Abstract
We developed a method for the reconstruction of a 100 kb DNA fragment into a bacterial artificial chromosome (BAC). The procedure makes use of iterative rounds of homologous recombination in Escherichia coli. Smaller, overlapping fragments of cloned DNA, such as cosmid clones, are required. They are transferred first into a temperature-sensitive replicon and then into the BAC of choice. We demonstrated the usefulness of this procedure by assembling a 90 kb genomic segment into an E.coli–Streptomyces artificial chromosome (ESAC). Using this procedure, ESACs are easy to handle and remarkably more stable than the starting cosmids.
INTRODUCTION
The development of bacterial artificial chromosomes (BACs) has provided an important genetic tool for the cloning and mapping of complex genomes. Different cloning vectors based on the replicon of the bacterial F plasmid or on bacteriophage P1 have been developed (1,2). They are a powerful resource in molecular biology because of their ability to harbor foreign DNA sequences of up to 300 kb, their ease of handling and their stability. They are particularly useful for harboring functional genomic segments such as mammalian genes, pathogenicity islands (3) and antibiotic biosynthesis clusters (4), which are too large to be cloned in other more conventional vectors. However, with the increasing size of the genomic segments cloned in BACs, the probability of finding convenient restriction sites decreases. This makes the manipulation of large BACs by in vitro methods very difficult. These methods rely on conveniently placed suitable restriction sites. Consequently, there has been a recent resurgence of interest in the use of in vivo methods for manipulating these large DNA segments (5–8).
We are interested in developing tools for the genetic manipulation of actinomycetes, a group of bacteria that are the major producers of pharmacologically active secondary metabolites. Recently we described the Escherichia coli–Streptomyces artificial chromosome (ESAC) vectors, BAC derivatives able to replicate in E.coli and to integrate site-specifically into the Streptomyces genome (9). We have also shown that it is possible to generate a large-insert library from an actinomycete DNA, such as Streptomyces coelicolor.
Here we report an alternative application of the ESAC vectors: the reconstruction of a 90 kb gene segment starting from pre-existing, smaller fragments of cloned DNA by the iterative use of homologous recombination in E.coli. This assembly approach could be advantageous when cosmid clones are already available and could overcome some of the problems encountered in the isolation of high molecular weight DNA from unusual actinomycete strains. Furthermore, while in vitro handling of large DNA fragments requires considerable technical skills, the in vivo manipulations described here are easy to perform. This methodology can, in principle, be applied to any DNA segment from any organism and does not require specialized BAC vectors.
MATERIALS AND METHODS
Bacterial strains, plasmids and DNA manipulations
The vectors pMAK705 (10) and pCYPAC2 (11) were kindly provided by Profs Sidney Kushner (University of Georgia, Athens, GA) and Pieter de Jong (Roswell Park Cancer Institute, Buffalo, NY), respectively. The ESAC vector pPAC-S1 has been described previously (9). Cosmids pRP16, pRP31 and pRP58 were isolated from a cosmid library (12) made in the vector Lorist6 (13). Escherichia coli strains C600, MC1061, LE392, DH1, DH10B, DH5α, JM103, JM109, XL1blue, NM554, NM522 and TG1 were from commercial sources; JC13031, JC8111, AB1157 and AB2463 were from the E.coli Genetic Stock Center (www.cgsc.biology.yale.edu) and PMC104 (14) was kindly provided by Prof. Woodcock (MacCallum Cancer Institute, Melbourne, Victoria, Australia).
