Abstract
We demonstrate a systematic and rational approach to create a library of natural and modified, dialkylated amino acids based upon arginine for development of an efficient small interfering RNA (siRNA) delivery system. These amino acids, designated DiLA2 compounds, in conjunction with other components, demonstrate unique properties for assembly into monodisperse, 100-nm small liposomal particles containing siRNA. We show that DiLA2-based liposomes undergo a pH-dependent phase transition to an inverted hexagonal phase facilitating efficient siRNA release from endosomes to the cytosol. Using an arginine-based DiLA2, cationic liposomes were prepared that provide high in vivo siRNA delivery efficiency and are well-tolerated in both cell and animal models. DiLA2-based liposomes demonstrate a linear dose–response with an ED50 of 0.1 mg/kg against liver-specific target genes in BALB/c mice.
Introduction
A robust, well-tolerated delivery system is critical to establishing small interfering RNA (siRNA)-based therapeutics. Cationic lipid delivery agents have played a dominant role in systemic delivery of nucleic acids due to their ability to be formulated into small liposomes. Historically, the major cationic lipids have been DOTAP and DOTMA, which have a quaternary ammonium head group attached via ester or ether linkages, respectively, to dioleyl chains.1,2,3 These cationic lipids have been formulated with cholesterol and PEGylated lipids into bilayer liposomal structures.
Typically, these lipids have resulted in relatively modest in vitro and in vivo activity.4,5 Recently, cationic lipids have been designed around the DOTMA motif whereby the alkyl chain unsaturation was increased from one to two double bonds and the quaternary amine head group was replaced by a tertiary amine to enable pH-dependent ionization in a biologically relevant pH range.6 Utilizing this approach, MacLachlan et al. demonstrated effective knockdown of several liver-based targets using stable nucleic acid lipid particles or SNALP.7 Additionally, several different classes of cationic lipids have been created from polymer libraries in a combinatorial fashion to allow for wide structural diversity.8,9 These so-called lipidoid libraries have similarities with lipids, but have various degrees of branching and dendritic-like structures, and exhibit a variety of primary, secondary, and tertiary amines which enable broad pH-dependent ionization behavior.
We describe here an approach that leverages the chemical diversity of amino acids to create dialkylated amino acids, termed DiLA2, as the basis of a new liposomal siRNA delivery system. We hypothesized that amino acids could serve as core structures for the design of lipophilic molecules which would readily form liposomal structures. In particular, the α-amino and α-carboxyl groups offer sites for the attachment of aliphatic chains of various lengths, types, and geometries. In this arrangement, the side chains of the individual amino acids function as the head group defining the charge of the DiLA2 and subsequently the surface characteristics of the liposomal particle. The 20 common amino acids contain sufficient chemical diversity through their side chains to mediate various biological recognition events. Moreover, additional chemical diversity is achieved through the use of non-natural synthetic amino acids that are easily synthesized or readily available commercially. This approach results in a myriad of potential head groups including positively charged (amino, guanido, imidazole, pyridyl), neutral (hydroxyl, thiol), and negatively charged (carboxyl) moieties.
We focused on arginine as a key amino acid for DiLA2 development. The guanidinium head group is known to interact strongly with negatively charged proteoglycans that facilitate binding to cell surfaces and enables uptake into endocytic vesicles.10,11,12 Additionally, the guanidinium head group binds strongly to phosphate groups resulting in tight association between the cationic molecule and the siRNA, resulting in highly encapsulated particles.13 In this report, we describe the process to identify and select a lead arginine-based DiLA2 and its development into an efficacious in vivo formulation utilizing cholesteryl hemisuccinate (CHEMS) as a helper lipid. Potent knockdown of several liver-specific targets including factor VII, apolipoprotein B100 (ApoB), and Transthyretin (TTR) genes are described.
Results
Selection of norarginine
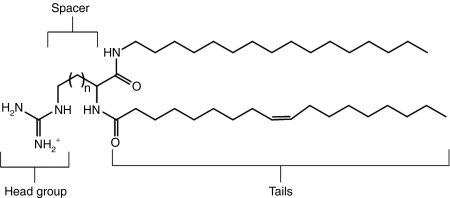
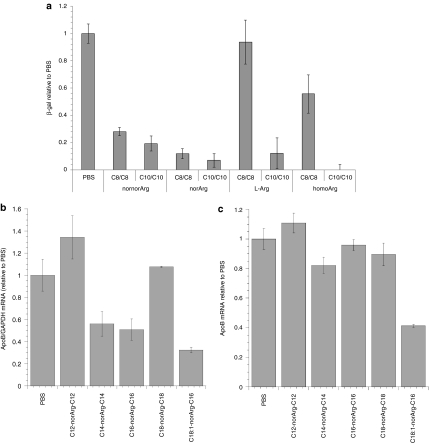
DiLA2 architecture utilizes side chains of amino acids as the head groups, whereas the α-amino and α-carboxyl groups serve as attachment sites for the hydrophobic tails (Figure 1). To develop a potent delivery molecule, our first objective was to select an appropriate head group and spacer that provided superior uptake and enable RNA interference (RNAi) activity in vitro. We chose arginine as a starting point for its ability to bind to nucleic acids, and provide favorable interactions with cell surface to facilitate uptake. A panel of arginine-based DiLA2 analogs was synthesized to evaluate the effect of the carbon spacer distance between the guanidinium head group and the α carbon. Spacers varying from one (2-amino-3-guanidopropionic acid, nornor arginine) to four (2-amino-6-guanidohexanoic acid, homoarginine) carbons were each made with two sets of symmetric short fusogenic alkyl tails (C8 and C10).14 Binary formulations of DiLA2:DOPE (1:1) formulated with LacZ siRNA were delivered to 9L/LacZ cells. The results of this study were used to rank order the performance of the different spacer configurations (Figure 2a). Although there was reasonable potency among the various spacers, Arg and homoArg configurations lost their activity with C8 tails, whereas only norarginine had good robustness with different tails. Additionally, in the absence of serum, greater cytotoxicity was observed with homoarginine than with norarginine. Because the efficacy and activity were similar, and norarginine exhibited good cell tolerability, we selected norarginine as the core amino acid to continue building a lead DiLA2 molecule.
Figure 1.
The generic structure of a guanidinium-based DiLA2 molecule. The head group, spacer, and linker regions are depicted. The lead guanidinium DiLA2 compound consists of a spacer region with n = 2 and C16 and C18:1 chains attached to the α-carboxyl and α-amino groups, respectively.
Figure 2.
Identification of a lead arginine-based DiLA2. (a) Spacer-length selection was performed by screening a binary lipid combination of DiLA2:DOPE (1:1) (N/P = 1.8) in 9L/LacZ cells. Unmodified LacZ small interfering RNA (siRNA) was transfected at 100 nmol/l and knockdown analysis was performed at 24 hours post-transfection. Comparison of arginine-based DiLA2's having 1–4 carbon spacers (1 = nornorArg, 2 = norArg, 3 = Arg, and 4 = homoArg) with dioctyl (C8/C8) and didecyl (C10/C10) tails were assessed. (b) Tail selection was performed by evaluating in vitro apolipoprotein B (ApoB) mRNA knockdown using a screening formulation [DiLA2:DSPC:CHOL:DMPE-PEG2K (30:20:49:1)]. Treatment groups included formulations with norArg DiLA2 having C12/C12, C14/C14, C16/C16, C18/C18, and C18:1/C16 aliphatic chain combinations, and phosphate-buffered saline (PBS) control. One hundred nmol/l formulated ApoB siRNA was transfected into HepG2 cells, cells were lysed at 24 hours, and quantitative reverse transcription (RT)-PCR analysis for mRNA expression was performed. Data are normalized to PBS-treated control, ±SD of n = 3 per group. (c) Final tail selection was performed by evaluating identical in vitro formulations in an in vivo ApoB mouse model. A single 8 mg/kg siRNA dose was administered via tail vein injection to BALB/c mice. Livers were harvested at 48 hours postdose and assayed using quantitative RT-PCR analyses. Data were normalized to PBS-treated mice and represented as group mean ± SE, n = 8/group. Student t-test (two-tailed) was performed on all groups. *P < 0.001 compared with PBS control.
