SUMMARY
Under fasting conditions, metazoans maintain energy balance by shifting from glucose to fat burning. In the fasted state, SIRT1 promotes catabolic gene expression by deacetylating the forkhead factor FOXO in response to stress and nutrient deprivation. The mechanisms by which hormonal signals regulate FOXO deacetylation remain unclear, however. We identified a hormone-dependent module, consisting of the Ser/Thr kinase SIK3 and the class IIa deacetylase HDAC4, which regulates FOXO activity in Drosophila. During feeding, HDAC4 is phosphorylated and sequestered in the cytoplasm by SIK3, whose activity is upregulated in response to insulin. SIK3 is inactivated during fasting, leading to the de-phosphorylation and nuclear translocation of HDAC4, and to FOXO deacetylation. SIK3 mutant flies are starvation-sensitive, reflecting FOXO-dependent increases in lipolysis that deplete triglyceride stores; reducing HDAC4 expression restored lipid accumulation. Our results reveal a hormone-regulated pathway that functions in parallel with the nutrient-sensing SIRT1 pathway to maintain energy balance.
INTRODUCTION
Obesity is a major risk factor in the development of insulin resistance, which is characterized by an inability for insulin to promote glucose uptake into muscle and to inhibit glucose production by the liver. Obesity-dependent increases in circulating free fatty acids have been associated with ectopic deposition of lipid in liver and muscle, where they interfere with insulin signaling (Kim et al., 2004).
Because of its short life cycle and ease of genetic manipulation, Drosophila has emerged as an important model organism for the study of obesity and diabetes (Baker and Thummel, 2007). A considerable percentage of all fly genes have clear mammalian orthologs, and over 75% of known human disease genes have functional orthologs in flies (Reiter and Bier, 2002). Indeed virtually all of the known components of the insulin signaling pathway are also present in the fly.
Glucose and lipid homeostasis in Drosophila is maintained by a group of neurosecretory cells in the brain that produce insulin-like peptides (Ikeya et al., 2002). Fasting metabolism is coordinated by a distinct group of cells in the ring gland that elaborate adipokinetic hormone (AKH) (Kim and Rulifson, 2004). Both hormones maintain energy balance through their actions on the fat body, the fly counterpart of mammalian liver and adipose tissue. Disruption of insulin-producing cells in Drosophila leads to increases in circulating glucose levels, mimicking certain features of type II diabetes.
Insulin has been shown to regulate glucose and lipid metabolism by triggering a cascade of lipid kinases that culminate in the activation of the Ser/Thr kinase AKT (Brazil and Hemmings, 2001). In turn, AKT regulates the expression of metabolic programs in part through the phosphorylation and cytoplasmic sequestration of the forkhead transcription factor FOXO (Barthel et al., 2005).
Superimposed on effects of AKT, FOXO activity is also inhibited through acetylation by the histone acetyl transferase paralogs P300 and CBP (Fukuoka et al., 2003; Matsuzaki et al., 2005). Acetylation has been shown to disrupt FOXO activity by reducing its DNA binding affinity, leading to increases in AKT-mediated phosphorylation (Matsuzaki et al., 2005). Conversely, FOXO is activated in part through deacetylation by the NAD+ dependent deacetylase SIRT1 (Brunet et al., 2004; Daitoku et al., 2004; Frescas et al., 2005) in response to nutrient deprivation, although the regulatory effects of feeding and fasting hormones on SIRT1 or other deacetylases have not been addressed.
Here we show that Drosophila SIK3, a member of the AMPK family of Ser/Thr kinases, plays a critical role in energy balance. We found that SIK3 is activated by AKT during feeding, and that upon activation it promotes lipid storage by blocking FOXO activity. We have further shown that SIK3 regulates FOXO activity by modulating its deacetylation by a class IIa HDAC. Taken together, these studies show that SIK3 links insulin signaling to FOXO-dependent changes in triglyceride storage.
RESULTS
SIK3 Promotes Lipid Storage
Within the AMPK family of Ser/Thr kinases, the Salt-inducible kinases (SIKs) have been shown to regulate hepatic glucose metabolism in mammals following their phosphorylation and activation by LKB1, a master kinase for the AMPK family (Dentin et al., 2007; Koo et al., 2005). The SIK kinases are highly conserved through evolution; there are two SIKs encoded by the Drosophila genome: CG4290 (fly SIK2) and CG15072 (fly SIK3) (Okamoto et al., 2004). We have investigated the physiological roles of SIK3 by mutating this gene using P element-mediated imprecise excision (see Experimental Procedures).
We generated a series of deletions affecting the SIK3 gene (Figure 1A). Because SIK3 null mutants (SIK372) die during early larval stages, we used a viable SIK3 hypomorphic allele (SIK348) (Figures 1A–C and S1A–C). SIK348 homozygous mutants showed markedly decreased lipid stores, and they were more sensitive to starvation than control (SIK3+51) flies (Figure 1D–E). By contrast with its effects on fasting, loss of SIK3 only modestly increased sensitivity (15–17%) to oxidative stress (Figure S1F–G). SIK348/SIK372 and SIK348/Df(2R)P34 trans-heterozygotes showed a similar lipid phenotype to SIK348 homozygotes, suggesting that SIK348 is a strong hypomorphic allele (Figure S1D–E). Further supporting this idea, SIK3 mRNA and protein levels are dramatically reduced in SIK348 flies (Figure 1B–C).
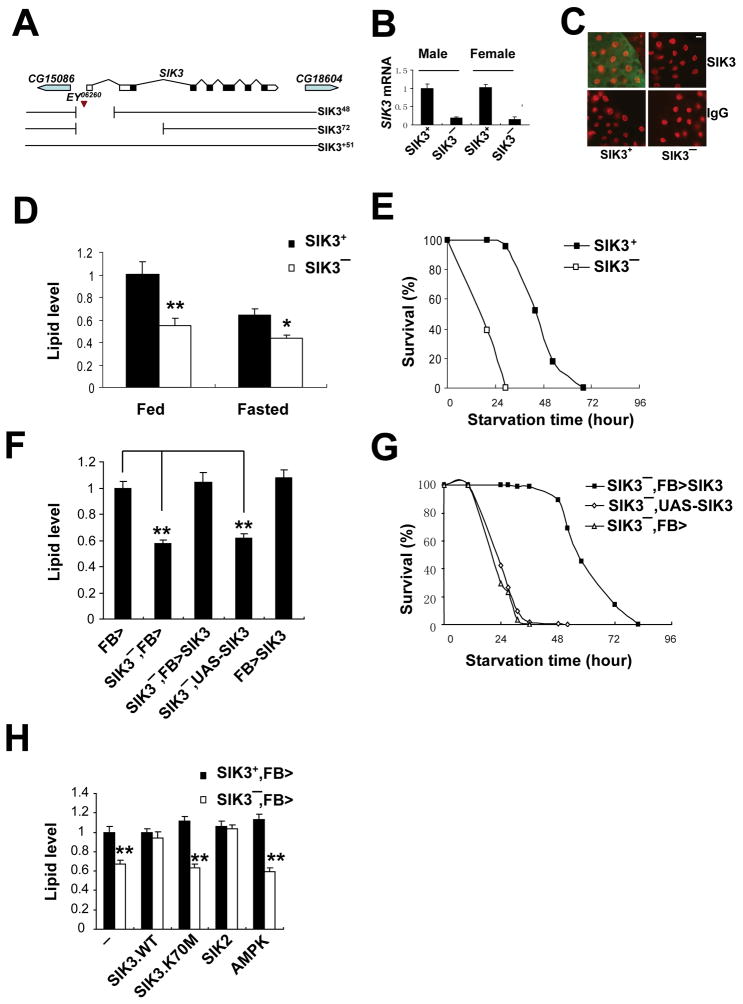
Figure 1. Reduced fat stores in SIK3 mutant flies, also see Figure S1.