In vitro plasmid constructions
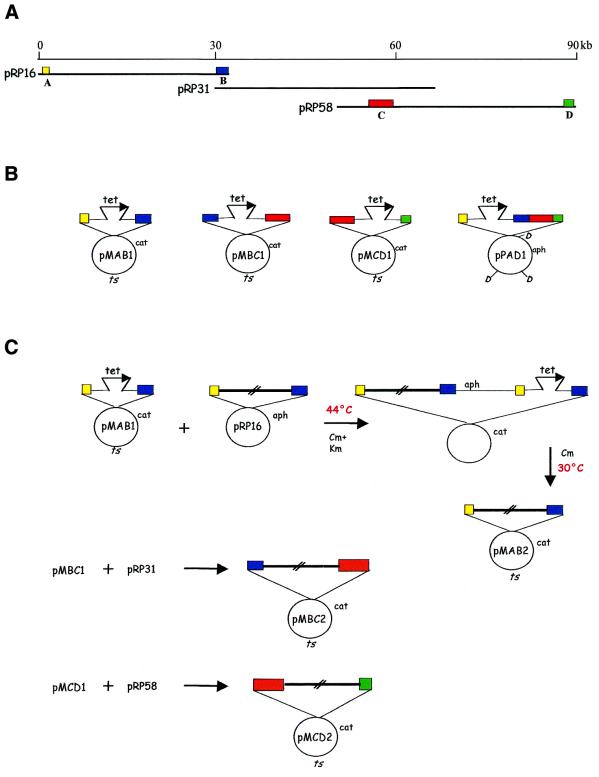
The starting material and the constructs used in this work are shown in Figure 1 and summarized in Table 1. Fragment A was a 0.9 kb SmaI–SstI segment ligated to an EcoRI–SmaI linker, while fragment B was a 1.8 kb SstI–BamHI segment ligated to a BamHI–XbaI linker (Fig. 1A). Both fragments were isolated from cosmid pRP16. Fragment C was a 4.0 kb BamHI–PstI segment ligated to a BamHI–XbaI linker, while fragment D was a 1.5 kb PstI–BamHI segment ligated to a BamHI–HindIII linker (Fig. 1A). Both fragments were isolated from cosmid pRP58. Fragments A and B, B and C, and C and D were ligated to pUC18 digested with EcoRI + BamHI, SstI + XbaI, and XbaI + HindIII, respectively, to yield plasmids pUAB1, pUBC1 and pUCD1 (Table 1). The tet fragment, obtained after PCR amplification from pBR322, was inserted into the unique SstI, XbaI and PstI sites of pUAB1, pUBC1 and pUCD1, respectively, to yield pUAB2, pUBC2 and pUCD2 (Table 1). The inserts from the resulting plasmids were excised after EcoRI + XbaI, EcoRI (complete) + PstI (partial), and XbaI (complete) + HindIII (partial) digestions of pUAB2, pUBC2 and pUCD2, respectively. These inserts were then blunt-ended and ligated to pMAK705, previously digested with HincII, to yield pMAB1, pMBC1 and pMCD1, respectively (Fig. 1B and Table 1).
Figure 1.
Starting genomic segment and constructs required for assembly. (A) The 90 kb P.rosea genomic fragment and the three cosmids pRP16, pRP31 and pRP58. Fragments A–D are color-coded. (B) The in vitro generated plasmids pMAB1, pMBC1, pMCD1 and pPAD1. (C) Scheme of cointegrate formation and resolution between each cosmid and the cognate ts construct. The interrupted empty bars designate the genomic segments comprised between fragments A and B, B and C, and C and D. Chloramphenicol and kanamycin are abbreviated to Cm and Km, respectively. cat, aph and tet represent Cm, Km and tetracycline resistance genes, respectively; ts, the temperature-sensitive replication origin. DraI sites in pPAD1 are indicated as D.
Table 1. Plasmids and artificial chromosomes.
| Name |
Replicon |
Markers |
Size (kb) |
Reference/obtained by |
| PAD2 | pPAC-S1 | KmR | 59 | resolution of pPAD1::pMAB2 |
| PAD21 | pPAC-S1 | KmR TcR | 57 | resolution of pPAD2::pMCD1 |
| PAD3 | pPAC-S1 | KmR TcR | 60 | resolution of pPAD21::pMCD3 |
| PAD4 | pPAC-S1 | KmR | 87 | resolution of pPAD3::pMCD2 |
| PAD5 | pPAC-S1 | KmR TcR | 89 | resolution of pPAD4::pMBC1 |
| PAD6 | pPAC-S1 | KmR | 108 | resolution of pPAD1::pMBC2 |
| pMAB1 | pMAK705 | CmR TcR ts | 10 | in vitro manipulation |
| pMAB2 | pMAK705 | CmR ts | 42 | resolution of pMAB1::pRP16 |
| pMAK705 | pSC101ts | CmR ts | 5.7 | (10) |
| pMBC1 | pMAK705 | CmR TcR ts | 13 | in vitro manipulation |
| pMBC2 | pMAK705 | CmR ts | 37 | resolution of pMBC1::pRP31 |
| pMCD1 | pMAK705 | CmR TcR ts | 13 | in vitro manipulation |
| pMCD2 | pMAK705 | CmR ts | 41 | resolution of pMCD1::pRP58 |
| pMCD3 | pMAK705 | CmR TcR ts | 10 | in vitro manipulation |
| pPAD1 | pPAC-S1 | KmR TcR | 30 | in vitro manipulation |
| pRP16 | Lorist6 | KmR | 42 | (12) |
| pRP31 | Lorist6 | KmR | 48 | (12) |
| pRP58 | Lorist6 | KmR | 45 | (12) |
| pUAB1 | pUC18 | ApR | 5.4 | in vitro manipulation |
| pUAB2 | pUC18 | ApR TcR | 7.1 | in vitro manipulation |
| pUBC1 | pUC18 | ApR | 8.3 | in vitro manipulation |
| pUBC2 | pUC18 | ApR TcR | 10 | in vitro manipulation |
| pUCD1 | pUC18 | ApR | 8.1 | in vitro manipulation |
| pUCD2 | pUC18 | ApR TcR | 10 | in vitro manipulation |
The 4.3 kb EcoRI–XbaI fragment from pUAB2 and the 5.5 kb XbaI–HindIII fragment from pUCD1 were ligated to pUC18 cut with EcoRI + HindIII, giving pUAD1. The 10 kb EcoRI–NdeI fragment from pUAD1 was blunt-ended and ligated to pPAC-S1, previously digested with ScaI, yielding pPAD1 (Fig. 1B and Table 1). The 1.4 kb fragment from pCYPAC2, obtained after digestion with XhoII, filling-in and digestion with KpnI, and the 3.2 kb fragment from pMCD1, obtained after XbaI digestion, filling-in and HindIII digestion, were ligated with pMAK705, previously digested with KpnI + HindIII, to yield pMCD3 (Table 1).