Selection of C18:1 and C16 tails
Using norarginine as the core amino acid, our focus next shifted to configuration of the aliphatic tails. A panel of norarginine DiLA2 was synthesized with symmetric linear tails ranging from C12 to C18 and an asymmetric tail of C18:1 combined with C16. Screening was performed in HepG2 cells using a base formulation comprised of the DiLA2 being evaluated formulated with cholesterol, DSPC, and a PEGylated lipid. The results showed that the shortest tail length (C12) did not have activity, and about a 50% reduction of ApoB message with C14 and C16 tails. At the C18 carbon length, all activity was lost. The most potent tail configuration was achieved with the asymmetric C18:1/C16 pairing (Figure 2b).15
Based on the in vitro results, we evaluated the in vivo performance of the same tail formulations in an ApoB mouse model (Figure 2c). BALB/c mice were administered 8 mg/kg of siRNA and ApoB message reduction was measured at 48 hours postdose. In comparison to the in vitro results, the in vivo results clearly demonstrated that mice treated with C18:1-norArg-C16 liposomes showed significant ApoB messenger knockdown of ~60%. The other alkyl chain lengths were not significantly different from the control. The decrease in ApoB mRNA levels observed with the C18:1-norArg-C16 liposome also led to a dose-dependent reduction in total serum cholesterol. In addition, significant inhibition of ApoB mRNA was found in the jejunum (data not shown), which is known to express ApoB, indicative of effective delivery beyond the liver. Based upon the in vitro and in vivo results we selected the C18:1/C16 configuration for formulation design
Development of an amphoteric formulation
After identifying a promising DiLA2 molecule that showed both in vitro and in vivo efficacy, we focused on further improving the activity of the formulation. We recognized that the guanidinium-based lead DiLA2 had a basic pKa (~12.2) resulting in a constitutively charged head group at neutral pH. This molecule did not exhibit pH-dependent ionizable behavior across the desired physiological pH range of 5–7.4 as would be experienced in the endosomal compartment. In order to impart pH-responsive ionizability to the liposomes, we pursued an amphoteric formulation approach, which requires partnering with an oppositely charged, ionizable compound.
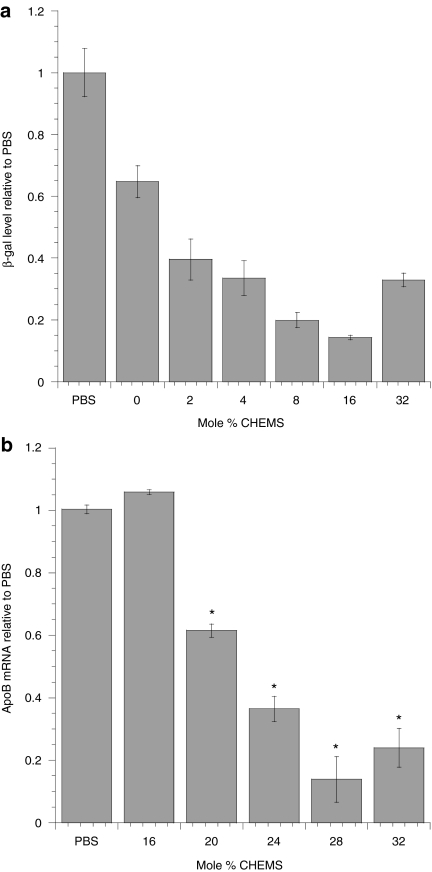
An amphoteric liposomal formulation was achieved by incorporating the negatively charged CHEMS in combination with the positively charged C18:1-norArg-C16 DiLA2. In order to reflect the total cationic charge contributed by the carrier components relative to the negative charge of the siRNA, we utilized a version of the N/P ratio we termed the “charged carrier to siRNA ratio.” This ratio is calculated by subtracting the mole fraction of the anionic component (charge = −1) from the mole fraction of the cationic component (charge = +1) and dividing by number of charges from the siRNA. We maintained the charged carrier to siRNA ratio constant at either 1.4 or 1.8 throughout the experiments. The optimal mole % of the CHEMS component was determined using LacZ siRNA in an in vitro selection assay measuring β-galactosidase protein activity. The formulations consisted of C18:1-norArg-C16 held at 50 mol%, and the PEG lipid held at 2 mol%. CHEMS was titrated from 0 to 32 mol%, with the remainder from cholesterol (total mol% cholesterol plus CHEMS = 48). Maximum knockdown was observed at 16 mol% CHEMS, resulting in a 70% reduction of the β-galactosidase protein activity (Figure 3a). A similar CHEMS range was tested in vivo by systemic administration of ApoB siRNA to BALB/c mice. A range of CHEMS from 16 to 32 mol% was evaluated. We maintained the range of CHEMS high enough to provide a shift in total net charge, but low enough to maintain a net cationic charge on the particles. The results indicated that 28 mol% was most active formulation with >80% inhibition of ApoB mRNA relative to buffer control after a dose of 2 mg/kg (Figure 3b).
Figure 3.
Identification of the optimal cholesteryl hemisuccinate (CHEMS) ratio. (a) In vitro assessment was performed with formulations comprised of C18:1-norArg-C16:cholesterol:CHEMS (50:50-X:X, X = 0–32 mol%) (CSSR = 1.8). LacZ small interfering RNA (siRNA) was transfected at 25 nmol/l in 9L/LacZ cells and harvested 48 hours later. β-Galactosidase was quantified in a colorimetric protein assay and data were normalized against control siRNA. Error bars represent the SD from n = 3 data points. (b) In vivo assessment was performed by titrating CHEMS into formulations containing C18:1-norArg-C16:CHOL:CHEMS:DMPE-PEG-2000 (48:50-X:X:2, X = 16 to 32) with apolipoprotein B (ApoB) siRNA (CCSR = 1.4). A single tail vein injection of ApoB siRNA at 2 mg/kg was administered to BALB/c mice. Serum cholesterol and liver ApoB mRNA levels were analyzed at 48 hours postdose. All message data were normalized to GAPDH and PPIA. Data were normalized to phosphate-buffered saline (PBS) control mice as mean ± SE, n = 8/group. Student t-test (two-tailed) was performed on all groups. *P < 0.05 compared with PBS control.
In vivo silencing against multiple targets
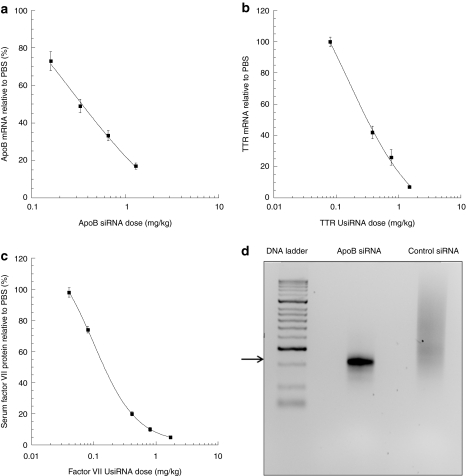
We selected a composition of C18:1-norArg-C16:CHOL:CHEMS: DMPE-PEG2K (50:20:28:2 mol%) as the lead formulation to continue our assessment of activity in different therapeutic models. To determine the potency and utility of our lead amphoteric formulation, we evaluated several hepatocyte-specific gene targets using different RNA-induced silencing complex and dicer length siRNA/UsiRNAs with different chemical modifications. To establish the ED50, we performed a dose titration with ApoB dicer length siRNA, factor VII, and TTR UsiRNAs in a dose range from 2.0 to 0.01 mg/kg. A dose-dependent decrease in liver ApoB mRNA serum factor VII protein activity, and TTR mRNA expression relative to buffer control was noted (Figure 4a–c). We established the ED50 to be 0.10, 0.16, and 0.25 mg/kg for factor VII protein activity and TTR and ApoB message, respectively. Additionally, we confirmed activity against the PCSK9 gene with 80% inhibition in PCSK9 mRNA levels at a dose of 2 mg/kg relative to buffer control (data not shown) in BALB/c mice.16
Figure 4.