(A) Diagram of SIK3 (CG15072) genomic locus is depicted. Location of P element EY06260 shown, along with SIK348 and SIK372 deletions generated by imprecise excision. SIK3+51 is a precise excision control line. SIK348 lacks the first (non-coding) exon, resulting in a significant reduction of SIK3 mRNA. The SIK372 deletion extends through the first two exons, removing sequences that encode part of the SIK3 kinase domain, and acts as a null allele.
(B) Q-PCR analysis of SIK3 mRNA levels in SIK3+ (SIK3+51) and SIK3− (SIK348) adult male and female flies under feeding conditions.
(C) Immunohistochemical analysis of SIK3 expression in fat bodies of WT (yw) and SIK3− (yw;SIK348/Df(2R)P34) L3 larvae. SIK3 protein shown in green, DNA shown in red. Scale bar; 10 μm.
(D, E) Relative fasting (24 hours) or ad libitum feeding lipid levels (D) and starvation sensitivities (E) of SIK3+ (SIK3+51) and SIK3− (SIK348) flies. (n=10 per genotype for lipid measurement and n=50 for starvation assays).
(F) Effect of fat body (FB) specific expression of SIK3 on lipid levels in SIK3− flies. FB>(FB-GAL4/+), SIK3−,FB>(FB-GAL4,SIK348/SIK348), SIK3−, FB>SIK3 (FB-GAL4, SIK348/SIK348;UAS-SIK3/+), SIK3−,UAS-SIK3(SIK348,UAS-SIK3/+) and FB>SIK3 (FB-GAL4/+; UAS-SIK3/+) flies are shown. (n=16 per genotype).
(G) Fat body specific expression of SIK3 rescues starvation sensitivity in SIK3− flies. Shown are survival curves for SIK3−, FB>SIK3 (FB-GAL4, SIK348/SIK348;UAS-SIK3/+), SIK3−, UAS-SIK3 (SIK348, UAS-SIK3/+) and SIK3−, FB> (FB-GAL4, SIK348/SIK348) flies. (n=97-127 per genotype).
(H) Lipid levels in SIK3 mutant flies with fat body specific expression of wild-type or catalytically inactive (K70M) SIK3, SIK2, and AMPK. Genotypes are: SIK3−, FB> (FB-GAL4, SIK348/SIK348), SIK3+, FB> (FB-GAL4/+), SIK3−, FB>SIK3.WT (FB-GAL4, SIK348/SIK348;UAS-SIK3/+), SIK3+, FB>SIK3.WT (FB-GAL4/+;UAS-SIK3/+), SIK3−, FB>SIK3.K70M (FB-GAL4, SIK348/SIK348;UAS-SIK3.K70M/+), SIK3+, FB>SIK3.K70M (FB-GAL4/+;UAS-SIK3.K70M/+), SIK3−, FB>SIK2 (FB-GAL4, SIK348/SIK348;UAS-SIK2/+), SIK3+, FB>SIK2 (FB-GAL4/+;UAS-SIK2/+), SIK3−, FB>AMPK (FB-GAL4, SIK348/SIK348;UAS-AMPKTD/+) and SIK3+, FB>AMPK (FB-GAL4/+;UAS-AMPKTD/+). (n=12 per genotype). Data are averages ± SEM (**, P<0.01; *, P<0.05).
SIK3 shows high levels of expression in the fat body, the fly equivalent of the mammalian liver and adipose tissue. Targeted transgenic expression of wild-type SIK3 in fat body restored lipid stores and starvation resistance and an associated developmental delay in SIK3 mutants (Figures 1F–G and data not shown). Transgenic expression of SIK3 in fat body also rescued the lethality of SIK372 null mutant flies (Figure S1C). By contrast, a kinase-dead form of SIK3 (SIK3.K70M) had no rescuing effect in this setting, indicating that SIK3 catalytic activity is required to maintain lipid balance in vivo (Figure 1H). Indeed, LKB1 phosphorylation-defective SIK3 (SIK3.T196A) also failed to rescue the metabolic phenotype in fat body (data not shown).
Because they share similar substrate specificities, other AMPK family members might be predicted to substitute for SIK3 in fat body. Supporting this idea, expression of SIK2 in fat body fully rescued lipid accumulation (Figure 1H); but AMPK did not, indicating that SIK kinases have distinct regulatory properties from other members of this family.
Insulin Regulates SIK3 Activity
Having found that SIK3 mutant flies are sensitive to starvation, we considered that SIK3 activity may be required for insulin effects on lipid accumulation during feeding. Supporting a genetic link between SIK3 and the insulin signaling pathway, fat body specific over-expression of the insulin-responsive Ser/Thr kinase AKT increased lipid levels in control flies (Verdu et al., 1999), but it had no effect in SIK3 mutants (Figure 2A).
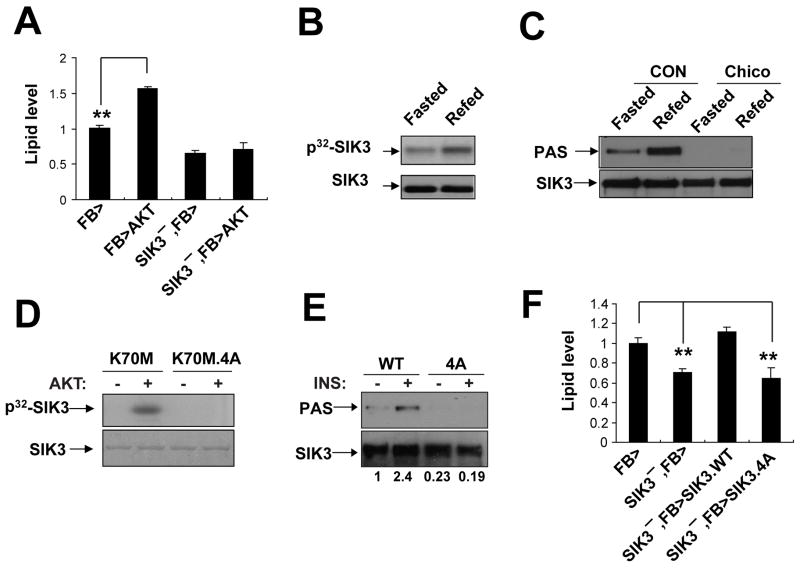
Figure 2. AKT stimulates SIK3 activity during feeding, also see Figure S2.
(A) Effect of fat-body specific AKT expression on lipid levels in wild-type and SIK3− flies. Genotypes are: FB> (FB-GAL4/+), FB>AKT (FB-GAL4/UAS-AKT), SIK3−, FB> (FB-GAL4,SIK348/SIK348) and SIK3−, FB>AKT (FB-GAL4,SIK348/UAS-AKT, SIK348). (n=16 per genotype).
(B) SIK3 catalytic activity in 24 hour fasted versus 0.5 hour refed flies, as determined by relative SIK3 autophosphorylation in fat bodies of flies (ppl-GAL4/UAS-SIK3.WT) expressing HA-tagged SIK3. Incorporation of γ32P-labeled ATP by in-vitro kinase assay of anti-HA immunoprecipitates shown. Immunoblot of total SIK3 protein amounts also indicated.
(C) Immunoblot showing relative amounts of phosphorylated SIK3 in fat bodies of wild-type and Chico mutant flies. Phospho-SIK3 levels determined using phospho-AKT substrate antibody (PAS). Total SIK3 levels measured with SIK3 antisera on HA-immunoprecipitates prepared from CON (ppl-GAL4,UAS-SIK3/+) and Chico (chico1;ppl-GAL4,UAS-SIK3/+) flies under 24 hour fasted or 0.5 hour refed conditions.
