Cdc42 or RhoA members of this family, which are known to be important for transformation by oncogenes such as Src and Ras [21,26,45], directly or indirectly promote the phosphorylation and activation of Stat3 [13,17,54]. Recent studies further showed that cadherin engagement causes a dramatic increase in the levels and activity of both mutant and wild-type Rac1 and Cdc42, which in turn leads to Stat3 activation [2,4]. Thus, there is compelling evidence to support the notion that members of the Rho family represent critical sources of signals that promote events leading to Stat3 activation in the context of malignant progression. The Rho family of small GTPases and their role in neoplasia have been recently reviewed [16,21,26,27,45] and will not be discussed here. In this review we summarize the prevailing evidence on the mechanism of Stat3 upregulation and neoplastic transformation following activation of wild-type or mutant members of the Rho family of small GTPases.
Cadherins activate Rho GTPases
Cadherin family of cell to cell adhesion receptors
The formation of cell–cell adhesion junctions is primarily modulated by the calcium-dependent family of cadherin receptors. These plasma membrane glycoproteins control the organization, specificity and dynamics of cell adhesion, which is crucial for the development and maintenance of tissue architecture. Classical, type I cadherins include the epithelial (E)-cadherin and neuronal cadherin, which are found in most tissues [56]. Type I cadherins share low amino acid homology with type II cadherins, e.g., cadherin-11. Classical cadherins consist of three domains, an extracellular, a single-pass transmembrane and an intracellular domain (Fig. 1A). The ectodomain consists of five modules of ~100 amino acids each with internal sequence homology [23]. In the presence of calcium, the extracellular segments expressed on the surface of opposing cells interact to form the cell to cell adherens junctions, which are stabilized by cytoskeletal elements inside the cell, through a homologous carboxy-terminal, cytoplasmic region for binding to β- or γ-catenin proteins, which are linked to actin filaments [6].
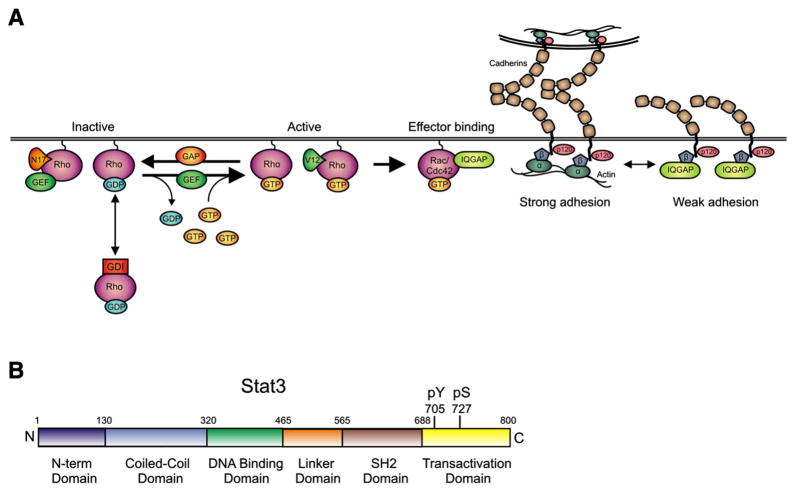
Fig. 1.
Activation of Rho GTPases (A) and Stat3 (B). (A) Rho GTPases are inactive when complexed to GDP and one of three known Rho-GDIs. Upon stimulation by extracellular factors, Rho is released from Rho-GDI and associates with the membrane through its C-terminal prenyl group. Rho-GEFs promote Rho-GTP exchange leading to the activation of various effector proteins. A Rho-GAP will then catalyze GTP hydrolysis and Rho-GDI will extract the GTPase from the membrane locking it once again in an inactive state. A substitution mutation of Thr for Asn at position 17 (Rac1 numbering) allows binding to GEF’s but inhibits interactions with effectors, so that the mutant titrates out GEFs and acts as dominant-negative. On the other hand, mutation to V12 or L61 cannot hydrolyse GTP and is constitutively active. IQGAP, a Rac1 and Cdc42 effector, negatively regulates adhesion by binding to β-catenin which causes β-catenin to dissociate from the β-catenin/α-catenin complex. Activated Rac1 or Cdc42 bind IQGAP and force it to release β-catenin thus strengthening cell to cell adhesion. (B) Structure of the Stat3 transcription factor. The N-terminal, coiled-coil, DNA binding, SH2 and transactivation domains are shown, along with the tyr705 and ser727 phosphorylation sites.
The Rho GTPases
The small GTPases of the Ras (Rat sarcoma) superfamily (Ras, Rho, Arf, Rab and Ran) are ~21 kDa proteins that function as molecular switches in signaling pathways which are initiated by a variety of membrane triggers [21]. The Rho (Ras homologous) proteins are a subfamily of the Ras superfamily which are highly conserved from lower eucaryotes to plants and mammals [7]. They differ from other members of the group by the presence of a Rho-specific insert in the GTPase domain (Rac1 amino acids, 123–135), which has been suggested to be involved in the recognition of downstream effector proteins. Over 20 Rho GTPases are known, and 3 members, RhoA, Rac1 and Cdc42, are ubiquitously expressed, can stimulate cell cycle progression [41] and are crucial for Ras-induced transformation [46], while Rac1 was shown to be required for transformation by oncogenes such as Src [53,62] and Ras [33].
The Rho GTPases are best known as master regulators of the actin cytoskeleton: Rac1 and Cdc42 remodel the actin cytoskeleton at the leading edge of the cell, resulting in filopodial (Cdc42) or lamellipodial (Rac1) protrusions. RhoA, B and C on the other hand are largely responsible for orchestrating focal adhesion assembly and actomyosin-mediated cell contraction at the rear of the cell, thus permitting cell movement across these adhesive contacts and subsequent detachment by the trailing end of the cell [67]. Increases in the levels of Rho family proteins have been observed in a number of cancers [16,26].