ED50 analysis of different gene targets. Apolipoprotein B (ApoB) small interfering RNA (siRNA), TTR UsiRNA, and factor VII UsiRNA formulated in C18:1-norArg-C16:CHEMS:CHOL:DMPE-PEG2K liposomes. Dose-dependent decrease in liver (a) ApoB mRNA with ApoB siRNA, (b) TTR mRNA with TTR UsiRNA, and (c) serum factor VII protein activity with factor VII UsiRNA. Formulated siRNA was administered from 2 to 0.01 mg/kg, and quantitative reverse transcription (RT)-PCR analyses were performed at 48 hours postdose. Data are normalized to phosphate-buffered saline (PBS)-treated control mice as mean ± SE, n = 8 or 5/group. (d) siRNA-induced inhibition of ApoB mRNA in mouse liver using 5′RACE assay. Arrow depicting 250 bp RNA-induced silencing complex (RISC)-mediated cleavage product from 5′RACE-PCR using gene-specific primers GSP2 and anchor primer. Treatment groups include ApoB siRNA and a nonspecific control siRNA. First lane: 100 bp DNA ladder; second lane: ApoB siRNA-treated liver; third lane: control siRNA-treated liver.
To confirm that the ApoB mRNA silencing occurred via a specific RNA-induced silencing complex-mediated mechanism of action, we performed 5′ rapid amplification of complementary DNA ends (RACE) using liver homogenates from treated and control animals. A PCR product of expected molecular size, ~250 bp, was detected and confirmed RNA-induced silencing complex-mediated activity. No specific PCR products were seen in the control animals (Figure 4d). Sequencing analysis of the cloned PCR products confirmed that the cleavage product was specific to the target gene cleavage site for the ApoB siRNA, further supporting an RNAi-mediated mechanism of action (data not shown).
ApoB duration of effect
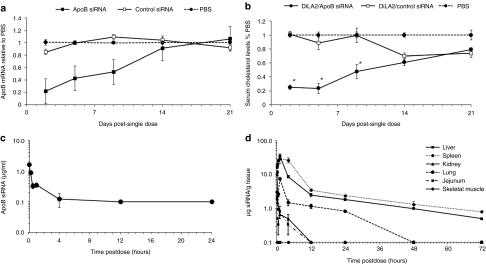
Duration of knockdown mediated by the C18:1-norArg-C16-based formulation with ApoB siRNA was evaluated at a single siRNA dose of 2 mg/kg and gene silencing and phenotypic effects were followed for 3 weeks postdose. Maximal inhibition of ApoB mRNA levels (~80%) was observed at day 2 postdose, followed by a sustained mRNA reduction of 50% through day 9, and a reduction of 20% or greater at day 14 (Figure 5a). Concomitantly, maximal reduction in serum cholesterol levels was observed at day 5 postdose (~80%) and a gradual decrease in cholesterol inhibition was observed at day 9 (~50%) followed by restoration of cholesterol to baseline levels by day 14 (Figure 5b).
Figure 5.
Pharmacokinetics and biodistribution. (a) Duration of knockdown effect on apolipoprotein B (ApoB) mRNA injected with a single small interfering RNA (siRNA) dose of 2 mg/kg ApoB UsiRNA in DiLA2 liposomes, and followed through day 21. Livers were harvested at days 2, 5, 9, 14, and 21 and analyzed for reduction in ApoB mRNA expression using quantitative reverse transcription-PCR. Values are represented as group mean ± SE, n = 5 per time point. Student t-test (two-tailed) was performed on all groups. *P < 0.05 suggesting statistical significance of treated group as compared to phosphate-buffered saline (PBS) control. (b) Duration of effect on total serum cholesterol levels with a dose of 2 mg/kg ApoB siRNA through day 21. Each data point represent mean serum cholesterol level as a percent of the PBS control ± SE at days 2, 5, 9, 14, and 21 postdose. Student t-test (two-tailed) was performed on all groups. *P < 0.05 suggesting statistical significance of treated group as compared to PBS control. (c) Pharmacokinetic profile of ApoB siRNA encapsulated in DiLA2 liposomes in serum. Sera was collected from 2 minutes to 72 hours post-single dose and analyzed using a dual-probe hybridization assay with biotin and fluorescein-labeled probes. Values are represented as group mean ± SE, n = 3 per time point. (d) Biodistribution profile of ApoB siRNA encapsulated in DiLA2 liposomes in tissues. Liver, spleen, lung, kidneys, and jejunum were harvested at various time points post-single intravenous administration. The siRNA concentration in tissues was analyzed using a dual probe hybridization assay with biotin and fluorescein-labeled probes. Values are represented as group mean ± SE, n = 3 per time point.
Pharmacokinetics and biodistribution
Pharmacokinetic and biodistribution analysis was performed using a dual-hybridization assay to quantitate the antisense (guide) strand of the ApoB siRNA. In serum, the siRNA was found to be rapidly cleared, with a half-life (T1/2) of ~20 minutes and was undetectable at 4 hours postdose (Figure 5c). The siRNA dose that left the blood compartment was found to distribute to several organs, with liver showing ~80% of the total siRNA dose, followed by spleen. A lower fraction of the total siRNA was identified in kidneys, lung, and jejunum.
Skeletal muscle was included as a negative control tissue and no detectable levels were found (Figure 5d). The pharmacokinetic profile of siRNA in liver showed a peak concentration (Cmax) of ~40 µg siRNA/g 1 hour postdose. The elimination from the liver occurred over a more prolonged time with levels of ~0.5 µg siRNA/g 72 hours postdose. The pharmacokinetic profile in the spleen was similar to the liver. The other tissues showed peak concentrations of siRNA ranging from 2 to 10 µg siRNA/g and decreased to below the lower limit of quantitation (0.1 µg/g) within 12 hours postdose. Of note, the concentrations in the jejunum suggested low exposure to siRNA whereas the mRNA knockdown was significant (data not shown).
Formulation tolerability
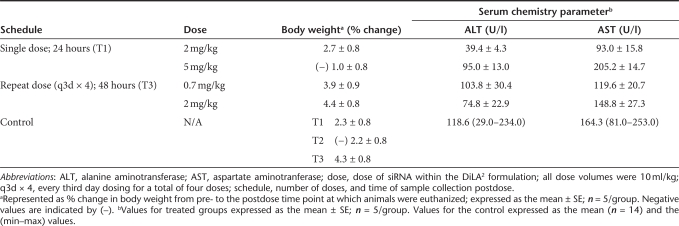
There were no clinical signs associated with dosing of the liposomes at dose levels up to 5 mg/kg, and treated mice exhibited changes in body weight postadministration that were only slightly different or equal to the buffer-treated controls for each time point (Table 1). Monitoring of aspartate aminotransferase and alanine aminotransferase values, which are indicative of liver function, showed comparable levels to the phosphate-buffered saline (PBS) control group. Single dose administrations of 2 or 5 mg/kg indicated no or only a slight increase, respectively, in liver enzymes. At the highest doses, mean values for the treated groups were less than twofold higher than the mean value of the controls (and within the control range observed). Furthermore, there was no increase in alanine aminotransferase or aspartate aminotransferase following repeat administration of four doses over 12 days (once every third day) with analysis at 48 hours post-final dose. To further evaluate tolerability, histopathology was conducted on liver sections from mice in the highest dose groups for single and repeat dose, and demonstrated no indications of hepatotoxicity or disruption of the normal architecture of the liver (data not shown).
Table 1. Tolerability profile with single and repeat dose in mice.