(D) In vitro kinase assays showing effect of recombinant AKT on phosphorylation of catalytically inactive (K70M) SIK3. Effect of mutating four putative AKT phosphorylation sites (T281/486A.S293/401A; 4A) in SIK3 on 32P incorporation shown. Total protein amounts for wild-type and 4A mutant SIK3 proteins indicated by Coomassie staining.
(E) Immunoblot showing effect of Ala substitutions at putative AKT phosphorylation sites in SIK3 (4A) on SIK3 phosphorylation in Drosophila S2 cells. Immunoblot with PAS antiserum was performed on HA-immunoprecipitates prepared from transfected cells. Exposure to insulin (0.5 hr) indicated. Total amounts of SIK3 shown. Relative amounts of phospho-SIK3, determined by densitometric analysis, indicated below each lane.
(F) Lipid levels in SIK3 mutant flies following fat body specific expression of wild-type or AKT phosphorylation-defective (4A) SIK3. Genotypes are: FB>(FB-GAL4/+), SIK3−, FB> (FB-GAL4, SIK348/SIK348), SIK3−, FB>SIK3.WT (FB-GAL4, SIK348/SIK348;UAS-SIK3/+) and SIK3−, FB>SIK3.4A (FB-GAL4, SIK348/SIK348;UAS-SIK3.4A/+). (n=16 per genotype). Data are averages ± SEM (**, P<0.01; *, P<0.05).
Because SIK3 appears to be required for lipid accumulation, we tested whether insulin regulates SIK3 activity. Supporting this notion, SIK3 catalytic activity in fat body was elevated during refeeding and decreased in response to fasting by in vitro kinase assay of HA-tagged SIK3 immunoprecipitates prepared from HA-SIK3 transgenic flies (Figure 2B). Moreover, exposure of Drosophila S2 cells to insulin increased the phosphorylation of SIK3 at potential AKT phosphorylation sites, as evaluated using a phospho-AKT substrate antibody (PAS); these effects were diminished when cells were depleted of AKT with a double-stranded AKT oligonucleotide (Figure S2A). Consistent with the proposed role of insulin signaling on SIK3 activity, disruption of chico, the fly homolog of insulin receptor substrate (Bohni et al., 1999), also blocked SIK3 phosphorylation (Figure 2C). Although it had no effect on fat stores by itself, disruption of one chico allele reduced lipid levels in SIK3 hypomorphic mutant flies, demonstrating a genetic interaction between these effectors (Figure S2B).
To identify residues in SIK3 that undergo phosphorylation during feeding, we performed mass spectrometry studies on HA-SIK3 immunoprecipitates from transgenic flies expressing HA-tagged SIK3 in the fat body. This analysis revealed a number of phosphorylation sites, including the LKB1 site at Thr196, and two residues (Ser401, Thr 486) that, along with Thr 281 and Ser293, correspond to consensus (RXRXXS/T) or near-consensus AKT phosphorylation sites in SIK3 and in its mammalian homolog Sik2 (Figure S2D,E). In keeping with these results, recombinant active AKT appeared to phosphorylate each of the four residues in SIK3 in vitro, by mutational analysis of a kinase-dead (K70M) form of SIK3 (Figure S2C). Furthermore, mutant SIK3.4A containing alanine substitutions at all 4 sites, was not phosphorylated by AKT in vitro or in cells exposed to insulin (Figure 2D–E). We next compared the rescuing activities of wild-type and phosphorylation-defective SIK3 in fat body. Although expression of wild-type SIK3 rescued lipid accumulation in SIK3 mutant flies, AKT phosphorylation defective SIK3.4A did not (Figure 2F), indicating that insulin promotes lipid accumulation during feeding in part through the AKT-mediated phosphorylation of SIK3. Although these studies support the notion that AKT is the physiological upstream kinase of SIK3 in fly fat body, we cannot exclude the involvement of other AGC kinases that may regulate SIK3 in a similar fashion.
SIK3 Inhibits Expression of Brummer Lipase
In principle, the loss of SIK3 expression could reduce lipid accumulation by blocking lipogenesis or by increasing lipolysis. Arguing against an effect on lipid absorption or on adipogenesis, high-fat feeding with soy oil rescued the fat storage defects and accompanying developmental delay in SIK3 mutants (Figure S3A and data not shown). Moreover, lipogenic gene expression (SREBP, ACC and FAS) in SIK3 mutants was comparable to wild-type, suggesting that triglyceride synthesis is not altered in SIK3 mutants (Figure S3B).
Under fasting conditions, increases in circulating AKH stimulate the mobilization of triglycerides by triggering the AKH receptor (AKHR) and stimulating cAMP signaling in fat body (Gronke et al., 2007). In parallel, decreases in insulin signaling lead to upregulation of Brummer (bmm) lipase, the fly homolog of adipose triglyceride lipase (ATGL) in mammals (Gronke et al., 2005). To evaluate the regulatory contributions of each pathway, we crossed SIK3 mutant flies to AKHR or bmm mutants.
Deletion of AKHR only partially rescued lipid accumulation in SIK3 mutant flies (Figure 3A), arguing against a direct effect of SIK3 on the AKH pathway per se. By contrast, SIK3/bmm double mutant flies were as obese as bmm single mutants, indicating that bmm likely acts downstream of SIK3. Supporting this notion, levels of bmm mRNA were upregulated in SIK3 mutant flies compared to wild-type (Figure 3B); elevated bmm mRNA levels were reduced following fat body specific expression of wild-type but not kinase-dead (K70M) SIK3. Collectively, these results indicate that SIK3 promotes lipid accumulation during feeding by down-regulating the bmm gene.
Figure 3. SIK3 blocks brummer lipase-dependent lipolysis, see also figure S3.
(A) Relative effect of AKH receptor (AKHR) and brummer (bmm) deficiency on lipid levels in wild-type and SIK3 mutant flies. Genotypes are: SIK3+,w− (w), SIK3−,w− (SIK348), SIK3+, AKHR− (AKHR1), SIK3−, AKHR− (AKHR1, SIK348), SIK3+,bmm− (bmm1), SIK3−,bmm− (SIK348;bmm1). (n=16 per genotype).
(B) Q-PCR analysis of bmm mRNA amounts in wild-type and SIK3 mutant flies under feeding conditions. Effect of expressing wild-type or kinase-dead (K70M) SIK3 in fat body on bmm mRNA amounts indicated. Genotypes are: SIK3+ (FB-GAL4/+), SIK3−: (FB-GAL4, SIK348/SIK348), SIK3+, FB>SIK3.WT (FB-GAL4/+;UAS-SIK3/+), SIK3−, FB>SIK3.WT (FB-GAL4,SIK348/SIK348;UAS-SIK3/+), SIK3+, FB>SIK3.K70M (FB-GAL4/+;UAS-SIK3.K70M/+), SIK3−, FB>SIK3.K70M (FB-GAL4,SIK348/SIK348;UAS-SIK3.K70M/+) flies. Data are averages ± SEM (**, P<0.01).
(C–E) Time course analysis of lipid depletion (C, n=14–20 per genotype), catabolic gene expression (D), and FOXO as well as AKT and SIK3 dephosphorylation (E) during fasting in adult wild-type flies. For panel E, AKT phosphorylated FOXO migrates slower than the unphosphorylated protein (see Figure S4D). Densitometric analysis of phospho-SIK3 amounts from two independent experiments shown.
(F) Q-PCR analysis of mRNA amounts for bmm and other fasting-inducible genes (PEPCK, lip3 and 4E-BP) in wild-type (w) and FOXO (FOXO21/FOXO25) mutant flies under fasting (24 hour) or ad libitum feeding conditions.