Screening E.coli hosts for cosmid stability
For testing the stability of cosmids in different E.coli strains we essentially followed the procedure of Ishiura et al. (15). Six independent colonies, obtained after transformation of each strain with a single cosmid, were grown overnight (first round culture), diluted 1:100 in fresh medium and grown for 8–10 h (second round culture), then further diluted 1:100 and grown for 8–10 h (third round culture). All cultures were grown in the presence of Km. After each round, cosmid DNA was prepared and, after restriction endonuclease digestion, analyzed by agarose gel electrophoresis.
Homologous recombination
We essentially followed a procedure kindly provided by Prof. Sidney Kushner (10). For insert exchange between a pMAK705 derivative (resistant to chloramphenicol, CmR, and temperature sensitive, ts, replicon) and a cosmid, E.coli cells carrying pMAB1, pMBC1 and pMCD1 were transformed with the cognate cosmids pRP16, pRP31 and pRP58, respectively. The resulting CmR KmR transformed colonies were pooled and grown at 30°C in LB broth containing Km + Cm, appropriately diluted and plated at 44°C. Single CmR KmR colonies growing at 44°C, and presumably carrying the desired pMAK705::cosmid cointegrate, were inoculated into LB broth containing Km and Cm, grown at 44°C and analyzed for plasmid content. For resolution of the cointegrates, individual CmR KmR colonies were inoculated into LB broth containing Cm and grown at 30°C for 24–72 h, diluting the cultures into fresh medium every 8–16 h. Appropriate dilutions were plated on Cm at 30°C, and a few hundred colonies were scored for KmS and TcS. Colonies with the desired CmR KmS TcS phenotype were analyzed for plasmid content. The procedure described above was also used for insert exchange between a ts construct and a pPAC-S1 derivative (KmR): E.coli cells carrying the appropriate pPAC-S1 derivative were transformed with the cognate pMAK705 construct. Selection of the cointegrate was as before, while resolution was conducted at 30°C in the presence of Km, followed by a 4–8 h incubation at 44°C before plating appropriate dilutions on Km. The resulting colonies were screened for CmS TcS or for CmS TcR as appropriate. Colonies with the desired KmR CmS TcR or KmR CmS TcS phenotype were analyzed for plasmid content.
RESULTS AND DISCUSSION
Strategy for assembly
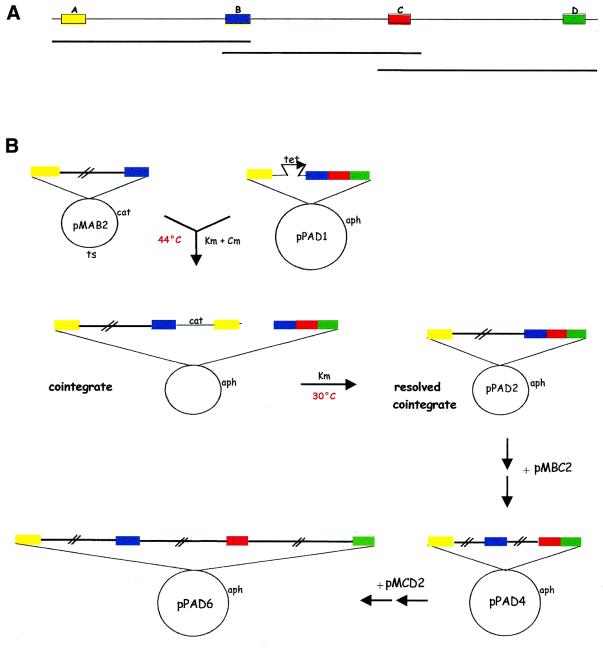
The objective of this work was to reconstruct a large genomic segment of ∼90 kb, available as a set of overlapping cosmids, into a single DNA fragment carried on a BAC. In order to do this, we utilized pPAC-S1, one of the pESAC vectors we constructed (9). This vector confers KmR in E.coli. Previous to the advent of BACs, genomic segments were often isolated from cosmid libraries and a 100 kb segment would usually be defined by three or more cosmids with small regions of overlaps (Fig. 2A). In order to drive homologous recombination, it is necessary to identify four small fragments in the genomic segment of interest (Fig. 2A): fragments A and D represent the left and right distal regions of the genomic segment, respectively; while fragments B and C lie within each of the two regions of overlap between adjacent cosmids. These four fragments are used to construct the A–B, B–C and C–D fragment cassettes in a ts replicon (e.g. pMAB1, pMBC1 and pMCD1 in Fig. 1B), and