Like Ras, most Rho family proteins act as molecular switches cycling between a GTP-bound, active form and a GDP-bound, inactive state [24]. GTP binding induces conformational changes which are localized within two surface loops (switch I and switch II, Rac1 amino acids 25–49 and 59–76, respectively [27]), which play an important role in GTP catalysis (Fig. 1A). The activity of Rho family proteins is regulated by three classes of proteins: guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs). GEFs catalyze the release of GDP from the Rho GTPases, which is the rate-limiting step in Rho activation. The free GTPase then associates with GTP, which alters the conformation of the switch regions of the enzyme to increase its affinity for the effectors (Fig. 1A). To date, over 70 Rho-GEFs have been described in humans. A substitution mutation of Thr for Asn at position 17 (Rac1 numbering) allows binding to GEFs but inhibits interactions with downstream effectors, so that these mutants titrate out the GEFs and act as dominant-negative [18]. In contrast to GEFs, the GAP proteins enhance the inherently low, intrinsic ability of the GTPases to hydrolyse the bound GTP to GDP. Therefore, GAPs promote inactivation and reverse the binding of effectors. There are over 80 mammalian Rho GAP proteins identified so far. Constitutively active mutants (e.g. Leu61 or Val12) cannot hydrolyse GTP; therefore, they signal continuously to their effectors.
The Rho family members invariably have a C-terminal sequence ending with a -CAAX motif. Lipid modification at the C-terminus such as farnesylation, geranylgeranylation or palmitoylation promotes their membrane attachment, where they can be activated by GEFs. The GDI proteins (3 members) are cytosolic proteins that form a complex with GDP-bound Rho GTPases, so that they inhibit attachment to the membrane, hence activation by GEFs. GDIs are also able to associate with the active form and prevent binding to downstream effectors [14]. Therefore, the activation state of a given Rho family GTPase is tightly regulated and occurs in a cell-type and pathway-dependent manner, depending upon the balance of these regulators at any given moment, and this determines its downstream signaling.
Rho family proteins do not usually exert their effects directly, but instead operate through a multitude of effector proteins. The main region of Rho binding to its effectors is switch I, although regions outside switch I have been implicated in the binding of certain effectors. Over 70 effectors have been described [11]. Most Rac1 and Cdc42 effectors contain a conserved, GTPase-binding consensus site (Cdc42/Rac interactive binding or CRIB domain), while many RhoA, B, and C effectors possess an N-terminal Rho effector homology domain (REM). Many effector molecules are serine/threonine kinases. The best characterised Rac1 and Cdc42 effectors are the p21-activated kinases (PAKs [22]), while the Rho-associated coiled-coil domain kinases (ROCK-I and II) represent the best characterised RhoA effectors [49].
It is interesting to note that the small GTPases can regulate each other’s activity via crosstalk. For example, oncogenic Ras needs to activate both Raf and Rac1 to transform [46]; the Tiam1, Rac1 GEF binds the Ras effector domain to activate Rac1 [33]. Similar to Tiam1, Cool-2 is activated by binding the Cdc42 effector domain, to act as a GEF for Rac1, while in a feedback loop, Rac-GTP inhibits the GEF activity of Cool-2 [19]. An additional mechanism whereby Rac1 downregulates Rho activity is through Rac1-mediated production of reactive oxygen species (ROS). ROS inhibit the low-molecular-weight phosphatase (LMwtPTPase) which normally dephosphorylates and inhibits the p190RhoGAP. Consequently, Rac1 activation reduces the activity of RhoA [40].
Cadherin engagement increases Rac1 and Cdc42 protein levels and activity
In addition to providing structure and integrity to the cell, cadherin adhesive engagement initiates intracellular signals that are communicated through the conserved cadherin tail domain to different cytoplasmic pathways. In fact, results from a number of labs indicated that Cdc42 and Rac1 are required for E-cadherin-mediated, cell–cell adhesion in MDCK cells [20], and are believed to contribute to the stability of cell–cell adhesion via their effect on cytoskeletal organization [9]: Expression of activated Rac1val12 in canine epithelial MDCK cells induces the accumulation of E-cadherin, β-catenin and actin filaments at sites of cell–cell contact, whereas overexpression of the dominant-negative, Rac1N17 mutant reduces their accumulation [59]. Rac1 and Cdc42 can regulate E-cadherin activity through their effector, IQGAP. IQGAP is localized to sites of cell–cell contact and negatively regulates adhesion by binding to β-catenin, which causes β-catenin to dissociate from α-catenin [32]. Rac1 and Cdc42 bind IQGAP and remove it from the β-catenin/α-catenin complex. As a result, activated Rac1 inhibits cell dissociation and scattering, and strengthens cell–cell adhesion. Interestingly, in a positive feedback mechanism, E-cadherin engagement also results in the rapid activation of Rac1 and Cdc42, in part through a PI3-kinase-mediated mechanism, while, as expected, it inhibits RhoA, through the production of ROS by Rac1 [25].