Additional serum chemistry parameters were also examined, e.g., blood urea nitrogen and creatinine for kidney function, with the results similar to that of alanine aminotransferase and aspartate aminotransferase with no or only slight elevation at the highest doses (data not shown). As a nonspecific measure of immune response, spleen weights were measured and were found to be within the expected range. Serum cytokine levels from mice treated with formulated unmodified and 2′-OMe end-modified ApoB siRNAs at 2 mg/kg were measured at 4 hours and 24 hours postdose. The unmodified ApoB siRNA demonstrated a transient increase in proinflammatory cytokines, interferon-α, interleukin (IL)-6, and IL-12p40 at 4 hours that returned to baseline levels by 24 hours postdose. In contrast, the 2′-OMe end-modified ApoB siRNA in DiLA2 liposomes provided no immune activation at 4 and 24 hours postdose relative to buffer-treated control suggesting that DiLA2 formulated modified ApoB siRNAs do not trigger immune response and are well tolerated (Supplementary Figure S1).
Physicochemical characterization of the formulations
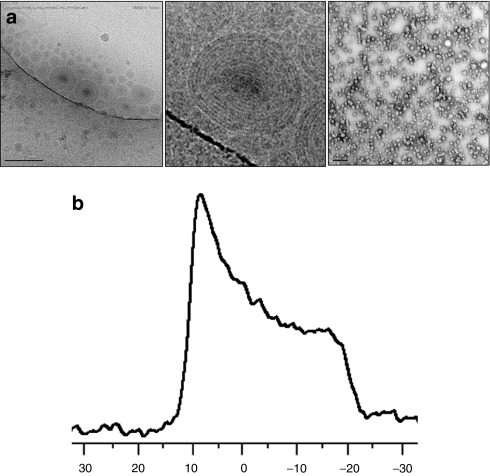
In vivo formulations were prepared using an ethanolic impinging stream process resulting in an average particle size from 100 to 125 nm in diameter, depending on the formulation, and exhibiting a low polydispersity index of <0.15. Liposomes exhibited multilamellar structure and were monodisperse as assessed by transmission electron microscopy and cryo-transmission electron microscopy microscopy (Figure 6a).
Figure 6.
Imaging and 31P-NMR data. (a) Cryo electron microscopy (EM) and transmission electron microscopy (TEM) microscopic analysis of liposomal particles. Particles assemble into multilamellar structures with low polydispersity. Bar = 200 nm on the left panel; Bar = 0.5 µm on the right panel. The center panel is an enlargement of one of the liposomes shown on the left panel cryo EM. (b) 31P-NMR. Left-sided spectral shift with peak centered at 10 p.p.m. demonstrates presence of hexagonal phase formation for C18:1-norArg-C16.
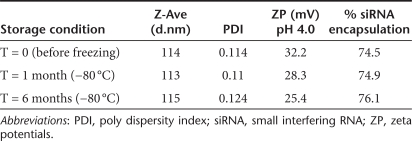
Dye measured encapsulation of siRNA ranged from 70 to 90%. Formulations were positively charged at pH 7.4 and exhibited a shift to higher positive values at pH 4 as the CHEMS carboxyl group underwent protonation and lost charge. Physical stability of the particles was assessed at various conditions and the formulation maintained all baseline (t = 0) physical properties, including size, encapsulation, and zeta-potential through a 6-month evaluation period at storage conditions up to 25 °C. With storage at −80 °C, formulations exhibited no changes in physical properties or loss in activity and were stable to numerous freeze thaw cycles (Table 2).
Table 2. DiLA2-based liposome physical characteristics.
In order to gain a mechanistic understanding of the properties of the DiLA2 formulations that supported in vivo activity, we investigated whether our lead DiLA2, C18:1-norArg-C16, could undergo a lamellar to hexagonal phase transition by performing 31P-NMR. A mixture of C18:1-norArg-C16 with DOPS (1:1 mole ratio) was evaluated at 5 and 25 °C. The resulting line shape of the 25 °C scan was a left-sided peak that had a high-field shoulder consistent with reported inverted hexagonal (HII) phases (Figure 6b).15 This line shape contrasted with the distinctly right-sided peak exhibiting a low-field shoulder of −20 p.p.m. observed at 5 °C, which is indicative of a lamellar phase (Supplementary Figure S2).
Discussion
The data presented in this study demonstrate the novelty and utility of using amino acids as core structures to create highly potent molecules for siRNA delivery. While a number of different structures can be envisioned, we present DiLA2 molecules, which consist of aliphatic hydrocarbon chains attached via amide bonds to the α-carboxyl and α-amino groups of an amino acid, allowing the amino acid side chain to form the spacer and head group. These molecules have amphiphilic character and readily partition into a lipid membrane to form liposomes.
To demonstrate our DiLA2 platform for siRNA delivery, we set out to develop an arginine-based DiLA2 that can form stable particles, exhibit phase transitions, and demonstrates a high degree of RNAi-mediated mRNA knockdown in vivo. To facilitate lead identification, a small compound library was developed that evaluated two core features of the molecule: (i) the spacer length and (ii) and tail configuration. The head group is defined as the terminal portion, usually containing the charged or ionizable group covalently attached to a spacer, which is a short-linear aliphatic carbon chain connected to the α-carbon of the amino acid. In the studies presented here, we maintained the head group constant by focusing on arginine-based amino acids due to their ability to bind nucleic acids and their role in enabling cellular uptake.12,17
In order to directly affect the shape parameter of the molecule, we modified the steric properties of the head group relative to the tails by varying the spacer length. Guanidinium-based analogs with spacers from 1 to 4 methylene carbons were evaluated. The shorter single carbon spacer had an increased tendency to form aggregates, whereas guanidinium head groups with three or four carbon spacers were active, but with lower tolerability. Norarginine was active, well-tolerated, and readily formed small, unimodal particle size distributions when partnered with a variety of different helper lipids, leading to selection of norarginine as the core amino acid.
After selecting the norarginine head group, we identified the appropriate tail structures to balance stability and efficacy. Before finalizing the selection of C18:1-norArg-C16, additional tail combinations with a higher degree of unsaturation were tested (data not shown). Unexpectedly, higher unsaturation in the tails did not improve the performance of those DiLA2 molecules compared to the asymmetric C18:1/C16 tail combination.6
It has been well documented that one of the key properties for transfection competent membrane active agents is that they undergo a phase transition from a lamellar to an inverted hexagonal phase in an endosomal environment.6,18,19 The structural attributes necessary to accomplish this transition can be summarized under the umbrella of the “shape theory” which postulates that amphiphilic molecules possessing broad “splayed” tails and small head groups have the capability to exhibit the necessary polymorphic transitions from a cylindrical-shape favoring lamellar phase formation, to a cone-shape favoring hexagonal phase formation.18,20 Additionally, the charge state of the head group, with either a constitutively cationic or ionizable functionality is important to enable polymorphic transitions, which by binding with anionic phosphatidylserine in the inner compartment of an endosome is regarded as the basis for membrane fusion and endosomal escape.21,22
In order to better understand the mechanism behind the effectiveness of the lead DiLA2, we carried out 31P-NMR. The analysis showed that C18:1-norArg-C16 formed a hexagonal phase at room temperature when mixed with DOPS, an anionic phospholipid that acts as a mimic for an endosomal membrane, and existed in a lamellar phase in the same system at 5 °C. Previous research has demonstrated that charge pairing of lipid head groups can promote hexagonal phase formation.21,23 This hexagonal phase has been shown to promote membrane fusion, resulting in endosomal escape.24 The ability for C18:1-norArg-C16 to exhibit a lamellar phase at lower temperatures and transition to a hexagonal phase form is believed to provide the basis for stability of the liposomes during circulation.