the A–B–C–D cassette in a BAC vector (e.g. pPAD1 in Fig. 1B). In the first stage of the assembly process, the insert from each cosmid is transferred, through homologous recombination, into the ts replicon. This is exemplified in Figure 1C for the isolation of pMAB2 from pRP16 and pMAB1. Similarly, pMBC2 and pMCD2 were obtained (Fig. 1C; details given below). In the second stage of the assembly process, the pPAC-S1 derivative is elongated in a step-wise fashion through sequential homologous recombination with pMAB2, pMBC2 and pMCD2 (Fig. 2B). Therefore, each round of homologous recombination makes use of the properties of a ts plasmid, which at the non-permissive temperature can exist only when fused to another replicon. Formation of a two-plasmid cointegrate, through a single crossover event in either homologous fragment, can thus be selected by plating E.coli cells at 44°C (Figs 1C and 2B). When the cells are brought back to the permissive temperature, the cointegrate is readily resolved by a second crossover event. If this occurs through the other homologous fragment, the genomic region of interest is transferred into the replicon of choice (Figs 1C and 2B). At the end of this process, the desired genomic segment has been reconstructed in the vector of choice without the need for in vitro manipulation of large DNA constructs.
Figure 2.
Strategy for assembly into an ESAC. (A) A genomic segment, covered by three overlapping cosmids, with the positions of fragments A–D. (B) Schematic of the recombination steps. The first crossover event between pMAB2 and pPAD1 leads to formation of the cointegrate (after recombination via fragment A), while the second crossover event (via fragment B) leads to the resolved cointegrate. The starting pPAC-S1 derivative is then enlarged in a step-wise fashion by cointegrate formation and resolution with the appropriate ts plasmids. Fragments A–D are color-coded as in Figure 1. Abbreviations are as in Figure 1.
Model system
To test this methodology, we made use of three overlapping cosmids previously isolated from a library of the actinomycete Planobispora rosea. These cosmids (designated pRP16, pRP31 and pRP58) define a genomic region of ∼90 kb (Fig. 1A), containing genes encoding peptide synthetases (12). These cosmids were made in the vector Lorist6, which confers KmR. As a ts replicon, we used the pSC101 derivative pMAK705 (10), which confers CmR. The four fragments necessary for this work are illustrated in Figure 1A: fragment A is unique to cosmid pRP16; fragment B is common to pRP16 and pRP31; fragment C is common to pRP16 and pRP58; and fragment D is unique to pRP58. For transferring the insert from each cosmid into the ts replicon, we constructed pMAB1, pMBC1 and pMCD1 (Fig. 1B) by cloning the A–B, B–C and C–D fragment cassettes in pMAK705. In each of these constructs, the two fragments are separated by the tet marker, conferring TcR. This facilitates the identification of the desired construct after resolution of the cointegrate. The insert from each cosmid was transferred into the ts replicon by selecting for the interplasmid cointegrate. This was achieved by plating the cells at 44°C on Km + Cm. The cointegrate was resolved by growing the cointegrate-harboring cells at 30°C in the presence of Cm only. Cells carrying the cosmid’s insert in the ts replicon were scored by their CmR TcS KmS phenotype.