In addition to E-cadherin-mediated activation of Rac1 and Cdc42, previous reports revealed another mechanism of Rac1 regulation involving protein stability, namely, degradation through the proteasome pathway [36,44]. In fact, cadherin engagement, e.g., achieved by high cell density, led to a dramatic increase in Rac1/Cdc42 protein levels through inhibition of proteasomal degradation [2,4]. Conversely, epithelial cell scattering brought about by hepatocyte growth factor (HGF) can induce the proteasome-mediated degradation of Rac1 [36]. Moreover, a mutational analysis further indicated that constitutive activation of Rac1, as well as binding of effectors, which might be acting as ubiquitin E3 ligases, are necessary for Rac1 degradation [44]. Still, results by Arulanandam et al. indicated that although permanently activated by mutation, protein levels of Rac1V12 and Rac1L61 are increased dramatically with cell density [2], indicating that cell density can overcome the degradative effect of activation. The effect of cadherin engagement upon Rac1/Cdc42 levels was found to be independent from direct cell to cell contact; plating HC11 mouse breast epithelial cells sparsely on surfaces coated with a fragment encompassing the two outermost domains of E-cadherin caused a dramatic increase in Rac1 protein levels and activity, compared to cells growing on plastic [4], indicating that E-cadherin engagement per se is responsible for the increase in Rac1 and Cdc42 levels. Such a mechanism could hold true for Cdc42, which mirrored Rac1 levels and stability increases with cell density. Similarly, cadherin-11 and N-cadherin were also found to activate Rac1 in different cell lines (Arulanandam et al., in preparation). The above data taken together indicate that cadherin engagement can abolish Rac1/Cdc42 proteasomal degradation, which leads to a dramatic increase in their levels and consequently their activity, even in the absence of direct cell to cell contact.
Rho GTPases activate Stat3
The Stat3 pathway in neoplasia
Stat3 was originally discovered as the acute phase response factor (APRF) that mediates the acute phase response in the liver via the induction of the C-reactive protein [50,71]. Stat3 is activated not only by cytokine receptors, such as the receptor for the interleukin-6 (IL6) family cytokines, but also growth receptor tyrosine kinases, such as the EGFR family including Her2/Neu, and non-receptor tyrosine kinases such as Src and Abl [63], and is also activated in response to stimulation of G-protein-coupled receptors [43]. Classically, the receptor stimulation by ligand induces Stat3 binding to phosphotyrosine residues of activated receptors through its SH2 domain and its phosphorylation on a critical tyr705 residue by the receptor itself, or by associated Janus kinase (JAK, Jak1–3, Tyk2) or Src family tyrosine kinases [68], and the phosphorylation is known to mediate dimerization between two Stat3 monomers through reciprocal SH2 domain–ptyr interactions [68]. However, studies have also identified pre-existing complexes between non-phosphorylated Stat3 monomers [51]. Stat3:Stat3 dimers translocate to the nucleus where they bind to target sequences in specific promoters, although Stat3 monomers have also been detected in the nucleus. Known Stat3 upregulated genes include Bcl-xL, Mcl1, survivin, Akt, vascular endothelial growth factor (VEGF), HGF, myc, cyclinD, and HIF1, while the p53 tumor suppressor is downregulated by Stat3 activity [69].
In contrast to normal Stat3 signaling, which is transient, hyperactive Stat3 is associated with malignant transformation and tumorigenesis. Constitutively active Stat3 is present in a large number of cancers, and studies show that aberrant Stat3 activity promotes tumor cell growth and survival, tumor angiogenesis and metastasis, and induces tumor immune evasion [68]. It was also shown that a constitutively active form of Stat3, Stat3C, is able to transform cultured cells, which further points to an etiologic role for Stat3 in cancer [10]. Of therapeutic importance, disruption of hyperactive Stat3 signaling in tumor xenografts induces tumor cell apoptosis and tumor regression with little effect upon normal tissues [65,68,70], which points to Stat3 as an important player in tumor progression.
Rac1 and MgcRacGAP mediate Stat3 ptyr705 phosphorylation and nuclear transport
Emerging evidence suggests a more complex mechanism for Stat3 activation than initially thought; recent studies indicate that Rac1 plays an important role in Stat3 (as well as Stat5) tyrosine phosphorylation and nuclear translocation:
The male germ cell RacGAP (MgcRacGAP) is an evolutionarily conserved protein which binds directly to and serves as a GAP against Rac1, Rac2 and Cdc42 in vitro [61]. Studies in murine M1 leukemia cells, which differentiate into macrophages upon IL6 stimulation, show that MgcRacGAP can bind through its cysteine-rich and GAP domains to the DNA binding domain of Stat3 (aa 338–362), and that the MgcRacGAP-Stat3 association is required for Stat3, tyr705 phosphorylation following cytokine stimulation. That is, besides having Rac1-GAP activity, the MgcRacGAP, GAP domain is required for IL6-induced, Stat3 ptyr705 phosphorylation, acting as an effector of Rac1. Although MgcRacGAP is constitutively associated with Rac1, the association with Stat3 is increased upon IL6 stimulation [60]. In addition, MgcRacGAP phosphorylation at ser-387 was implicated in transformation by Src, although the exact mechanism is unclear [15].