SIK3 Modulates FOXO Activity
Under fasting conditions, decreases in insulin signaling have been found to stimulate catabolic gene expression through the dephosphorylation and nuclear entry of the forkhead activator FOXO (Junger et al., 2003). Based on the presence of consensus FOXO binding sites at −56 and −46 in the bmm promoter (Figure S3C), we considered that FOXO may regulate bmm gene expression.
We examined the kinetics of FOXO activation and lipid utilization in fasted flies (Figure 3C–E). Amounts of phosphorylated AKT fell over a 48 hour period after food withdrawal, leading to corresponding decreases in amounts of phosphorylated SIK3. Amounts of dephosphorylated, active FOXO increased in parallel with SIK3 inactivation in response to starvation; these were accompanied by increases in bmm mRNA levels and in lipid disposal. Consistent with a regulatory role for FOXO, fasting-associated increases in bmm gene expression were blocked in FOXO mutant flies (Figure 3F). Conversely, FOXO over-expression enhanced bmm promoter activity in reporter assays; these effects were blocked by mutation of the −46 FOXO binding site on the bmm promoter or by over-expression of SIK3 (Figure S3C–D). FOXO over-expression in fat body was also sufficient to enhance bmm gene expression and to promote lipid depletion (Figure S3E–F).
Having seen that bmm gene expression is elevated in SIK3 mutant flies, we wondered whether SIK3 negatively regulates FOXO activity. Supporting this idea, amounts of nuclear-localized active FOXO were increased 5-fold in ad libitum fed SIK3 mutant larvae compared to controls (Figure 4A–B). As a result, mRNA amounts for other FOXO target genes (4E-BP, PEPCK) were upregulated in SIK3 mutants relative to control flies (Figure S4A–B).
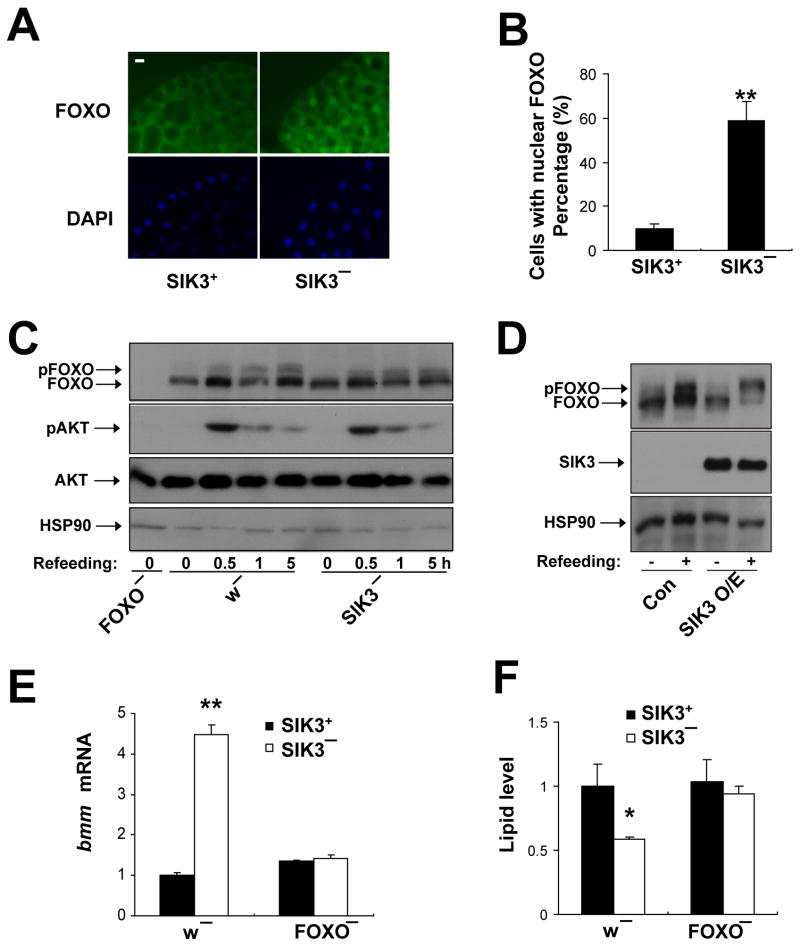
Figure 4. SIK3 inhibits FOXO activity, also see Figure S4.
(A–B) Immunohistochemical analysis and quantification of FOXO localization in SIK3+ (SIK3+51/Df(2R)P34) and SIK3− (SIK348/Df(2R)P34) L3 larvae. Arrows point to nuclear-localized FOXO protein. Scale bar; 10 μm. (B) Percentage of cells with nuclear FOXO staining in (A) shown graphically. At least 400 cells were counted for each genotype.
(C,D) Effect of SIK3 depletion (C) or fat body-specific over-expression (D) on FOXO phosphorylation in flies. Genotypes are: w− (w), SIK3− (SIK348), FOXO− (FOXO21/FOXO25), Con (r4-GAL4,UAS-HAFOXO/+), and SIK3 over-expression (O/E) (r4-GAL4,UAS-HA-FOXO/UAS-SIK3). In panel C, flies were fasted for 24 hours (0) and then refed for 0.5, 1, or 5 hours. In panel D, flies were fasted for 24 hours and refed for 0.5 hours.
(E, F) Effect of FOXO gene disruption on brummer mRNA amounts (E) and lipid levels (F) in SIK3 mutant flies. Genotypes are: SIK3+,w− (w), SIK3−,w− (SIK348), SIK3+,FOXO− (FOXO21/FOXO25), and SIK3−, FOXO− (SIK348; FOXO21/FOXO25). (n=24 per genotype). Data are averages ± SEM (**, P<0.01).
In the fed state, increases in insulin signaling reduce catabolic gene expression through AKT-mediated phosphorylation and cytoplasmic translocation of FOXO (Teleman et al., 2008). In principle, the increase in FOXO activity in SIK3 mutant flies could reflect decreases in either insulin expression or in insulin signaling. Arguing these possibilities, mRNA amounts for insulin like peptide 2 (Ilp2) were similar in wild-type and SIK3 mutant flies (Figure S4C); and refeeding upregulated AKT phosphorylation comparably in both groups (Figure 4C).
By contrast with its effects in wild-type flies, refeeding increased FOXO phosphorylation only modestly in SIK3 mutants (Figures 4C and S4D). Conversely, refeeding stimulated FOXO phosphorylation more robustly in transgenic flies over-expressing SIK3 in fat body relative to controls (Figure 4D). Pointing to a genetic link between SIK3 and FOXO, disruption of the FOXO gene restored both bmm expression and lipid accumulation in SIK3 mutant flies (Figure 4E–F). Taken together, these results suggest that SIK3 inhibits lipolysis during feeding by reducing FOXO activity and thereby lowering bmm gene expression in fat body.
SIK3 Regulates a Class IIa HDAC
We considered that SIK3 could modulate FOXO activity directly through phosphorylation. Although purified SIK3 was capable of phosphorylating recombinant FOXO in vitro, it did so with low stoichiometry and at non-consensus sites (Ser66, Ser531), which, when mutated to alanine, did not appear to enhance FOXO activity (Figure S4E–G). These results suggest that SIK3 likely regulates FOXO activity through an intermediary factor.
To identify this putative regulator, we performed a genetic screen using a set of 270 chromosomal deletions covering 95% of the Drosophila genome. By capitalizing on an ectopic wing vein phenotype caused by wing-specific knockdown of SIK3 function as an easily scorable output for SIK3 activity, we identified one deletion, Df(X)BSC713, which strongly suppressed the loss of SIK3 function in wing (Figure S5A–B). One of the genes deleted in Df(X)BSC713 is HDAC4, which has previously been identified as a substrate for SIK phosphorylation in both C. elegans and mammals (Berdeaux et al., 2007; van der Linden et al., 2006). Supporting a functional link in Drosophila, removal of one copy of HDAC4 (HDAC4e04575) suppressed effects of SIK3 loss of function (LOF) in the wing. Conversely, gain of HDAC4 function (GOF) by over-expression in wing enhanced SIK3 LOF, and it suppressed SIK3 GOF (Figure S5C). Taken together, these results suggest that HDAC4 acts downstream of SIK3 in Drosophila.