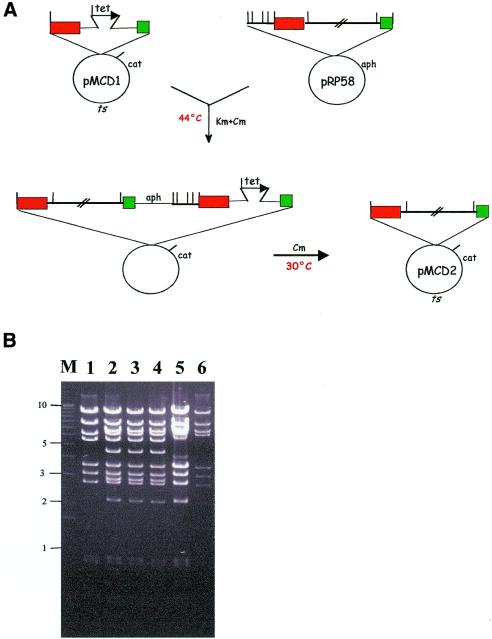
This procedure is exemplified in Figure 3A for pMCD1 and pRP58, leading to the isolation of pMCD2 (Table 1), a construct carrying the desired portion of the pRP58 insert. The fidelity of the insert exchange between pRP58 and pMCD1was evaluated by a careful comparison of the BamHI, PstI and SacI profiles of pRP58, pMCD2 and the pMCD1::pRP58 cointegrate. This is illustrated in Figure 3A for the BamHI profiles. It can be seen that all fragments present in pRP58 (lanes 5 and 6) are also present in pMCD2 (lane 1), except for the 2.3 and 0.7 kb fragments, which lie to the left side of fragment C, and the 5.8 kb Lorist6-containing fragment. Instead, pMCD2 shows a 5.5 kb pMAK705-containing fragment. Thus, the BamHI profile of pMCD2 is that expected from a pMAK705 derivative carrying the genomic segment comprised between fragments C and D. Three independent pMCD1::pRP58 cointegrates are also shown in Figure 3B (lanes 2–4). They all show a profile consistent with a homologous recombination event, where the 4.2 and 2.8 kb fragments (absent in pRP58 and pMCD2) represent the tet-bearing fragments present in pMCD1 and lost in pMCD2.
Figure 3.
Isolation and resolution of a cointegrate. (A) Selection for the pMCD1::pRP58 cointegrate at 44°C (non-permissive temperature) and resolution at 30°C to yield pMCD2. The antibiotics used for selection are indicated. BamHI sites are indicated by bars. Other symbols and abbreviations are as in Figure 1. (B) Analysis of cointegrates and resolved cointegrates. The BamHI profiles of pMCD2 (lane 1), of three independent pMCD1::pRP58 cointegrates (lanes 2–4) and of pRP58 (lanes 5–6) are shown. M, molecular weight marker, with relevant sizes (in kb) on the left.
In three separate experiments we were able to obtain the ts derivatives pMAB2, pMBC2 and pMCD2 after cointegrate formation and resolution between pRP16 and pMAB1, pRP31 and pMBC1, and pRP58 and pMCD1, respectively (Fig. 1C and Table 1). The frequency of cointegrate formation (expressed as CmR KmR over KmR colonies) was ∼10–2 and was apparently independent of the growth time in liquid medium, i.e. most cointegrates had probably formed during overnight incubation on solid medium. For resolution of the cointegrate, 8–24 h growth at 30°C led to >90% KmS colonies. The frequency of TcS CmR colonies varied from cointegrate to cointegrate, ranging between 10–2 and 10–1. After the correct cointegrate had been isolated, resolution always led to the faithful transfer of the desired segment into pMAB2 or pMBC2, as verified similarly to pMCD2 (data not shown). However, the major difficulty we encountered was in the isolation of the correct cointegrate. This is discussed in further detail below.
Construction of a 120 kb ESAC
The inserts from the ts constructs pMAB2, pMBC2 and pMCD2 were sequentially exchanged with pPAD1, the ESAC construct carrying the A–tet–B–C–D cassette (Fig. 1B) according to the scheme outlined in Figure 2B. Construct PAD2 (Table 1), which carries most of the pRP16 insert, was readily obtained by interplasmid insert exchange between pPAD1 and pMAB2. In this case, cells harboring the cointegrate were selected as in Figure 3A. Resolution and segregation of PAD2 was facilitated by counter-selecting the ts derivative obtained after cointegrate resolution by growing cells at the non-permissive temperature. KmR CmS colonies were isolated at >90% frequency and TcS colonies were readily identified.
In order to facilitate selection of the desired ESAC after cointegrate resolution with a ts construct, we decided to insert the tet marker into the appropriate pPAC-S1 construct before the next elongation cycle. To introduce the tet gene between fragments C and D, we introduced PAD2 and pMCD1 in the same host, leading to the isolation of the correct cointegrate PAD2::pMCD1, after homologous recombination via fragment C. However, when the cointegrate was resolved, all the KmR CmS TcR colonies we isolated carried the construct PAD21 (Table 1). Careful analysis of the restriction profile of this construct indicated that PAD21 had suffered a 4.0 kb deletion that included the entire fragment D and part of pPAC-S1. This deletion is likely to have resulted from a homologous recombination event that occurred between the tet gene, present between fragments C and D, and a residual tet 3′-end located in pPAC-S1 (data not shown). This event restored a functional tet gene. The deletion in PAD21 was repaired by homologous recombination with the ts construct pMCD3 (Table 1), leading to PAD3, which now contains an ∼1 kb deletion in the vector (Table 1). Insert exchange between PAD3 and pMCD2 led to the expected construct PAD4 (Fig. 2B and Table 1) carrying a 68 kb insert. Two further rounds of recombination led to PAD5 (containing the tet gene) and then to the final construct PAD6, which carries the desired 90 kb insert (Fig. 2B and Table 1).