The higher activity for the asymmetric chains (C18:1–C16) compared to the symmetric C12, C14, and C16 DiLA2 or lipids may be explained by the presence of the unsaturated bond in the C18 chain, which forms a kink in the alkyl chain contributing to a decrease in the lipid-melting temperature and a concomitant increase in membrane fluidity and hexagonal phase formation potential.25,26 In addition to the membrane fluidity generated by the C18:1 chain, the presence of an asymmetric alkyl chain helps to accommodate the curvature stress, which is induced by the interaction between the negatively charged siRNA and the positively charged guanidinium head group on the DiLA2.27
In order to neutralize the charge on the liposome, which has been implicated in complement activation, we explored the incorporation of a negatively charged ionizable cholesterol derivative, CHEMS.28 This lipid has been used to generate pH-sensitive liposomes in combination with a cationic lipid.29 CHEMS has pKa of 5.7, is negatively charged at physiological pH, and becomes more neutral at an acidified endosomal pH. The protonation of CHEMS in acidic conditions favors hexagonal phase formation, and lowers the lamellar to hexagonal phase transition temperature of membranes.29,30
We reasoned that the combination of our hexagonal phase competent guanidine-based DiLA2, which provides RNA-binding and cell interaction properties, coupled with an anionic, pH-responsive phase-transitioning lipid would result in a highly efficacious and well-tolerated delivery system. Together, these properties enable modulation of charge on the liposomal surface as a function of pH. The ability to mask cationic charge with a negative lipid is a key attribute of amphoteric delivery systems, which provides for low or negative charge during circulation in blood while allowing for a greater positive charge on the surface of the particle as the endosomal pH acidifies. As reflected by our measured zeta-potential transition, the particles exhibited a low-cationic surface charge at neutral pH and underwent a charge transition to a more charged state as the pH was lowered from 7.4 to 4.0.31
Upon intravenous administration, the biodistribution results showed that particles were present in a variety of endothelial cells, whereas the muscle and connective tissues appeared to receive little or no siRNA. This can be partly explained by the slight cationic charge of the particle which was intended to provide ubiquitous uptake through cells via nonspecific macropinocytosis.32 This charge-mediated uptake pathway is prevalent in cells which display negatively charged glycosaminoglycans, and differs from neutral particle uptake recently reported to enhance delivery to the liver by liposomal association with apolipoprotein E.33 Because of the ability to transfect a variety of tissues comprised of epithelial and endothelial cells, we have successfully utilized our amphoteric delivery system in topical models, and have additional data in the treatment of bladder cancer.34
The in vivo tolerability and efficacy of the formulation is of paramount importance in RNAi-based therapeutics. The modified amino acid-based DiLA2 liposomes presented in this study demonstrated significant RNAi activity in a mouse model. Maximal mRNA inhibition of >90% was achieved with factor VII UsiRNA at 1 mg/kg dose and low ED50 values (50% inhibition in gene silencing) were obtained with factor VII and TTR UsiRNA (0.10 and 0.17 mg/kg, respectively). This DiLA2 delivery formulation also provided efficacy against multiple gene targets, demonstrating the potential utility of this formulation against significant diseases in the liver and other organs such as small intestine. The presence of sequence-specific 5′ RACE-PCR cleavage product further confirmed RNAi-mediated mechanism of action. The persistence of ApoB mRNA inhibition and reduction in serum cholesterol with a half-life of 14 days was found to be similar to previously reported siRNA delivery systems.2,3,7 Notably, the delivery system was well tolerated up to 5 mg/kg in a single dose or a repeat dose of 2 mg/kg every third day for a total of four doses and had an ~30-fold therapeutic window with either no or minimal liver disruption.
In summary, we have identified a molecule, C18:1-norArg-C16, which enabled formulation and development of an efficacious, well-tolerated systemic delivery system. Additionally, a versatile platform of amino acid-derived amphipathic molecules has been introduced. To date, only a small percentage of these molecules have been studied to determine their properties and how they may be used for siRNA delivery. We believe that these DiLA2 molecules will have superior properties to lipids due to their ease of manufacture and potential for metabolism, which could lead to improved long-term tolerability. This broad platform can be used for future design of liposomal formulations for siRNA delivery that can be potentially used to treat various diseases.
Materials and Methods
UsiRNA and DiLA2 analogs were synthesized at Marina Biotech (Bothell, WA). siRNA was purchased from IDT (Coralville, IA). The synthesis of C18:1-norArg-C16, which is representative of the synthesis of all the DiLA2 molecules, is described. CHEMS and cholesterol were obtained from Anatrace (Maumee, OH). DMPE-PEG2K was obtained from Genzyme (Boston, MA). All cells were obtained from ATCC (Manassas, VA). All animal studies were carried out at Marina Biotech using protocols approved by the Institutional Animal Care and Use Committee (IACUC).
Methods
Synthesis of siRNA: Synthesis of oligonucleotides was carried out using the standard 2-cyanoethyl phosphoramidite method on a polystyrene support (GE Healthcare, Waukesha, WI; Glen Research, Sterling, VA). All phosphoramidite building blocks were purchased directly from suppliers (Sigma-Proligo, St Louis, MO; Glen Research; RiboTask, Odense, Denmark). All oligonucleotides were synthesized using an ABI 3400 DNA/RNA synthesizer (Foster City, CA), cleaved from the solid support using concentrated NH4OH, and deprotected using a 3:1 mixture of NH4OH:EtOH at 55 °C. The deprotection of 2′-TBDMS protecting groups was achieved by incubating the base-deprotected RNA with a solution of N-methylpyrrolidone/triethylamine/triethylamine Tris(hydrofluoride) (6:3:4 by volume) at 65 °C. Cleaved and deprotected oligonucleotides were purified by anion exchange chromatography (SourceTM 15Q; GE Healthcare) followed by desalting (SephadexTM; GE Healthcare).
The sequence of factor VII UsiRNA used in these studies is:
Sense: 5′-unaUCACCCUGUCUUGGUUUCAAunaUunaU-3′
Antisense: 5′-UUGAAACCAAGACAGGGUGunaUunaU-3′
The sequence of ApoB siRNA is:
Sense: 5′-GGAAUCmUmUAmUAmUmUmUGAUCmCAsA-3′
Antisense: 5′-mUmUGGAUmCAAAmUAmUAAGAmUUCmCsmCsU-3′
The sequence of TTR UsiRNA is:
Sense: 5′-unaUGACUGGUAUUUGUGUCUGAunaUunaU-3′
Antisense: 5′-UCAGACACAAAUACCAGUCunaUunaU-3′
The sequence of control UsiRNA is:
Sense: 5′-CCAGGGUCUCCCAGUACAUunaUunaU-3′
Antisense: 5′-AUGUACUGGGAGACCCUGGunaUunaU-3′
Synthesis of C18:1-norArg-C16: Nα-Fmoc-Nγ-Boc--2,4-diaminobutyric acid was suspended in dichloromethane (DCM) to which was added two equivalents of diisopropylethyl amine and the resulting solution was added to 2-chlorotrityl chloride resin. After 1 hour, the reaction mixture was filtered and washed three times with dimethylformamide. The Fmoc group was removed by treating the resin for 20 minutes with 20% piperidine in dimethylformamide yielding the free α-amine. Oleic acid was preactivated with 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate and two equivalents of diisopropylethyl amine and added to the resin and the reaction was deemed complete by negative Kaiser test. The compound was cleaved from the resin by multiple treatments with 1% trifluoroacetic acid in DCM followed by evaporation under reduced pressure yielding free carboxylate intermediate. The second alkyl chain was attached by preactivating the free carboxyl group with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride and N-hydroxybenzotriazole in a 1:1 mixture of dimethylformamide and DCM for 10 minutes followed by addition of hexyldecyl amine in same solvent and subsequent stirring for 30 minutes. Excess of hexyldecylamine was removed by treatment with Amberlite cation exchange (Dow, Midland, MI) resin and crude compound was purified by flash chromatography. The pure dialkylated intermediate was dissolved in 1 mol/l HCl/ethyl acetate solution and the γ-Boc group was removed within 1 hour followed by removal of the solvent under reduced pressure. The resulting white solid was taken up in DCM to which TEA was added to facilitate dissolution followed by treatment with 1, 3 Di-Boc-2-(trifluoromethylsulfonyl) guanidine for 1 hour. Upon completion of the reaction DCM was washed with 2 mol/l sodium bisulfate, saturated sodium bicarbonate and dried over MgSO4 and removed under reduced pressure. The resulting residue was dissolved in absolute ethanol and two Boc groups were removed by adding dissolved compound drop wise to 12 N HCl. The final product precipitated during reaction and was recrystallized from EtOH and dried under vacuum providing C18:1-norArg-C16 as white solid, TLC, Rf = 0.25 [DCM/MeOH (90:10), iodine vapor exposure]. 1H NMR (CD3OD, 400 MHz) Δ 5.34 (m, 2H), 4.37 (q, J = 5.6 Hz, 1H), 3.17–3.34 (m, 4H), 2.28 (t, J = 7.2 Hz, 2H), 2.02–2.10 (m, 6H), 1.83–1.90 (m, 1H), 1.61–1.63 (m, 2H), 1.48–1.52 (m, 6H), 1.28–1.33 (m, 46H), 0.90 (t, J = 6.8, 6H). 13C NMR (CD3OD, 100 MHz) Δ 176.53, 173.39, 158.73, 130.92, 130.80, 52.17, 40.51, 39.40, 36.85, 33.11, 32.34, 30.84–30.32 (18 C), 28.22, 28.19, 28.00, 26.91, 23.78, 14.51. HPLC-ELS/MS: Rt 14.27 minutes (TIC) (2–98%, 20 minutes), m/z 649 for (M+H)+ (calculated 648.06 for C39H77N5O2). HPLC-ELS: Rt 20.829 minutes (99.47%). HPLC chromatograms are shown in Supplementary Figures S3 and S4.