Following their synthesis in the cytoplasm, transcription factors such as the STAT proteins have to cross the nuclear envelope in order to enter the nucleus. The exact mechanism of Stat3 translocation to the nucleus is just now beginning to emerge. Recent evidence suggests that, besides tyr705 phosphorylation, the Rac1/MgcRacGAP complex may be involved in Stat3 translocation. As a general mechanism for nuclear import, proteins larger than ~50 kDa require specific sequences, the nuclear localisation signals (NLS), or binding to NLS-containing chaperones. The best characterised NLS is the mono- or bi-partite, polybasic NLS, which is usually recognized by a family of proteins, the importin-α carriers. These associate with importin-β which docks the complex to the nuclear pore, so that the complex can migrate into the nucleus. The small GTPase, Ran-GTP, then binds importin-β, and the complex is disassembled inside the nucleus [12]. In the case of Stat1 it was recognized that it must be phosphorylated first on tyr701 and dimerized, in order to reveal a conditional NLS which can associate with importin-α5 [48]. Despite the initial assumption, however, that all STATs need to be phosphorylated to enter the nucleus, Stat3 was found to be nuclear and to shuttle between nucleus and cytoplasm, independent of tyrosine phosphorylation. Nevertheless, phosphorylation is still required to bind to specific DNA target sites [35]. It was further demonstrated that a sequence within the coiled-coil domain of Stat3 (amino acids 150–162) is necessary for nuclear translocation and is critical for the recognition of unphosphorylated as well as phosphorylated Stat3 by importins α3 and α6 [29,30,35]. However, although the 150–162 sequence was found to be indispensable for nuclear translocation of full-length Stat3, substitution of the basic amino acid cluster in this sequence did not hamper nuclear accumulation, indicating that aa 150–162 may only be part of a conformational structure that is required for nuclear import rather than a bona fide NLS [35].
In addition to this mechanism, recent evidence brought forth the role of MgcRacGAP as a nuclear chaperone for translocation of Stat3 (as well as Stat5) to the nucleus. In fact, the ternary complex between ptyrStat3, Rac1-GTP and MgcRacGAP facilitates the association with importin-α/β and nuclear translocation; mutation of the polybasic region of the MgcRacGAP NLS inhibited the nuclear translocation and transcriptional activity of Stat3 and Stat5 [28]. Therefore, Rac1 is emerging as an important regulator of Stat3 phosphorylation, nuclear import and function, acting through the MgcRacGAP/Rac/Stat3 complex (Fig. 2).
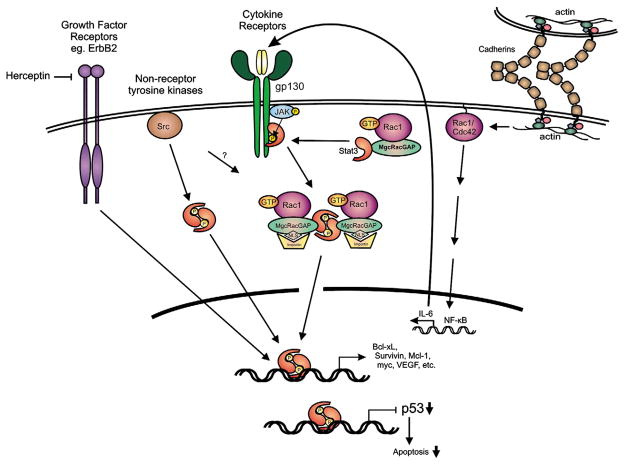
Fig. 2.
Stat3 is activated by growth factor receptors such as Her2/ErbB2 (that can be inhibited by drugs such as Herceptin), non-receptor tyrosine kinases such as Src and cytokine receptors such as the IL6 family. Activated Rac1-GTP, in a complex with MgcRacGAP and Stat3, facilitates Stat3 phosphorylation by the IL6-R/Jak complex, which results in targeting of the complex to the nuclear envelope, driven by the NLS of MgcRacGAP. The Stat3 dimer then binds specific DNA sequences to initiate transcription of Stat3 responsive genes or downregulation of other genes such as the p53 anti-oncogene. MgcRacGAP may also play a role in Stat3 activation by Src. In addition to this mechanism, cadherin engagement was shown to cause a dramatic increase in the levels of Rac1 and Cdc42 proteins and activity, which results in a transcriptional activation of IL6 through NFκB, hence Stat3 activation.
Mutationally activated Rho GTPases activate Stat3 through IL6 secretion
Data from a number of labs have demonstrated a functional role for the Rho GTPases in the activation of Stat3. An earlier study first implicated RhoA in the phosphorylation of Stat3 on ser727 in Src-transformed cells. Moreover, in vitro kinase assays using purified Stat3 as the substrate revealed that both p38 and JNK kinases, which can be activated by the Rho GTPases, phosphorylate Stat3 on ser727, while inhibition of p38 activity suppressed Stat3 activation and vSrc-mediated transformation [62]. In a subsequent report, mutationally activated RhoA was shown to activate Stat3 (but not Stat1) through phosphorylation at both tyr705 and ser727 in human cells, while this Stat3 activation, mediated by the ROCK effector, is required for RhoA-mediated transformation [5]. Further results similarly demonstrated Stat3 activation by mutationally activated forms of Rac1, Cdc42 and RhoA [13,17,54].