Phosphorylation has been shown to sequester class IIa HDACs in the cytoplasm by stimulating their interaction with 14-3-3 proteins (Bassel-Duby and Olson, 2006). Following their de-phosphorylation, class IIa HDACs migrate to the nucleus where they modulate cellular gene expression by associating with relevant transcription factors. Consistent with the presence of three conserved consensus SIK phosphorylation sites known to be phosphorylated by SIKs in C. elegans and mammals (Berdeaux et al., 2007; van der Linden et al., 2006), wild-type fly HDAC4 was efficiently phosphorylated in vitro and in HepG2 and Drosophila S2 cells by SIK3 (Figure 5A–C and S5E). In contrast, SIK3 did not phosphorylate a mutant HDAC4 polypeptide containing alanine substitutions at all three putative Ser phospho-acceptor sites (Figure 5B–C). Indeed, phosphorylation-defective HDAC4 was more active than wild-type in promoting the ectopic wing vein phenotype (Figure S5D). In keeping with the ability of the cAMP dependent kinase PKA to disrupt the activity of the SIKs through phosphorylation at a conserved Ser residue corresponding to Ser563 in SIK3 (Screaton et al., 2004), exposure to cAMP agonist reduced amounts of phosphorylated HDAC4 in cells expressing wild-type SIK3 (Figures 5C and S5F). By contrast, exposure to cAMP agonist actually increased HDAC4 phosphorylation in cells expressing PKA phosphorylation-defective S563A mutant SIK3.
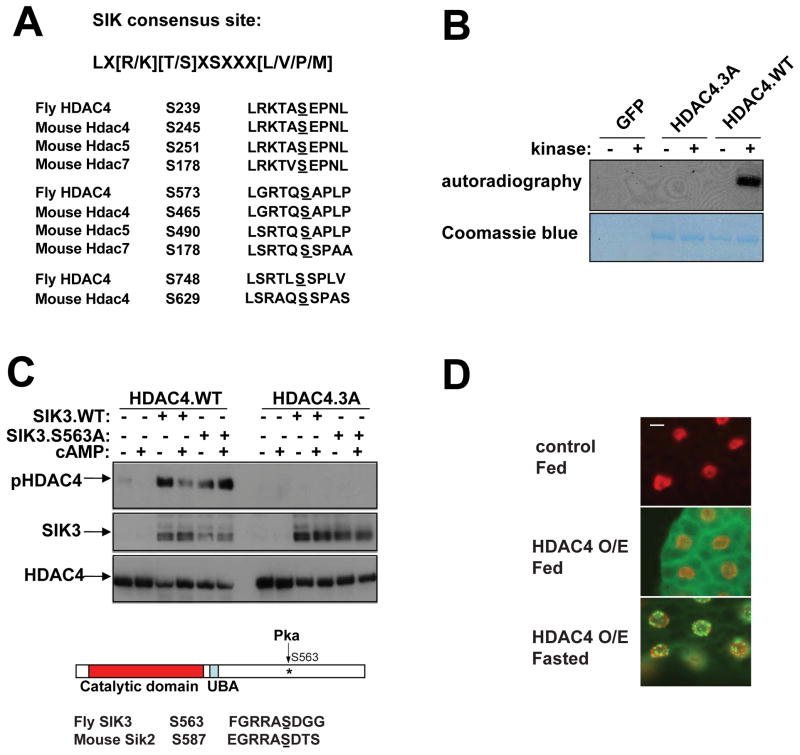
Figure 5. SIK3 phosphorylates HDAC4 and promotes its cytoplasmic translocation, also see Figure S5.
(A) Alignment of SIK consensus phosphorylation motif with conserved phospho-acceptor sites in HDAC4 and in mammalian class IIa HDACs (Hdac4/5/7).
(B) Autoradiogram showing relative phosphorylation of wild-type (HDAC4.WT) and phosphorylation defective (HDAC4.3A) HDAC4 polypeptides by SIK3 in in vitro kinase assays using 32P-labeled ATP. Lower panel shows total protein amounts for HDAC4.WT and HDAC4.3A by Coomassie staining.
(C) Immunoblot showing effect of wild-type and PKA phosphorylation defective SIK3 (S563A) on amounts of phospho (Ser239) and total HDAC4 in transfected HepG2 cells. Effect of cAMP agonists (0.5 mM IBMX, 0.5 mM 8-Br-cAMP; 4 hours) on phospho (Ser239) HDAC4 amounts shown. Bottom, diagram of SIK3 showing location of inhibitory PKA phosphorylation site relative to catalytic and ubiquitin binding (UBA) domains indicated. Below, alignment of PKA phosphorylation sites in Drosophila SIK3 and mouse SIK2.
(D) Immunohistochemical analysis of FLAG-HDAC4 localization in fed control (r4-GAL4/+), and ad libitum fed or 10 hour fasted HDAC4-over-expressing (r4-GAL4/UAS-HDAC4) L3 larvae. HDAC4 protein shown in green, DNA shown in red. Scale bar; 10 μm.
Realizing that HDAC4 phosphorylation promotes its nucleocytoplasmic shuttling, we tested whether HDAC4 subcellular localization is regulated by dietary status. Under feeding conditions, HDAC4 was largely confined to the cytoplasm of larval fat body cells; fasting triggered nuclear shuttling of HDAC4 (Figure 5D). To determine whether SIK3 is sufficient to promote HDAC4 shuttling, we employed Hela cells, which are defective in the AMPK family master kinase LKB1 (Hawley et al., 2003). Consistent with the absence of endogenous SIK-related activities, HDAC4 was primarily nuclear-localized in Hela cells. Over-expression of constitutively active SIK3T196E increased the cytoplasmic localization of HDAC4 (Figure S5G). Taken together, these results indicate that HDAC4 activity in fat body is regulated by phosphorylation or dephosphorylation in response to feeding and fasting signals.
HDAC4 Activates FOXO
Given our finding that SIK3 promotes the translocation of HDAC4 to the cytoplasm, we wondered whether nuclear HDAC4 modulates FOXO activity. Supporting this possibility, HDAC4 was found to associate with FOXO in co-immunoprecipitation studies of Drosophila S2 and Hela cells expressing epitope-tagged HDAC4 and FOXO (Figure 6A). Consistent with its proposed role in regulating HDAC4 localization, over-expression of constitutively active SIK3T.196E disrupted the FOXO:HDAC interaction (Figure S6A). Furthermore, FOXO target gene expression was down-regulated in HDAC4 hypomorphic mutant flies (Figure 6B, top). Conversely, mRNA amounts for PEPCK, bmm, and CPTI were increased in transgenic flies over-expressing wild-type HDAC4 in fat body (Figure S6B) whereas non-FOXO target genes (eg. HSL) were relatively unchanged.
Figure 6. HDAC4 activates FOXO, also see Figure S6.
(A) Immunoblot of FOXO recovered from immunoprecipitates of HDAC4 or control IgG prepared from Drosophila S2 cells. Input amounts of transfected HDAC4 and FOXO shown.
(B) Q-PCR analysis of mRNA amounts for fasting inducible genes (PEPCK, bmm, CPTI) in HDAC4 hypomorphic mutant flies (top) or in flies expressing phosphorylation-defective, constitutively active HDAC4.3A in fat-body (bottom). Genotypes are: FB>(FB-GAL4/+) and FB>HDAC4.3A (FB-GAL4/+;UAS-HDAC4.3A/+) for over-expression assays. For HDAC4 hypomorphic mutant flies, genotypes are HDAC4+ (HDAC4KG09091/+) and HDAC4− (HDAC4KG09091/HDAC4e04575). Effects of HDAC4− (top) examined in 24 hour fasted flies. Effects of HDAC4.3A examined under ad libitum feeding conditions.