Formulation of liposomes. Formulations for in vitro testing were prepared by dissolving the liposome components in ethanol at a total DiLA2 and lipid concentration of 5 mmol/l. The siRNA was diluted in 10 mmol/l HEPES, 5% dextrose pH 7.4. One volume of ethanolic stock solution was added to 100 volumes of siRNA then vortexed for a final siRNA concentration of 400 nmol/l siRNA. For transfection, 25 µl of formulation-siRNA complex was added to 75 µl of cells/well in 96-well format for a final siRNA concentration of 100 nmol/l. The final ethanol concentration in the cell culture media was 0.25 % (vol/vol) when dosed for in vitro knockdown activity.
Liposomes for in vivo testing were prepared using an impinging stream method. Briefly, separate stock solutions consisting of DiLA2 and lipids (50 mol% C18:1-norArg-C16, 28 mol% CHEMS, 20 mol% cholesterol, and 2 mol% DMPE-PEG2K, 24 mmol/l total concentration) in ethanol and siRNA (72 µmol/l) in 215 mmol/l sucrose, 20 mmol/l phosphate (SUP buffer (pH 7.4)) were prepared. The two solution streams were impinged to form the liposomes which were subsequently incubated for 1 hour followed by reduction in ethanol to 10%. Ethanol was removed and liposomes concentrated using tangential flow filtration on a PES 100 kDa membrane Vivaflow 50 (Sartorius, Goettingen, Germany). The dosing solution were filtered through 0.2-µm membranes and stored frozen at −80 °C. The liposome solution was then diluted to the appropriate dosing concentration using the SUP buffer.
Liposomes for 31P-NMR analysis were formulated by hydration of lipid/DiLA2 thin films. Equimolar amounts of lipid and DiLA2 were dissolved in chloroform or ethanol, followed by evaporation of solvent under a stream of nitrogen while heating at 60 °C. Forty micromole of total lipid/DiLA2 were used. Thin films were placed under vacuum to remove residual solvent, and then rehydrated with 1 ml of AD buffer (10 mmol/l acetate + 5% dextrose, pH 4.0). Liposomes were alternatively heated and vortexed until components were well dispersed. The liposomes were then exposed to three freeze thaw cycles (liquid nitrogen/room temperature).
Physical characterization of the liposomes. Size and zeta-potential were measured using a Malvern Nano ZS particle sizer (Malvern, Worcestershire, UK). Encapsulation was measured using a fluorescent dye-based assay to measure the internal and external siRNA. Briefly, an siRNA standard curve was obtained from 1 to 50 µg/ml using a 1.5× solution of the fluorescent dye SYBR-Gold (Ex:495/Em:538) (Invitrogen, Carlsbad, CA) with and without 0.5% Triton X-100. Samples were diluted to a target 20 µg/ml concentration and were treated with 1.5× SYBR-Gold to measure the external siRNA and a similar treatment was performed with liposomes dissolved in 0.5 % (vol/vol) Triton X-100 in 1.5× SYBR-Gold to quantify total siRNA. The difference between the external fluorescence, quantified against a standard curve, and the total fluorescence, provides the total siRNA encapsulated inside of the liposome. Cryo-transmission electron microscopy was performed at NanoImaging Systems, San Diego, CA.
31P-NMR analysis. Proton decoupled 31P-NMR spectra were obtained with a Bruker AV-500 spectrometer (Bruker, Madison, WI) with an operating frequency of 202.41 MHz. Operating parameters included 30° pulses, a sweep width of 200–400 p.p.m., and 1–2 second interpulse delay. A minimum of 2,000 free induction decays were obtained for each formulation, and line broadening of 200 Hz was applied before Fourier transformation. All measurements were done at 5 and 25 °C.
Assessment of in vitro knockdown activity. In vitro knockdown activity was evaluated in two different cell lines: the human HepG2 hepatocarcinoma cell line for mRNA knockdown and the rat gliobastoma 9L/LacZ cell line for β-galactosidase protein knockdown. HepG2 and 9L/LacZ cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum in a humidified incubator at 5% CO2.
HepG2 cells were trypsinized, washed, and 75 µl of cells in Dulbecco's modified Eagle's medium with or without 10% fetal bovine serum were added to wells containing 25-µl transfection liposomes in 10 mmol/l HEPES/5% dextrose, pH 7.4, at 15,000 cells/well for a total volume of 100 µl. After 5 hours, 100 µl of Dulbecco's modified Eagle's medium with 20% fetal bovine serum was added to the cells transfected in the absence of serum to achieve a final 10% serum concentration. At 24 hours, media was removed, cells were lysed, and RNA was prepared (Invitrogen). Quantitative reverse transcription-PCR for ApoB and 36B4 mRNA was performed using a qScript 1-Step quantitative reverse transcription-PCR kit (Quanta Biosciences, Gaithersburg, MD). ApoB mRNA levels compared to 36B4 mRNA levels for normalization and as an indicator of toxicity. Primers for ApoB were 5′-ATCAAGAGGGGCATCATTTCT-3′ and 5′-GAGTGGAGCAGTTTCCATACAC-3′ and for 36B4, 5′-TCTATCATCAACGGGTACAAACGA-3′ and 5′-CTTTTCAGCAAGTGGGAAGGTG-3′. Control siRNA was transfected with Lipofectamine RNAiMAX reagent (Invitrogen). Transfections were performed in triplicate.
9L/LacZ cells, after trypsinization and washing, were plated in 96-well plates, 7,500 cells/well, and allowed to adhere overnight in Dulbecco's modified Eagle's medium with 10% serum. The following day, the media was removed, replaced with 75 µl/well serum-free OptiMEM plus 25 µl of siRNA delivery complexes and incubated for 5 hours. After incubation, 100 µl of 20% serum media was added to achieve a final concentration of serum of 10%. At 24 hours, media was removed and fresh medium added. Cells were lysed and assayed for β-galactosidase activity using M-PER and Pierce Mammalian β-galatosidase assay reagent (Thermo Scientific, Rockford, IL).
In vivo mouse dosing. Six- to eight-week-old female BALB/c mice (Charles River Laboratories, Wilmington, MA) were dosed intravenously with buffer, target-specific, or control siRNA encapsulated liposomes via tail vein injection under normal pressure at a final dosing volume of 0.2 ml/20 g body weight. For ApoB studies, mice were fasted for ~12 hours before serum collection and necropsy.