Despite extensive efforts, the exact mechanism of Stat3 activation by Rho’s has been a matter of controversy. Simon et al. reported a direct binding between Rac1 and Stat3 in transiently transfected, Cos1 cells, while Rac1N17 inhibited EGF-mediated, Stat3 activation [54]. On the other hand, using neutralising antibodies to inhibit IL6 binding, Faruqi et al. showed that Rac1 activates Stat3 indirectly through autocrine induction of IL-6 [17]. These results were contradicted in a later study where, using Stat3 null cells, it was demonstrated that Stat3 activation by the Rho GTPases could occur without the formation of a stable complex, and in the absence of IL6 secretion [13]. Notably, a recent report indicated that the engagement of E-cadherin, by growing the mouse breast epithelial HC11 cells to high densities, causes a dramatic increase in the levels and activity of Rac/Cdc42 ([4], see next section). Cell density was not taken into account previously, but would significantly impact studies the observations regarding the effect of Rho GTPases upon Stat3 activity. Stat3 ptyr705 levels in HC11 mouse breast epithelial cells over-expressing activated RacV12 (or RacLeu61) or Cdc42V12 mutants were compared to the parental HC11 line, both grown to different densities (from 50% confluence to 5 days post-confluence), and found to be higher at all cell densities, indicating that the activated forms of the GTPases promoted the activation of Stat3, in agreement with previous data [13,17,54]. In a similar experiment, activated RhoAV13 was also found to activate Stat3, although to a lesser extent than Rac1V12 or Cdc42V12 (Arulanandam et al., unpublished). Inhibition experiments indicated that Stat3 activation by RacV12/Cdc42V12 required NFκB and Jak activity [2]. Further examination of the mechanism showed that the addition of medium conditioned by RacV12-expressing cells to the parental line caused an increase in Stat3 activity, suggesting the presence of autocrine factor (s) that in turn promote Stat3 activation. Screening for mRNA of 86 cytokines by RT-PCR in RacV12-expressing cells indicated a significant increase in at least 3 cytokines of the IL6 family compared to untransfected HC11 cells. Furthermore, downregulation of the gp130, common receptor subunit of the family, blocked the induction of Stat3 activity in RacV12-expressing cells, indicating an important role for this family in Stat3 activation [2,4] (Fig. 2). The fact that more than one cytokine of the IL6 family appears to be involved explains the previous results where addition of neutralising antibodies to IL6 alone did not block Stat3 activation by RacV12[13]. The above results taken together demonstrate that activated forms of members of the Rho family can activate Stat3 through nuclear factor (NF)κB, gp130 and JAKs and provides a basis to explain the apparent discrepancy in the potential that Rho family GTPases promote Stat3 activation.
Cadherin engagement activates Stat3 through Rac1/Cdc42 and IL6
The dramatic activation of Rac1/Cdc42 following cadherin engagement [4], coupled with the ability of Rac1/Cdc42 to activate Stat3 [2], leads to the conclusion that cadherin engagement may activate Stat3 through an increase in activity of the Rho GTPases. In fact, results from a number of labs revealed that cell density causes a striking increase in the activity of Stat3 in breast carcinoma [67], melanoma [31], head and neck squamous cell carcinoma [42,55], as well as normal epithelial cells [4,57,58] and fibroblasts ([67], reviewed in [47]). Although tumor cells or cells transformed by Src or other oncogenes had higher Stat3 activity than their normal counterparts when sparse, cell density caused a further Stat3 activation [66,67]. Moreover, growing HC11 mouse breast epithelial cells on surfaces coated with E-cadherin caused a dramatic increase in Stat3 activity, demonstrating that E-cadherin engagement is sufficient to directly activate Stat3, in the absence of cell to cell contact [4]. The density-dependent Stat3 activation was strikingly greater than that brought about by serum or EGF stimulation and was not affected by inhibition or ablation of the cellular Src, Fyn, Yes, Abl, EGFR, Fer or IGF1-R kinases. As expected, this Stat3 activation is triggered by a dramatic increase in the levels of the Rac1 and Cdc42, Rho family GTPases, brought about by inhibition of proteasomal degradation [2], and a corresponding increase in their activity, following E-cadherin engagement [4]. The Rac1/Cdc42 upregulation, in turn, causes a dramatic increase in the expression of a number of cytokines of the IL6 family, which are responsible for the Stat3 activation observed.
Specificity of Stat3 activation
A key question is how and why cadherin engagement would promote the activation of Stat3, but not other pathways, such as Erk1/2. Notably, although IL-6/IL-6R typically activates extracellular-signal activated kinase 1/2 (Erk) in subconfluent cells, post-confluent cultures do not respond with Erk activation upon IL6 stimulation [4]. In the same vein, E-cadherin engagement does not lead to Erk activation. This demonstrates a rather specific Stat3 response of cells to E-cadherin engagement, despite the fact that the two pathways, Erk and Stat3, are often coordinately regulated by oncogenes and growth factor or cytokine receptors including IL-6 [4]. It is conceivable that Erk-specific phosphatases such as Cdc25A [34] may be activated at high densities or that other adaptors required for Erk activation by IL6 might be down-regulated following cadherin engagement. It is interesting to note in this context that cell density also increases Stat5, tyr694 phosphorylation and activity in K562 human chronic myelogenous leukemia cells, although the role of Rho GTPases or cadherins in this is unknown [39].
Phenotypic effects of activated Rho GTPases require Stat3
Rac1 activation was shown to stimulate survival signals through activation of the PI3k/Akt pathway [37]. However, although Rac1/Cdc42 activity is dramatically increased following cadherin engagement, no increase in Akt-473 phosphorylation was seen with cell density in HC11 cells (Arulanandam et al., unpublished results). In sharp contrast, Stat3 is dramatically upregulated at high densities and known to provide a major survival mechanism to cultured epithelial cells and fibroblasts [47,66,67]. Given the propensity of over-confluent cells to undergo growth arrest and apoptosis, it is conceivable that the Stat3 activation serves to provide a survival signal as a last effort to rescue from an impending apoptosis. Interestingly, Stat3 also enhances the resistance of tumor cells to chemotherapeutic agents [8]. The observations that cells are more resistant to inhibition of Src [1] and to chemotherapy when grown to high densities [38] are consistent with the induction of Stat3 signaling at post-confluence, which would provide a survival mechanism.
Stat3 could mediate the proliferative and migration signals provided by activated Rac1, consistent with the known function of Stat3. Thus, expression of activated Rac1V12 or Cdc42V12 mutants in epithelial cells was shown to increase cell migration in a “wound-healing,” cell culture assay, and this process was found to require Stat3 [13] and gp130 [2]. In the same vein, Stat3 is required for the neoplastic conversion of HEK-293T cells expressing activated RhoA to anchorage independence [5].