(C) Immunoblot showing time course of FOXO phosphorylation and acetylation during refeeding in wild-type (w), SIK3 mutant (SIK348). Ac-FOXO protein detected using anti ac-FOXO1 (K242/245) antibody after immunoprecipitation with anti- acetyl-lysine antibody. Extract from FOXO mutant (FOXO21/FOXO25) flies (FOXO−) included to show specificity of ac-FOXO1 and non-discriminating FOXO antisera. (D) Top: schematic showing domain organization of FOXO, including DNA-binding domain (DBD), nuclear localization sequence (NLS), nuclear export sequence (NES), and trans-activation domain (TA). Middle: alignment of conserved lysine residues in the DBDs of Drosophila FOXO and human FOXO1. Conserved lysine residues that undergo acetylation (Lys 179,182 in Drosophila; Lys 242, 245 in human) shown. Bottom, Immunoblot showing Ac-FOXO, and unphosphorylated (FOXO) or phospho-FOXO (pFOXO) protein amounts in 24 hour fasted or 1 hour refed wild-type (HDAC4+) and HDAC4-mutant (HDAC4−) flies. Relative amounts of acetylated FOXO, determined by densitometric analysis, indicated above each lane.
Genotypes are: HDAC4+ (HDAC4KG09091/+) and HDAC4− (HDAC4KG09091/HDAC4e04575).
(E) In vitro deacetylation assay showing effect of purified HDAC4 on deacetylation of a GST-FOXO (aa.95–195) polypeptide acetylated in vitro with the histone acetyl transferase P300. Immunoblot shows amounts of acetylated FOXO (Ac-FOXO) using anti-acetyl lysine antibody. Input protein amounts for GST-FOXO (aa. 95–195) and HDAC4 proteins determined by Ponceau S staining.
(F) Effect of oral supplementation with class I/II HDAC inhibitor trichostatin A (TSA) in the food (10μM) on fat body lipid levels in SIK3 mutant flies. (n=10 per genotype). Genotypes are: w− (w) and SIK3− (SIK348). Data are averages ± SEM (**, P<0.01; *, P<0.05).
To test whether HDAC4 regulates FOXO target gene expression in the fat body itself, we measured bmm mRNA levels in fat bodies isolated from early L3 larvae expressing a constitutively active HDAC4.3A transgene in fat body. bmm mRNA amounts were elevated in fat bodies from transgenic compared to control larvae, although the extent of bmm upregulation in larval fat bodies was more modest than in adult flies (Figure S6C).
Arguing against a general effect of this deacetylase on cellular gene transcription, GAL4 driven expression of a GFP transgene was unaffected by HDAC4 over-expression (Figure S6D). Pointing to the importance of nuclear HDAC4, phosphorylation-defective HDAC4 was more active than wild-type HDAC4 in stimulating FOXO target gene expression in fat body (Figure 6B, bottom). Consistent with the substantial upregulation of bmm in adult flies, HDAC4.3A transgenic flies also had lower lipid stores, and they were more sensitive to starvation compared to controls (Figure S6E–F). Taken together, these results indicate that HDAC4 is both necessary and sufficient to activate FOXO.
Under feeding conditions, P300 and CBP have been shown to inhibit catabolic gene expression by acetylating FOXO1 at conserved lysine residues within the forkhead domain (Daitoku et al., 2004). Acetylation appears to reduce FOXO activity by reducing its DNA binding affinity and thereby enhancing its phosphorylation by AKT (Frescas et al., 2005; Matsuzaki et al., 2005). Conversely, stress and nutrient deprivation have been found to increase FOXO activity during starvation through deacetylation by the NAD+ dependent deacetylase SirT1 and by trichostatin A-sensitive class I/II histone deacetylases (Brunet et al., 2004; Matsuzaki et al., 2005).
In parallel with its stimulatory effects on FOXO phosphorylation, refeeding also enhanced FOXO acetylation in wild-type flies, but not in SIK3 mutants (Figure 6C). By contrast, amounts of acetylated FOXO were substantially increased in HDAC4 hypomorphic mutants after 24 hours of fasting, and they remained elevated during refeeding (Figure 6D).
In principle, HDAC4 could reduce amounts of acetylated FOXO by blocking its association with P300/CBP or by catalyzing its deacetylation. Arguing for the latter scenario, purified HDAC4 was capable of deacetylating FOXO, which had been acetylated in vitro by the HAT coactivator P300 (Figure S6G and 6E). Superimposed on its direct effects, HDAC4 may also regulate FOXO activity through an association with HDAC complexes. In mass spectrometry studies to identify HDAC4-associated proteins in fat body, we recovered SMRTER, EBI, and HDAC3, all components of a deacetylase complex that have been shown to interact with class IIa HDACs in mammalian cells (Fischle et al., 2002) (Figure S6H). Consistent with the ability of HDAC4 to promote FOXO deacetylation, either directly or through its association with the SMRTER complex, addition of the Class I/II HDAC inhibitor trichostatin A (TSA) to the food increased lipid levels in SIK3 mutant flies (Figure 6F).
SIK3 Inhibits HDAC4-Mediated Deacetylation of FOXO
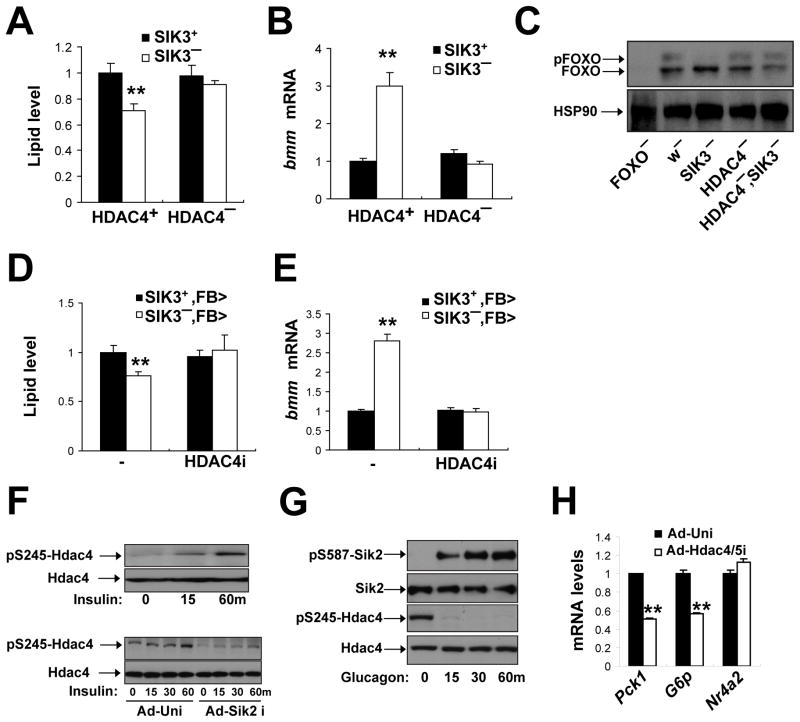
Having observed that HDAC4 regulates FOXO activity, we tested whether SIK3 modulates FOXO target gene expression via an HDAC4-dependent mechanism. Disrupting HDAC4 expression, either through P-element mediated insertion (Figure 7A–C) or through RNAi-mediated depletion in fat body (Figure 7D–E), restored lipid levels in SIK3 mutant flies. Consistent with the increase in fat stores, bmm gene expression was also reduced to wild-type levels in HDAC4/SIK3 double-mutants. In keeping with the proposed role of FOXO in this process, amounts of phosphorylated, inactive FOXO were increased in HDAC4/SIK3 double mutants under refed conditions (Figure 7C). Taken together, these results suggest that SIK3 inhibits lipolysis during feeding by phosphorylating HDAC4 and blocking the deacetylation of FOXO.