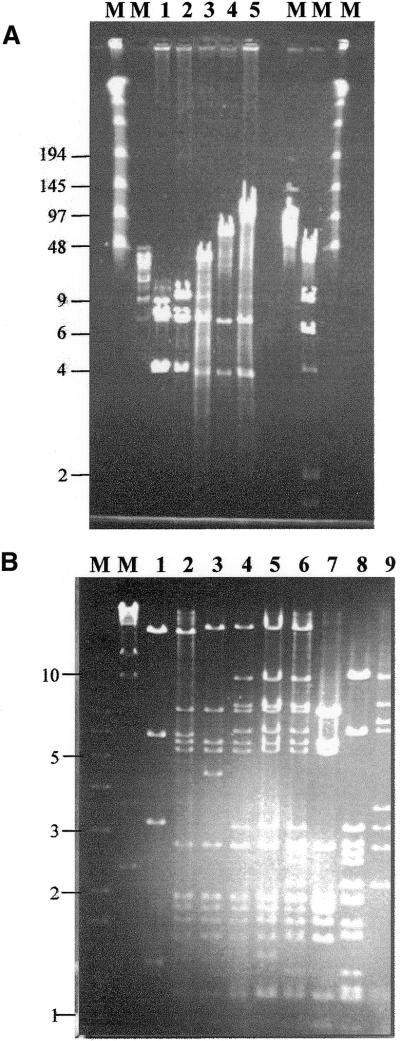
The progression of pPAC-S1 derivatives, from the 10 kb insert in PAD1 to the 90 kb insert in PAD6, can be visualized by their DraI profiles (Fig. 4A), which range from 10 (pPAD1, lane 2) to 39 (PAD2, lane 3), 68 (PAD4, lane 4) and 90 kb (PAD6, lane 5). The fidelity of the entire assembly process is illustrated in Figure 4B. It can be seen that PAD6 (lane 6) contains all the BamHI fragments present in pRP16 (lane 7), pRP31 (lane 8) and pRP58 (lane 9). The only exceptions are the vector-containing fragments (7.5, 10.5 and 6.7 kb for pRP16, pRP31 and pRP58, respectively), replaced in PAD6 by a 20 kb fragment; and fragment A- and D-containing fragments (1.9 and 3.4 kb from pRP16 and pRP58, respectively), whose distal sites were lost during construction of pMAB1 and pMCD1. The profiles of PAD2 (lane 2), PAD4 (lane 4) and PAD 5 (lane 5) are consistent with the expected structures. Similar analyses with PstI and SacI indicated that all fragments expected from the restriction map of the genomic segment could be accounted for in PAD6. Thus, the reconstruction of a 90 kb segment starting from the original cosmids had occurred without any rearrangements detectable by this type of analysis. The stable integration of PAD6 into the Streptomyces lividans chromosome has already been described (9).
Figure 4.
Analysis of the growing ESACs. (A) Pulse field gel electrophoresis analysis of ESACs after DraI digestion: pPAC-S1 (lane 1), pPAD1 (lane 2), PAD2 (lane 3), PAD4 (lane 4) and PAD6 (lane 5). M, molecular weight markers, with relevant sizes (in kb) on the left. Running conditions: 1% agarose gel in 0.5× TBE, 6–9 V/cm for 20 h at 14°C. (B) Comparisons of the BamHI profiles. Lanes 1–6 contain pPAD1, PAD2, PAD3, PAD4, PAD5 and PAD6, respectively; pRP16, pRP31 and pRP58 are in lanes 7–9, respectively. M, molecular weight marker, with relevant sizes (in kb) on the left.