Conclusions
In summary, while the mechanisms continue to be investigated, there is strong evidence that certain Rho GTPases are important Stat3 activators. (1) Mutational activation of Rac1, RhoA and Cdc42 leads to Stat3 activation. (2) Rac1 and Cdc42 mediate the cadherin signal that leads to activation of Stat3 through NFκB and gp130. (3) Rac1 binds MgcRacGAP and Stat3 and this complex promotes interaction with the IL6 receptor for tyr705 phosphorylation and the activation of the MgcRacGAP - NLS, thereby facilitating nuclear translocation for Stat3-mediated transcription of specific genes (Fig. 2).
The importance of cadherin engagement upon levels and activity of proteins such as Rho and Stat3 is just beginning to emerge. Cadherin engagement was also shown to activate Jak1, which is required for Stat3 activation [67]. In sharp contrast, Src was not required for Stat3 activation following cadherin engagement and its activity was unaffected by cell density [3].
Overall, the potential for the Rho GTPases to promote the prosurvival Stat3 signaling in the context of malignant transformation underscores the importance of cell–cell interactions in the tumor microenvironment in facilitating tumor progression. Due to the generally higher level of expression of E2F transcription factors which are potent apoptosis inducers [52], many tumor cells may have higher requirements for survival signals, such as provided by the cadherin/Rac1/gp130 axis. The increased dependence on Stat3 for survival would explain the increased sensitivity of cells transformed by oncogenes such as Src [64] or the large tumor antigen of simian virus 40 [66] to Stat3 inhibition, a finding which could have significant therapeutic implications.
Acknowledgments
Special thanks are due to Dr. Richard Jove for valuable advice. The financial assistance of the Canadian Institutes of Health Research (CIHR), the Canadian Breast Cancer Foundation (Ontario Chapter), the Natural Sciences and Engineering Research Council of Canada (NSERC), the Ontario Centers of Excellence, the Breast Cancer Action Kingston, the Clare Nelson bequest fund and the Canadian Breast Cancer Research Alliance (L.R.) is gratefully acknowledged. R.A. was supported by a Canada Graduate Scholarships Doctoral award from CIHR, the Ontario Women’s Health Scholars Award from the Ontario Council of Graduate Studies, a Queen’s University Graduate Award and a MITACS Elevate Postdoctoral fellowship. M.G. was supported by a postdoctoral fellowship from the Ministry of Research and Innovation, a postdoctoral award from the province of Ontario, and a US Army breast cancer program. JT was supported by a grant from the National Cancer Institute (CA128865).
Abbreviations
- STATs
signal transducers and activators of transcription
- MgcRacGAP
male germ cell RacGAP
- E-cadherin
epithelial cadherin
- MDCK
Madin–Darby canine kidney cells
- Rho
Ras homologous GTPases, Rho
- Ras
Rat sarcoma
- GEF
guanine nucleotide exchange factor
- GAP
GTPase activating proteins
- GDI
guanine nucleotide dissociation inhibitor
- CRIB
Cdc42/Rac interactive binding domain
- REM
Rho effector homology domain
- PAK
p21-activated kinase
- ROCK
Rho-associated coiled-coil domain kinases
- ROS
reactive oxygen species
- LMwtPTPase
low-molecular-weight phosphatase
- Cool-2
cloned out of library-2
- Tiam-1
T-cell lymphoma invasion and metastasis-1
- PI3-kinase
phosphatidylinositol —3 kinase
- APRF
acute phase response factor
- IL6
interleukin-6
- EGFR
epidermal growth factor receptor
- HGF
hepatocyte growth factor
- VEGF
vascular endothelial growth factor
- ptyr
phosphorylated tyrosine
- NLS
nuclear localisation signal
- Erk
extracellular-signal activated kinase 1/2
- SH2
Src homology 2
- JAKs
Janus kinases
- NFκB
nuclear factor-kappaB
References
- 1.Anagnostopoulou A, Vultur A, Arulanandam R, Cao J, Turkson J, Jove R, Kim JS, Glenn M, Hamilton AD, Raptis L. Differential effects of Stat3 inhibition in sparse vs confluent normal and breast cancer cells. Cancer Lett. 2006;242:120–132. doi: 10.1016/j.canlet.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 2.Arulanandam R, Geletu M, Feracci H, Raptis L. RacV12 requires gp130 for Stat3 activation, cell proliferation and migration. Exp Cell Res. 2010;316:875–886. doi: 10.1016/j.yexcr.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Arulanandam R, Geletu M, Raptis L. The simian virus 40 large tumor antigen activates cSrc and requires Src for full neoplastic transformation. Anticancer Res. 2010;30:47–54. [PubMed] [Google Scholar]
- 4.Arulanandam R, Vultur A, Cao J, Carefoot E, Truesdell P, Elliott B, Larue L, Feracci H, Raptis L. Cadherin–cadherin engagement promotes survival via Rac/Cdc42 and Stat3. Mol Cancer Res. 2009;17:1310–1327. doi: 10.1158/1541-7786.MCR-08-0469. [DOI] [PubMed] [Google Scholar]