Figure 7. SIK3 inhibits FOXO activity via an HDAC4-dependent mechanism.
(A–C) Effect of HDAC4 gene disruption on lipid levels (A) bmm mRNA amounts (B), and FOXO phosphorylation (C) in SIK3 mutant relative to control flies. Data panels A and B is from ad libitum fed flies. Data in panel C was from 1 hour refed flies. Genotypes are: SIK3+(w), SIK3− (SIK348), SIK3+, HDAC4− (HDAC4KG09091/HDAC4e04575), and SIK3−, HDAC4− (HDAC4KG09091/HDAC4e04575; SIK348), FOXO− (FOXO21/FOXO25). (n=24 per genotype).
(D, E) Effect of fat body-specific depletion of HDAC4 (HDAC4i) on lipid levels (D) and bmm mRNA amounts (E) in SIK3 mutant relative to control flies under feeding conditions. Genotypes are: SIK3+,FB>(FB-GAL4/+), SIK3−,FB> (FB-GAL4,SIK348/SIK348), SIK3+,FB>HDAC4i(FB-GAL4/+;UAS-HDAC4 RNAi/+), and SIK3−, FB>HDAC4i (FB-GAL4,SIK348/SIK348; UAS-HDAC4 RNAi/+). (n=14–16 per genotype).
(F) Immunoblot showing amounts of phosphorylated mouse HDAC4 in extracts from primary hepatocytes following RNAi-mediated depletion of SIK2. Effect of exposure to insulin (100nM) for different times (in minutes) indicated.
(G) Immunoblot showing effect of glucagon (100nM) on HDAC4 phosphorylation at Ser245 and on inhibitory phosphorylation of SIK2 at Ser587 in primary mouse hepatocytes.
(H) Q-PCR analysis of PEPCK, Glucose 6 phosphatase, and NR4A2 mRNA amounts in primary hepatocytes following exposure to glucagon (100nM) for 1 hour. Effect of adenovirus encoded HDAC4/5 RNAis shown.
Based on the proposed role of SIK3 and HDAC4 in fasting metabolism, we tested whether this regulatory module in also conserved in mammals. Exposure to insulin triggered the phosphorylation of mammalian HDAC4 at Ser245 in wild-type mouse hepatocytes; these effects were disrupted following RNAi-mediated depletion of SIK2, the mammalian homolog of fly SIK3, which is expressed at highest levels in adipose and liver (Dentin et al., 2007; Horike et al., 2003) (Figure 7F and S7A). Conversely, exposure of hepatocytes to glucagon promoted the de-phosphorylation of HDAC4 at Ser245, reflecting the inhibitory PKA-mediated phosphorylation of SIK2 at Ser587 (Figures 7G and S7A).
Having seen effects of insulin and glucagon on phosphorylation of a mammalian class IIa HDAC, we tested whether fasting or feeding signals also promote shuttling of these deacetylases. Exposure of HepG2 hepatocytes to insulin promoted cytoplasmic translocation of a mammalian HDAC5-GFP fusion protein (Figure S7B). Conversely, exposure to inhibitors of SIK activity, such as PI3K inhibitor (LY294002) or FSK, stimulated the nuclear translocation of HDAC5-GFP (Figure S7B).
We examined whether mammalian class IIa HDACs also regulate FOXO activity. Similar to Drosophila, endogenous mouse HDAC4 was found to associate with endogenous Foxo1 in primary hepatocytes by co-immunoprecipitation assay (Figure S7C). Inhibiting class IIa HDACs, either by treatment with trichostatin A or by RNAi-mediated depletion of HDAC4 and HDAC5, disrupted the induction of gluconeogenic FOXO target genes (Pck1, G6p) in response to glucagon (Figures 7H and S7D–E). Collectively, these studies demonstrate that the SIK/class IIa HDAC module is conserved in mammals, where it promotes catabolic gene expression through the upregulation of FOXO activity (Figure S7F).
DISCUSSION
During fasting, metazoans maintain energy balance through increases in triglyceride lipolysis by Hormone Sensitive Lipase (HSL) and ATGL in response to hormonal signals. This process is reversed during feeding, when increases in insulin signaling promote fat storage in part through the inhibition of both lipases. Although insulin has been shown to down-regulate HSL activity by upregulating phosphodiesterases that terminate cAMP signaling (Kitamura et al., 1999), the mechanism by which it inhibits ATGL expression is unknown.
We have shown that SIK3 is activated by AKT during feeding, when it promotes lipid storage in adult flies by inhibiting FOXO activity and thereby reducing the expression of brummer lipase, the fly homolog of ATGL. Although insulin promotes fat storage in mammals, loss of insulin signaling in chico null flies actually leads to an increase in lipid stores. Loss of chico is thought to promote broad changes in intracellular signaling in multiple tissues (Bohni et al., 1999; Clancy et al., 2001); these changes likely compensate for loss of SIK3 activity in the fat body. Nevertheless, SIK3 activity is critical for lipid storage in a wild-type background.
SIK3 did not appear to inhibit FOXO activity directly through phosphorylation, but rather by blocking its deacetylation by HDAC4. We found that class IIa HDACs are insulin and cAMP regulated deacetylases that promote FOXO activation and catabolic gene expression during fasting (Figure S7F); they are inhibited through SIK-mediated phosphorylation and cytoplasmic translocation. In addition to SIK3, other Ser/Thr kinases including calmodulin-dependent kinases and other members of the AMPK family may also regulate class IIa HDAC shuttling through phosphorylation at the same sites under different conditions.
Fasting hormones disrupt SIK activity through PKA-mediated phosphorylation, leading to the de-phosphorylation and nuclear translocation of class IIa HDACs. The subsequent upregulation of FOXO activity by class IIa HDACs was unexpected because these enzymes are thought to function in muscle and brain as signal-dependent repressors of gene expression (Haberland et al., 2009).
Acetylation has been proposed to modulate FOXO activity by decreasing its DNA binding affinity and thereby enhancing its phosphorylation by AKT (Brent et al., 2008; Matsuzaki et al., 2005). Consistent with this view, we found that the acetylation and phosphorylation of FOXO increased contemporaneously during refeeding; both FOXO modifications were decreased in SIK3 mutant animals, reflecting the upregulation of HDAC4 activity. Indeed, disruption of HDAC4 restored FOXO phosphorylation and target gene expression in SIK3 mutant flies. Although fat body-specific rescue experiments demonstrate a SIK3 requirement in the fat body and argue for cell autonomous effects of HDAC4 on FOXO target gene expression, we cannot rule out additional non-cell autonomous effects of SIK3/HDAC4 activity. For example, FOXO target gene induction in response to fat body-specific HDAC4 overexpression, as detected in whole fly RNA, could additionally be occurring in tissues other than the fat body.
Supporting a substantial role for these enzymes in glucose and lipid metabolism, the effects of SIK3 and HDAC4 in Drosophila appear to be conserved in mammals. We found that Sik2, the mouse homolog of fly SIK3, mediates the phosphorylation and inactivation of Hdac4 in mouse hepatocytes in response to insulin. Conversely, exposure to glucagon increased Hdac4 activity through the PKA-mediated inhibition of Sik2. In an accompanying paper, Mihaylova and colleagues (Mihaylova et al., 2011) also show that mammalian class IIa HDACs are fasting regulated deacetylases, which increase FOXO target gene expression in mice.
Consistent with our results in Drosophila, mutations in the HDAC4 gene that result in haploinsufficiency have been associated with obesity in humans, although the underlying mechanism remains unclear (Williams et al.).