Analysis of mRNA from mice. At 48 hours post-single siRNA dose, liver and jejunum (small intestine) were harvested from siRNA and PBS-treated mice (n = 5–8), and homogenized in lysis buffer using FastPrep-24 sample preparation system (MP Biomedicals, Solon, OH). Total RNA was extracted from tissue homogenates using the PureLink 96 RNA Isolation Kit according to the manufacturer's protocol (Invitrogen). Total RNA isolated was then quantified using the NanoDrop spectrophotometer and qualitatively analyzed for RNA integrity using a Bioanalyzer 2100 system (Agilent, Santa Clara, CA). Total RNA (50–100 ng) was reverse transcribed into complementary DNA using Applied Biosystems High Capacity complementary DNA Archive Kit according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). Quantitative real-time PCR to detect mRNA expression levels was carried out in a 384-well format using the PerfeCta SYBR Green FastMix (Quanta Biosciences), using the Applied Biosystems 7900HT platform (Applied Biosystems, Foster City, CA). Data was then analyzed using ΔΔCT method in qBASE relative quantification software (Biogazelle, Belgium). The endogenous genes, GAPDH and PPIA, were selected for use in mRNA normalization based on geNorm analysis (http://medgen.ugent.be/~jvdesomp/genorm). Relative ApoB mRNA levels using two ApoB-specific primers were normalized against GAPDH and PPIA, and fold change in ApoB, factor VII, TTR, and PCSK9 mRNA levels were ascertained for the siRNA-treated group normalized to the PBS-treated control. Primers were:
ApoB-10365 F: 5′-TGGCCAACATGATTGCCGATGT-3′
ApoB-10365 R:5′AACCAGCGCTCCAAGTGACATT-3′
ApoB-12211 F:5′-AGCAATGTGCCCAAGGCTTCTA-3′
ApoB-12211 R: 5′-GGCATGTTCAGCATTGACCTGT-3′
GAPDH F: 5′-TGGCAAAGTGGAGATTGTTGCC-3′
GAPDH R: 5′-AAGATGGTGATGGGCTTCCCG-3′
PPIA F: 5′-CAGACGCCACTGTCGCTTT-3′
PPIA R: 5′-TGTCTTTGGAACTTTGTCTGCAA-3′
PCSK92246 F: 5′-TGCAAAATCAAGGAGCATGGG-3′
PCSK92246 R: 5′-CAGGGAGCACATTGCATCC-3′
TTR F: 5′-AATCGTACTGGAAGACACTTGG-3′
TTR R: 5′-TGGTGCTGTAGGAGTATGGG-3′
Factor VII F: 5′-GCTTCTCTGCTTTCTGCTCC-3′
Factor VII R: 5′-TTTGCCTGTGTAGGACACCA-3′
Analysis of cholesterol. Total cholesterol levels were measured on serum collected at 48 hours postdose using the Amplex Red Cholesterol Assay Kit (Invitrogen) according to manufacturer's protocol. Cholesterol levels were then represented as mg/dl and changes in serum cholesterol levels were assessed by comparing the treated groups with the PBS control group.
Analysis of factor VII protein activity in serum. Factor VII protein activity levels was measured using Biophen FVII chromogenic assay Kit (Aniara, Mason, OH) on serum collected at 48 hours postdose according to manufacturer's protocol. Decrease in factor VII activity level of the treated group is represented as percentage of the PBS control activity.
Dose response. In dose–response experiments, mice were injected with DiLA2 liposomes with siRNAs at 2, 1, 0.5, 0.1, and 0.05 mg siRNA/kg dose levels. Animals were euthanized at 48 hours postdose and assayed for ApoB mRNA levels in liver and cholesterol levels in serum. Duration of effect studies involved dosing the mice with 2 mg/kg of siRNA/DiLA2 liposomes or PBS and a subset of animals (n = 5) in each group were euthanized at days 2, 5, 9, 14, and 21 postdose. Liver ApoB mRNA and serum cholesterol levels were evaluated for persistence of gene silencing activity and phenotypic effect.
5′ RACE assay. Total RNA was isolated from mouse liver tissues weighing ~50–100 mg using Trizol RNA isolation kit (Invitrogen) followed by phenol:choloroform:isoamyl alcohol extraction and DNAse treatment (Turbo DNAse; Invitrogen) to remove contaminating proteins and DNA, respectively. One microgram of total RNA was reverse transcribed using a gene-specific primer (GSP: 5′-TGGAAGAAGTTGGTGTTCATCTGGA-3′) and reverse transcriptase Superscript TM II, a derivative of M-MLV reverse transcription with reduced RNase H activity provided in the RACE kit (Invitrogen). The original RNA was removed from the duplex by treatment with RNase H and cleaned using a SNAP column. To PCR-amplify the specific cleavage product, a homopolymeric tail was added to the 3′ end of the complementary DNA and two rounds of PCR amplification were carried out using GSP1 (5′-TCTGTAGTATAGCCAAAGTGGTCCA-3′) and the deoxyinosine-containing anchor primer (Invitrogen) and the nested PCR used the gene-specific primer GSP2 (5′-CAAAGCCTTGTTGACACTGTCTGGGAAG-3′) and the adaptor primer. PCR products were resolved on a 2% agarose gel and visualized with ethidium bromide.
Dual-probe hybridization assay. Freshly isolated or frozen tissue samples were homogenized in ice-cold 3 mol/l GETN buffer [3 mol/l guanidine thiocyanate, 0.5 mol/l NaCl, 0.01 mol/l EDTA, 0.1 mol/l Tris, pH 7.5] using a FastPrep-24 homogenizer (MP Biomedicals) and diluted to identical concentrations (1:5–1:500 in 1 mol/l GETN). Homogenates were assayed for siRNA content by means of a hybridization assay to detect the antisense strand. Briefly, 50 µl of biotinylated ApoB capture probe (CN5709: biotin-mGmGlnaAmAlnaTfluCfluUfluUlnaAfluUmA; where m = 2′ OMe, flu = 2′ Fluoro, lna = locked nucleic acid; 1 µmol/l) was trapped on streptavidin-coated 96-well assay plates (Roche, Indianapolis, IN) and the excess washed away with high-salt wash buffer (1 mol/l NaCl, 2 mmol/l MgCl2, 0.1% Tween-20, 0.1 mol/l Tris, pH 9). This was followed by addition of 50 µl of fluorescein-labeled ApoB reporter probe (CN5711: lnaTfluUfluUmGlnaAfluUlnaCfluCmAmA-fluorescein; 0.1 µmol/l) in 1 mol/l GETN buffer (same as 3 mol/l GETN buffer except guanidine thiocyanate was at 1 mol/l concentration) was added to each well. Diluted tissue samples or serum were heated to 90 °C for 10 minutes, and volume equal to that of the reporter oligo was rapidly mixed with the reporter probe in the 96-well plates and allowed to hybridize for 5 minutes at room temperature. Wells were then washed with high-salt wash buffer and incubated with a horseradish peroxidase antifluorescein Fab antiserum (Roche #1 426 346; Roche), rewashed and siRNA detected after incubation with TMB substrate (KPL, Gaithersburg, MD) on a SpectraMax M5 spectrophotometer (Molecular Devices, Sunnyvale, CA). Levels were determined by comparison to a standard curve generated by spiking in known concentrations of ApoB siRNA in the buffer-treated tissue background.
Serum chemistry. Blood was collected via the vena cava immediately following euthanasia, and the blood was placed into serum separator tubes and allowed to clot at room temperature for a minimum of 20 minutes. Samples were processed to serum and chilled until analyzed (Phoenix Central Laboratories, Everett, WA).
Detection of cytokine response. BALB/c mice were injected intravenously with 2 mg/kg dose of unmodified and 2′OMe modified ApoB siRNAs or PBS and serum samples were collected at 4 hours and 24 hours postdose to assess the cytokine levels in treated animals. Serum levels of mouse interferon-α were measured by a sandwich ELISA immunoassay (PBL Biomedical, Piscataway, NJ) according to manufacturer's instructions. Upregulation of proinflammatory cytokines, Th1 and Th2 polarizing response in serum was assessed using Procarta's cytokine profile kit that uses the xMAP technology (multianalyte profiling beads) to detect multiple cytokines in a 96-well format. The xMAP system combines a flow cytometer, fluorescent-dyed microspheres (beads), lasers and digital signal processing to multiplex proteins within a single sample. A Procarta TM custom 10-plex cytokine panel (Panomics, Fremont CA) including IL-2, IL-6, IL-10, IL-12p40, IL-12 p70, IL-13, IL-1β, tumor necrosis factor-α, and interferon-γ was used to enable detection of cytokines in the serum.