- 5.Aznar S, Valeron PF, del Rincon SV, Perez LF, Perona R, Lacal JC. Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in Rho A GTPase oncogenic transformation. Mol Biol Cell. 2001;12:3282–3294. doi: 10.1091/mbc.12.10.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin JM, Nelson WJ. Bench to bedside and back again: molecular mechanisms of alpha-catenin function and roles in tumorigenesis. Semin Cancer Biol. 2008;18:53–64. doi: 10.1016/j.semcancer.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourguignon LY, Peyrollier K, Xia W, Gilad E. Hyaluronan–CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283:17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braga VM, Del Maschio A, Machesky L, DeJana E. Regulation of cadherin function by Rho and Rac: modulation by junction maturation and cellular context. Mol Biol Cell. 1999;10:9–22. doi: 10.1091/mbc.10.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 11.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 13.Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J Biol Chem. 2005;280:17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 14.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Doki N, Kawashima T, Nomura Y, Tsuchiya A, Oneyama C, Akagi T, Nojima Y, Kitamura T. Constitutive phosphorylation of a Rac GAP MgcRacGAP is implicated in v-Src-induced transformation of NIH3T3 cells. Cancer Sci. 2009;100:1675–1679. doi: 10.1111/j.1349-7006.2009.01235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellenbroek SI, Collard JG. Rho GTPases: functions and association with cancer. Clin Exp Metastasis. 2007;24:657–672. doi: 10.1007/s10585-007-9119-1. [DOI] [PubMed] [Google Scholar]
- 17.Faruqi TR, Gomez D, Bustelo XR, Bar-Sagi D, Reich NC. Rac1 mediates STAT3 activation by autocrine IL-6. Proc Natl Acad Sci USA. 2001;98:9014–9019. doi: 10.1073/pnas.161281298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- 19.Feng Q, Baird D, Peng X, Wang J, Ly T, Guan JL, Cerione RA. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat Cell Biol. 2006;8:945–956. doi: 10.1038/ncb1453. [DOI] [PubMed] [Google Scholar]
- 20.Fukata M, Kaibuchi K. Rho-family GTPases in cadherin-mediated cell–cell adhesion. Nat Rev Mol Cell Biol. 2001;2:887–897. doi: 10.1038/35103068. [DOI] [PubMed] [Google Scholar]
- 21.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann C, Shepelev M, Chernoff J. The genetics of Pak. J Cell Sci. 2004;117:4343–4354. doi: 10.1242/jcs.01392. [DOI] [PubMed] [Google Scholar]
- 23.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 24.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 25.Jaffer ZM, Chernoff J. The cross Rho’ds of cell–cell adhesion. J Biol Chem. 2004;279:35123–35126. doi: 10.1074/jbc.R400010200. [DOI] [PubMed] [Google Scholar]
- 26.Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Karnoub AE, Symons M, Campbell SL, Der CJ. Molecular basis for Rho GTPase signaling specificity. Breast Cancer Res Treat. 2004;84:61–71. doi: 10.1023/B:BREA.0000018427.84929.5c. [DOI] [PubMed] [Google Scholar]
- 28.Kawashima T, Bao YC, Minoshima Y, Nomura Y, Hatori T, Hori T, Fukagawa T, Fukada T, Takahashi N, Nosaka T, Inoue M, Sato T, Kukimoto-Niino M, Shirouzu M, Yokoyama S, Kitamura T. A Rac GTPase-activating protein, MgcRacGAP, is a nuclear localizing signal-containing nuclear chaperone in the activation of STAT transcription factors. Mol Cell Biol. 2009;29:1796–1813. doi: 10.1128/MCB.01423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 30.Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreis S, Munz GA, Haan S, Heinrich PC, Behrmann I. Cell density dependent increase of constitutive signal transducers and activators of transcription 3 activity in melanoma cells is mediated by Janus kinases. Mol Cancer Res. 2007;5:1331–1341. doi: 10.1158/1541-7786.MCR-07-0317. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, Shoji I, Matsuura Y, Yonehara S, Kaibuchi K. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell–cell adhesion. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 33.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG, Der CJ. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4:621–625. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 34.Lazo JS, Nemoto K, Pestell KE, Cooley K, Southwick EC, Mitchell DA, Furey W, Gussio R, Zaharevitz DW, Joo B, Wipf P. Identification of a potent and selective pharmacophore for Cdc25 dual specificity phosphatase inhibitors. Mol Pharmacol. 2002;61:720–728. doi: 10.1124/mol.61.4.720. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch EA, Stall J, Schmidt G, Chavrier P, D’Souza-Schorey C. Proteasome-mediated degradation of Rac1-GTP during epithelial cell scattering. Mol Biol Cell. 2006;17:2236–2242. doi: 10.1091/mbc.E05-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murga C, Zohar M, Teramoto H, Gutkind JS. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappaB. Oncogene. 2002;21:207–216. doi: 10.1038/sj.onc.1205036. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T, Kato Y, Fuji H, Horiuchi T, Chiba Y, Tanaka K. E-cadherin-dependent intercellular adhesion enhances chemoresistance. Int J Mol Med. 2003;12:693–700. [PubMed] [Google Scholar]
- 39.Nam S, Williams A, Vultur A, List A, Bhalla K, Smith D, Lee FY, Jove R. Dasatinib (BMS-354825) inhibits Stat5 signaling associated with apoptosis in chronic myelogenous leukemia cells. Mol Cancer Ther. 2007;6:1400–1405. doi: 10.1158/1535-7163.MCT-06-0446. [DOI] [PubMed] [Google Scholar]
- 40.Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol. 2003;5:236–241. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- 41.Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 42.Onishi A, Chen Q, Humtsoe JO, Kramer RH. STAT3 signaling is induced by intercellular adhesion in squamous cell carcinoma cells. Exp Cell Res. 2008;314:377–386. doi: 10.1016/j.yexcr.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelletier S, Duhamel F, Coulombe P, Popoff MR, Meloche S. Rho family GTPases are required for activation of Jak/STAT signaling by G protein-coupled receptors. Mol Cell Biol. 2003;23:1316–1333. doi: 10.1128/MCB.23.4.1316-1333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pop M, Aktories K, Schmidt G. Isotype-specific degradation of Rac activated by the cytotoxic necrotizing factor 1. J Biol Chem. 2004;279:35840–35848. doi: 10.1074/jbc.M404346200. [DOI] [PubMed] [Google Scholar]