Taken together, these results suggest that the class IIa HDACs and SIRT1 represent parallel pathways, which regulate fasting programs following their activation by hormonal and nutrient signals, respectively. In this regard, it is interesting to speculate that mammalian class IIa HDACs may trigger catabolic gene expression during early fasting, when circulating concentrations of glucagon and catecholamines are typically high, while SIRT1 may function primarily at later “protein-sparing” stages of fasting, when cellular energy levels are low. Future studies should provide additional insight into the roles of SIRT1 and class IIa HDACs in the fasting adaptation.
EXPERIMENTAL PROCEDURES
Fly strains
P element insertion EY06260 (Bloomington #15962) was used to generate SIK3 deletions by imprecise excision. Two SIK3 deletion alleles were characterized: SIK348 is a hypomorphic allele that removes the first exon, resulting in a significant reduction of SIK3 mRNA; SIK372 is a null allele that deletes the first two exons, removing part of the kinase domain. Both SIK372 homozygotes and SIK372/Df(2R)P34 are lethal in larval stages. The deletion endpoints for each SIK3 mutant were determined by PCR and DNA sequencing. Flies were maintained on standard fly food at 25°C with light-dark cycle (Percival Incubator, Model: 136VL). 3–5 day old flies were used in each experiment. For refeeding studies, flies were fasted for 24 hours and then shifted to standard fly food for times indicated. For high-fat or trichostatin A feeding studies, 5% soy oil (Sigma #S7381) or 10 uM trichostatin A (Sigma # T1952) was mixed with the standard fly food.
Lipid measurement
TAG measurement was performed as described (Palanker et al., 2009). Samples were assayed using a Modulus microplate spectrophotometer at 560nm. Lipid levels were normalized to protein amounts in each homogenate using a Bradford assay (Bio-Rad). Measurements were performed in ad libitum fed adult flies unless otherwise indicated.
Starvation assay
3- to 5-day-old flies were transferred to vials of 1% agar/PBS with filter papers soaked with H2O, and dead flies were scored every 4–8 hours. For protein, RNA, and lipid analysis, flies are fasted for about 24 hours unless otherwise indicated.
Cell culture and transfection
Drosophila S2, HEK293T, and HepG2 cells were maintained as previously described (Wang et al., 2008). Hela cells were cultured in DMEM (Mediatech, #10-017-CV) containing 10% FBS at 37°C with 5% CO2. Transfections were performed as reported (Wang et al., 2008). Human GFP-HDAC5 was employed for cell localization studies as described (Berdeaux et al., 2007). Primary hepatocytes were cultured as described (Dentin et al., 2007). Ad-Hdac4/5i viruses (Mihaylova et al., 2011) and Ad-Sik2i virus (Dentin et al., 2007) were generated as reported. 10 p.f.u./cell of adenovirally encoded Ad-Uni, Ad-Sik2i, Ad-Hdac4i and Ad-Hdac5i were used for RNAi-mediated knockdown experiments. After 2–3 days infection, cells were collected for protein or RNA analysis.
Western blot
Cells or flies were lysed on ice in lysis-buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 6 mM EGTA, 20 mM NaF, 1% TritonX-100, and protease inhibitors) for 15–20 min. After centrifugation at 13000 rpm for 15 min, supernatants were reserved for protein determinations and SDS-PAGE analysis. The following antibodies were used: anti-phospho-AKT substrate, anti-AKT, anti-Foxo1, anti-pS239-HDAC4, anti-Ac-lysine, anti-ACC, and anti-phosphor-PKA substrate antibodies are from Cell Signaling Technology. Anti-Hsp90, anti-HA and anti-HDAC4 antibodies are from Santa Cruz Biotechnology, anti-FLAG antibody is from Sigma, and anti-SIK2 antibody is from Abgent. Drosophila FOXO antibodies are from O. Puig, and anti-Ac-FOXO1K242/245 antibody is from A. Fukamizu. Anti-SIK3 (Drosophila) rabbit polyclonal antiserum was prepared and affinity purified as described (Wagner et al., 2000) using recombinant GST-SIK3 (aa. 381–695) as immunogen. Densitometric analysis is carried out using Scion Image software.
Statistical analyses
Statistical significance was determined using an unpaired two-tailed Student’s t test with unequal variance. Data are presented as the average ± SEM.
Supplementary Material
Acknowledgments
This study was supported by NIH grants (R01-DK049777, R01-DK083834 to MM), (R01-DK077979 to JBT), (R01 DK080425 and P01CA120964 to RJS), an American Diabetes Association Junior Faculty Award (1-08-JF-47 to RJS), The Keickhefer Foundation, The Clayton Foundation for Medical Research, and The Helmsley Foundation. We thank Y. Imai and A. Fukamizu (Tsukuba) for gift of antisera. We also thank E. Hafen, J. Chung, R. Kulnlein, J. Scott, J. Park, Bloomington Stock Center, VDRC, Exelixis, and TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for fly stocks used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Berdeaux R, Goebel N, Banaszynski L, Takemori H, Wandless T, Shelton GD, Montminy M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Brazil DP, Hemmings BA. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem Sci. 2001;26:657–664. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008;16:1407–1416. doi: 10.1016/j.str.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, Fukamizu A. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med. 2003;12:503–508. [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike N, Takemori H, Katoh Y, Doi J, Min L, Asano T, Sun XJ, Yamamoto H, Kasayama S, Muraoka M, et al. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J Biol Chem. 2003;278:18440–18447. doi: 10.1074/jbc.M211770200. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Fillmore JJ, Sunshine MJ, Albrecht B, Higashimori T, Kim DW, Liu ZX, Soos TJ, Cline GW, O’Brien WR, et al. PKC-theta knockout mice are protected from fat-induced insulin resistance. J Clin Invest. 2004;114:823–827. doi: 10.1172/JCI22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Kitamura Y, Kuroda S, Hino Y, Ando M, Kotani K, Konishi H, Matsuzaki H, Kikkawa U, Ogawa W, Kasuga M. Insulin-induced phosphorylation and activation of cyclic nucleotide phosphodiesterase 3B by the serine-threonine kinase Akt. Mol Cell Biol. 1999;19:6286–6296. doi: 10.1128/mcb.19.9.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova M, Vasquez D, Ravnskjaer K, Denechaud P, Yu R, Alvarez J, Downes M, Evans RM, Montminy M, Shaw R. Class IIa Histone Deacetylases are Hormone-activated regulators of FOXO and Mammalian Glucose Homeostasis. Cell. 2011 doi: 10.1016/j.cell.2011.03.043. In the Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Takemori H, Katoh Y. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol Metab. 2004;15:21–26. doi: 10.1016/j.tem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Bier E. Using Drosophila melanogaster to uncover human disease gene function and potential drug target proteins. Expert Opin Ther Targets. 2002;6:387–399. doi: 10.1517/14728222.6.3.387. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- van der Linden AM, Nolan KM, Sengupta P. KIN-29 SIK regulates chemoreceptor gene expression via an MEF2 transcription factor and a class II HDAC. Embo J. 2006 doi: 10.1038/sj.emboj.7601479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- Wagner B, Bauer A, Schutz G, Montminy M. Stimulus-specific interaction between activator-coactivator cognates revealed with a novel complex-specific antiserum. J Biol Chem. 2000;275:8263–8266. doi: 10.1074/jbc.275.12.8263. [DOI] [PubMed] [Google Scholar]
- Wang B, Goode J, Best J, Meltzer J, Schilman PE, Chen J, Garza D, Thomas JB, Montminy M. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008;7:434–444. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Aldred MA, Der Kaloustian VM, Halal F, Gowans G, McLeod DR, Zondag S, Toriello HV, Magenis RE, Elsea SH. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.