Stability of cosmids and ESACs, and influence of recA
The procedure described above requires that the E.coli replicons are sufficiently stable to undergo the multiple rounds of growth (for preparation of competent cells, transformation, growth at the permissive temperature, plating at 44°C) necessary for the isolation of the cointegrates, and the further rounds of growth for the resolution of cointegrates and the segregation of cells carrying a single replicon. As mentioned above, the major difficulty we encountered was due to cosmid instability. We observed deletions of pRP16, pRP31 and pRP58 after a few rounds of propagation in the E.coli strain C600. While pRP16 was sufficiently stable to allow the isolation of pMAB2 after introduction of both pRP16 and pMAB1 in the same cells, pRP31 and pRP58 could not be recovered intact from C600 even after one round of growth (i.e. transformation and growth for plasmid analysis). It should be noted that C600 was initially chosen for the increased stability of pRP16 in this strain over that seen with MC1061, the strain recommended for homologous recombination (10). Consequently, we were unable to isolate the desired cointegrates after introduction of pRP31 or pRP58 in C600 cells carrying the cognate pMAK705 derivatives (data not shown). Deletions in cosmids during propagation in E.coli have been described to occur by recA-independent mechanisms (14,16). Following the approach described by Ishiura et al. (15), we screened a total of 17 E.coli strains (see Materials and Methods) for the stability of pRP58. Only with DH1 and DH5α could we recover intact pRP58 in ∼50 and 25% of colonies, respectively, after three rounds of growth. From the other strains tested, intact pRP58 was recovered only from XL1Blue (the original host of the cosmid library), but for no more than one round of growth. Similar results were seen with pRP31. Therefore, DH1 was the host used for the isolation of pMBC2 and pMCD2.
Notwithstanding that most of the ESACs have insert sizes considerably larger than the cosmids, they were far more stable. All the recombination work between the pPAC-S1 derivatives and the cognate pMAK705 constructs was carried out in C600, with no deletions observed. Even the pPAD5::pMBC2 cointegrate, an episome of ∼130 kb, was easily propagated for several rounds without any apparent instability. These results make it unlikely that the deletions observed with pRP31 and pRP58 were due to some idiosyncrasy between insert sequences and the E.coli host. Rather, they suggest that it is either the low copy number of pPAC-S1 derivatives or the vector Lorist6 that significantly contributes to insert stability.
It is worth mentioning that we could isolate products from interplasmid recombination (i.e. cointegrates) or from intraplasmid recombination (i.e. resolved cointegrates) either in a recA+ background (C600) or in a recA strain (DH1 carries the recA1 allele). The recA-dependence of homologous recombination has been extensively studied (reviewed in 17). recA-independent intraplasmid recombination has been observed, although it occurred at a 100-fold lower frequency than in a recA+ strain when ‘long’ (∼4000 bp) intervening sequences separated the direct repeats (18). No results were reported with intervening sequences of comparable size to those used here (30–40 kb). In our hands, when working with DH1 as the host strain, only 1–10% of the colonies isolated after plating at 44°C grew after overnight incubation in liquid medium at the same temperature. With C600, most of the colonies from solid medium could grow in liquid medium. This suggests that cointegrates in the DH1 background may be more difficult to select than in C600 after plating cells at 44°C, or less stable during further growth. We did not observe significant differences in the frequencies of cointegrate resolution using either C600 or DH1. However, cointegrate resolution involved selection for the antibiotic-resistance marker carried by the desired replicon. Thus, the segregational stability of the unselected replicon might have also influenced the observed frequencies. Other reports that have made use of homologous recombination for constructing or manipulating large constructs have mostly relied on a recA+ host (5,19,20). The use of DH1 for inter- and intraplasmid recombination has already been described (21). The possibility of using recA mutants for homologous recombination may facilitate the manipulation of the large constructs carried by BACs. Approaches using inducible recombination systems in a recA background have been described (7–8,22), as well as approaches that rely on homologous and site-specific recombination (23).
CONCLUSIONS
We have developed a procedure to assemble a large DNA fragment starting from partially overlapping, cloned DNA. This approach does not require the use of specialized vectors and we anticipate that it can be applied to any BAC of choice. In addition, it requires relatively simple in vitro DNA manipulations, while the large constructs are conveniently handled in vivo. While cloning in the low-copy number vectors pMAK705 and pPAC-S1 may be tricky, this is greatly facilitated by first constructing the required cassettes in a pUC-type vector. Since all cassettes carry the tet marker, cloning in pMAK705 and pPAC-S1 is facilitated by selection for TcR. This methodology may be applicable to all those cases where large genomic segments of interest have been isolated and characterized, but the original organism is not easily genetically manipulated.
We believe this procedure will be particularly useful with antibiotic-producing actinomycetes. Many genera are known and thousands of strains have been described (24). Furthermore, many antibiotic gene clusters are close to 100 kb in size. However, the DNA isolated from many of the actinomycete strains is readily degraded upon gel electrophoresis (25) and most actinomycetes cannot be genetically manipulated. These limitations may prevent the direct construction of BAC libraries or the rescue of desired genomic segment in vivo (26). Our results indicate that assembly from pre-existing cosmids is a feasible approach and that pPAC-S1 is a versatile tool for manipulation of large segments. Since large genomic fragments can be efficiently incorporated into the S.lividans genome (9), many of the antibiotic gene clusters isolated as overlapping cosmids from hard-to-deal-with strains can now be efficiently assembled and mobilized into different actinomycete backgrounds. This further expands the possibility of producing novel antibiotics by genetic manipulation.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Gianni Dehò for helpful discussions and for critical reading of the manuscript. We thank Anna Maria Puglia and Francesco Giusino for sharing unpublished results, and Pieter de Jong, Sidney Kushner and Alvin Clark (University of California, Berkeley, CA) for plasmids and strains.