- 45.Popoff MR, Geny B. Multifaceted role of Rho, Rac, Cdc42 and Ras in intercellular junctions, lessons from toxins. Biochim Biophys Acta. 2009;1788:797–812. doi: 10.1016/j.bbamem.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 47.Raptis L, Arulanandam R, Vultur A, Geletu M, Chevalier S, Feracci H. Beyond structure, to survival: Stat3 activation by cadherin engagement. Biochem Cell Biol. 2009;87:835–843. doi: 10.1139/o09-061. [DOI] [PubMed] [Google Scholar]
- 48.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 49.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 50.Ripperger JA, Fritz S, Richter K, Hocke GM, Lottspeich F, Fey GH. Transcription factors Stat3 and Stat5b are present in rat liver nuclei late in an acute phase response and bind interleukin-6 response elements. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- 51.Schroder M, Kroeger KM, Volk HD, Eidne KA, Grutz G. Preassociation of nonactivated STAT3 molecules demonstrated in living cells using bioluminescence resonance energy transfer: a new model of STAT activation? J Leukoc Biol. 2004;75:792–797. doi: 10.1189/jlb.1003496. [DOI] [PubMed] [Google Scholar]
- 52.Sears RC, Nevins JR. Signaling networks that link cell proliferation and cell fate. J Biol Chem. 2002;277:11617–11620. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- 53.Servitja JM, Marinissen MJ, Sodhi A, Bustelo XR, Gutkind JS. Rac1 function is required for Src-induced transformation. Evidence of a role for Tiam1 and Vav2 in Rac activation by Src. J Biol Chem. 2003;278:34339–34346. doi: 10.1074/jbc.M302960200. [DOI] [PubMed] [Google Scholar]
- 54.Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran BH, Guan KL. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 55.Steinman RA, Wentzel A, Lu Y, Stehle C, Grandis JR. Activation of Stat3 by cell confluence reveals negative regulation of Stat3 by cdk2. Oncogene. 2003;22:3608–3615. doi: 10.1038/sj.onc.1206523. [DOI] [PubMed] [Google Scholar]
- 56.Stemmler MP. Cadherins in development and cancer. Mol Biosyst. 2008;4:835–850. doi: 10.1039/b719215k. [DOI] [PubMed] [Google Scholar]
- 57.Su HW, Wang SW, Ghishan FK, Kiela PR, Tang MJ. Cell confluency-induced Stat3 activation regulates NHE3 expression by recruiting Sp1 and Sp3 to the proximal NHE3 promoter region during epithelial dome formation. Am J Physiol Cell Physiol. 2009;296:C13–C24. doi: 10.1152/ajpcell.00263.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su HW, Yeh HH, Wang SW, Shen MR, Chen TL, Kiela PR, Ghishan FK, Tang MJ. Cell confluence-induced activation of signal transducer and activator of transcription-3 (Stat3) triggers epithelial dome formation via augmentation of sodium hydrogen exchanger-3 (NHE3) expression. J Biol Chem. 2007;282:9883–9894. doi: 10.1074/jbc.M606754200. [DOI] [PubMed] [Google Scholar]
- 59.Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell–cell adhesion by rac and rho small G proteins in MDCK cells. J Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tonozuka Y, Minoshima Y, Bao YC, Moon Y, Tsubono Y, Hatori T, Nakajima H, Nosaka T, Kawashima T, Kitamura T. A GTPase-activating protein binds STAT3 and is required for IL-6-induced STAT3 activation and for differentiation of a leukemic cell line. Blood. 2004;104:3550–3557. doi: 10.1182/blood-2004-03-1066. [DOI] [PubMed] [Google Scholar]
- 61.Toure A, Dorseuil O, Morin L, Timmons P, Jegou B, Reibel L, Gacon G. MgcRacGAP, a new human GTPase-activating protein for Rac and Cdc42 similar to Drosophila rotundRacGAP gene product, is expressed in male germ cells. J Biol Chem. 1998;273:6019–6023. doi: 10.1074/jbc.273.11.6019. [DOI] [PubMed] [Google Scholar]
- 62.Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, Sekharam M, Frank DA, Holzman LB, Wu J, Sebti S, Jove R. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turkson J, Bowman T, Garcia R, Caldenhoven E, de Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turkson J, Zhang S, Mora LB, Burns A, Sebti S, Jove R. A novel platinum compound that inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J Biol Chem. 2005;280:32979–32988. doi: 10.1074/jbc.M502694200. [DOI] [PubMed] [Google Scholar]
- 65.Turkson J, Zhang S, Palmer J, Kay H, Stanko J, Mora LB, Sebti S, Yu H, Jove R. Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity. Mol Cancer Ther. 2004;3:1533–1542. [PubMed] [Google Scholar]
- 66.Vultur A, Arulanandam R, Turkson J, Niu G, Jove R, Raptis L. Stat3 is required for full neoplastic transformation by the Simian Virus 40 Large Tumor antigen. Mol Biol Cell. 2005;16:3832–3846. doi: 10.1091/mbc.E04-12-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vultur A, Cao J, Arulanandam R, Turkson J, Jove R, Greer P, Craig A, Elliott BE, Raptis L. Cell to cell adhesion modulates Stat3 activity in normal and breast carcinoma cells. Oncogene. 2004;23:2600–2616. doi: 10.1038/sj.onc.1207378. [DOI] [PubMed] [Google Scholar]
- 68.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 69.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang D, Sun M, Samols D, Kushner I. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem. 1996;271:9503–9509. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]