SUPPLEMENTARY MATERIAL Figure S1. Serum cytokine profile with DiLA2-encapsulated unmodified and 2′-OMe modified ApoB siRNA. Figure S2. 31P-NMR data showing lamellar form of C18:1-norArg-C16. Figure S3. HPLC-ELS chromatogram of C18:1-norArg-C16. Figure S4. HPLC-MS chromatogram of C18:1-norArg-C16.
Acknowledgments
The authors would like to acknowledge the contributions of Steven C. Quay for thoughtful discussions. We would also like to acknowledge Annemarie Cohen, Chad Kratochwill, and Jaya Giyanani for formulation support and Iwona Maciagiewicz for DiLA2 synthesis support. Additionally, we would like to acknowledge Brian Granger and Oleksandr Baturevych for in vivo support, and Gary Millen for vivarium support.
Supplementary Material
Serum cytokine profile with DiLA2-encapsulated unmodified and 2′-OMe modified ApoB siRNA.
31P-NMR data showing lamellar form of C18:1-norArg-C16.
HPLC-ELS chromatogram of C18:1-norArg-C16.
HPLC-MS chromatogram of C18:1-norArg-C16.
REFERENCES
- Simberg D, Weisman S, Talmon Y., and, Barenholz Y. DOTAP (and other cationic lipids): chemistry, biophysics, and transfection. Crit Rev Ther Drug Carrier Syst. 2004;21:257–317. doi: 10.1615/critrevtherdrugcarriersyst.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- Ren T, Song YK, Zhang G., and, Liu D. Structural basis of DOTMA for its high intravenous transfection activity in mouse. Gene Ther. 2000;7:764–768. doi: 10.1038/sj.gt.3301153. [DOI] [PubMed] [Google Scholar]
- Barron LG, Uyechi LS., and, Szoka FC., Jr Cationic lipids are essential for gene delivery mediated by intravenous administration of lipoplexes. Gene Ther. 1999;6:1179–1183. doi: 10.1038/sj.gt.3300929. [DOI] [PubMed] [Google Scholar]
- Sioud M., and, Sørensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochem Biophys Res Commun. 2003;312:1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- White PJ. Barriers to successful delivery of short interfering RNA after systemic administration. Clin Exp Pharmacol Physiol. 2008;35:1371–1376. doi: 10.1111/j.1440-1681.2008.04992.x. [DOI] [PubMed] [Google Scholar]
- Heyes J, Palmer L, Bremner K., and, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN.et al. (2006RNAi-mediated gene silencing in non-human primates Nature 441111–114. [DOI] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N.et al. (2008A combinatorial library of lipid-like materials for delivery of RNAi therapeutics Nat Biotechnol 26561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M.et al. (2009Development of lipidoid-siRNA formulations for systemic delivery to the liver Mol Ther 17872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuge M, Takeuchi T, Nakase I, Jones AT., and, Futaki S. Cellular internalization and distribution of arginine-rich peptides as a function of extracellular peptide concentration, serum, and plasma membrane associated proteoglycans. Bioconjug Chem. 2008;19:656–664. doi: 10.1021/bc700289w. [DOI] [PubMed] [Google Scholar]
- Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y.et al. (2004Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement Mol Ther 101011–1022. [DOI] [PubMed] [Google Scholar]
- Rothbard JB, Kreider E, VanDeusen CL, Wright L, Wylie BL., and, Wender PA. Arginine-rich molecular transporters for drug delivery: role of backbone spacing in cellular uptake. J Med Chem. 2002;45:3612–3618. doi: 10.1021/jm0105676. [DOI] [PubMed] [Google Scholar]
- Bayer TS, Booth LN, Knudsen SM., and, Ellington AD. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA. 2005;11:1848–1857. doi: 10.1261/rna.2167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner JH, Kumar R, Sridhar CN, Wheeler CJ, Tsai YJ, Border R.et al. (1994Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations J Biol Chem 2692550–2561. [PubMed] [Google Scholar]
- Ways P., and, Hanahan DJ. Characterization and quantification of red cell lipids in normal man. J Lipid Res. 1964;5:318–328. [PubMed] [Google Scholar]
- Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A.et al. (2008Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates Proc Natl Acad Sci USA 10511915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M.et al. (2007Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery Gene Ther 14682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez IM, Maurer N., and, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK.et al. (2010Rational design of cationic lipids for siRNA delivery Nat Biotechnol 28172–176. [DOI] [PubMed] [Google Scholar]
- Lafleur M, Bloom M., and, Cullis PR. Lipid polymorphism and hydrocarbon order. Biochem Cell Biol. 1990;68:1–8. doi: 10.1139/o90-001. [DOI] [PubMed] [Google Scholar]
- Lewis RN., and, McElhaney RN. Surface charge markedly attenuates the nonlamellar phase-forming propensities of lipid bilayer membranes: calorimetric and (31)P-nuclear magnetic resonance studies of mixtures of cationic, anionic, and zwitterionic lipids. Biophys J. 2000;79:1455–1464. doi: 10.1016/S0006-3495(00)76397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., and, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- Bailey AL., and, Cullis PR. Liposome fusion. Current Topics in Membranes. 1997;44:359–373. [Google Scholar]
- Xu Y., and, Szoka FC., Jr Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- Trauble H. The movement of molecules across lipid membranes: A molecular theory. J Membr Biol. 1971;4:193–208. doi: 10.1007/BF02431971. [DOI] [PubMed] [Google Scholar]
- Chapman D. Phase transitions and fluidity characteristics of lipids and cell membranes. Q Rev Biophys. 1975;8:185–235. doi: 10.1017/s0033583500001797. [DOI] [PubMed] [Google Scholar]
- Epand RM, Robinson KS, Andrews ME., and, Epand RF. Dependence of the bilayer to hexagonal phase transition on amphiphile chain length. Biochemistry. 1989;28:9398–9402. doi: 10.1021/bi00450a022. [DOI] [PubMed] [Google Scholar]
- Chonn A, Cullis PR., and, Devine DV. The role of surface charge in the activation of the classical and alternative pathways of complement by liposomes. J Immunol. 1991;146:4234–4241. [PubMed] [Google Scholar]
- Hafez IM., and, Cullis PR. Cholesteryl hemisuccinate exhibits pH sensitive polymorphic phase behavior. Biochim Biophys Acta. 2000;1463:107–114. doi: 10.1016/s0005-2736(99)00186-8. [DOI] [PubMed] [Google Scholar]
- Ellens H, Bentz J., and, Szoka FC. Destabilization of phosphatidylethanolamine liposomes at the hexagonal phase transition temperature. Biochemistry. 1986;25:285–294. doi: 10.1021/bi00350a001. [DOI] [PubMed] [Google Scholar]
- Jung S, Otberg N, Thiede G, Richter H, Sterry W, Panzner S.et al. (2006Innovative liposomes as a transfollicular drug delivery system: penetration into porcine hair follicles J Invest Dermatol 1261728–1732. [DOI] [PubMed] [Google Scholar]
- Sahay G, Alakhova DY., and, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN.et al. (2010Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms Mol Ther 181357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth S, Matsui Y, Fosnaugh K, Liu Y, Vaish N, Adami R.et al. (2011RNAi-based therapeutics targeting surviving and PLK1 for treatment of bladder cancer Mol Ther Epub ahead of print; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serum cytokine profile with DiLA2-encapsulated unmodified and 2′-OMe modified ApoB siRNA.
31P-NMR data showing lamellar form of C18:1-norArg-C16.
HPLC-ELS chromatogram of C18:1-norArg-C16.
HPLC-MS chromatogram of C18:1-norArg-C16.