References
- 1.Shizuya H., Birren,B., Kim,U.J., Mancino,V., Slepak,T., Tachiiri,Y. and Simon,M. (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA, 89, 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ioannou P.A., Amemiya,C.T., Garnes,J., Kroisel,P.M., Shizuya,H., Chen,C., Batzer,M.A. and de Jong,P.J. (1994) A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat. Genet., 6, 84–89. [DOI] [PubMed] [Google Scholar]
- 3.Groisman E.A. and Ochman,H. (1996) Pathogenicity islands: bacterial evolution in quantum leaps. Cell, 87, 791–794. [DOI] [PubMed] [Google Scholar]
- 4.Martìn J.F. and Liras,P. (1989) Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol., 43, 173–206. [DOI] [PubMed] [Google Scholar]
- 5.Yang X.W., Model,P. and Heintz,N. (1997) Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol., 15, 859–865. [DOI] [PubMed] [Google Scholar]
- 6.Muyrers J.P., Zhang,Y., Testa,G. and Stewart,A.F. (1999) Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res., 27, 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narayanan K., Williamson,R., Zhang,Y., Stewart,A.F. and Ioannou,P.A. (1999) Efficient and precise engineering of a 200 kb β-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther., 6, 442–447. [DOI] [PubMed] [Google Scholar]
- 8.Yu D., Ellis,H.M., Lee,E.C., Jenkins,N.A., Copeland,N.G. and Court,D.L. (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sosio M., Giusino,F., Cappellano,C., Bossi,E., Puglia,A.M. and Donadio,S. (2000) Artificial chromosomes for antibiotic-producing actinomycetes. Nat. Biotechnol., 18, 343–345. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton C.M., Aldea,M., Washburn,B.K., Babitzke,P. and Kushner,S.R. (1989) New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol., 171, 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ioannou P.A. and de Jong,P.J. (1996) Construction of bacterial artificial chromosome libraries using the modified P1 (PAC) system. In Dracopoli,N.C., Haines,J.N., Korf,B.R., Moir,D.T., Morton,C.C., Seidman,C.E., Seidman,J.G. and Smith,D.R. (eds), Current Protocols in Human Genetics. Wiley, New York, NY, pp. 5.15.1–5.15.24
- 12.Sosio M., Bossi,E., Bianchi,A. and Donadio,S. (2000) Multiple peptide synthetase gene clusters in actinomycetes. Mol. Gen. Genet., 264, 213–221. [DOI] [PubMed] [Google Scholar]
- 13.Gibson T.J., Rosenthal,A. and Waterston,R.H. (1987) Lorist6, a cosmid vector with BamHI, NotI, ScaI and HindIII cloning sites and altered neomycin phosphotransferase gene expression. Gene, 53, 283–286. [DOI] [PubMed] [Google Scholar]
- 14.Doherty J.P., Lindeman,R., Trent,R.J., Graham,M.W. and Woodcock,D.M. (1993) Escherichia coli host strains SURE™ and SRB fail to preserve a palindrome cloned in λ phage: improved alternate host strains. Gene, 124, 29–35. [DOI] [PubMed] [Google Scholar]
- 15.Ishiura M., Hazumi,N., Shinagawa,H., Nakata,A., Uchida,T. and Okada,Y. (1990) RecA-independent high-frequency deletion of recombinant cosmid DNA in Escherichia coli. J. Gen. Microbiol., 136, 69–79. [DOI] [PubMed] [Google Scholar]
- 16.Ishiura M., Hazumi,N., Koide,T., Uchida,T. and Okada,Y. (1989) A recB recC sbcB recJ host prevents recA-independent deletions in recombinant cosmid DNA propagated in Escherichia coli. J. Bacteriol., 171, 1068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd R.G. and Low,K.B. (1996) Homologous recombination. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 2236–2255.
- 18.Bi X. and Liu,L.F. (1994) recA-independent and recA-dependent intramolecular plasmid recombination: differential homology requirement and distance effect. J. Mol. Biol., 235, 414–423. [DOI] [PubMed] [Google Scholar]
- 19.Kao C.M., Katz,L. and Khosla,C. (1994) Engineered biosynthesis of a complete macrolactone in a heterologous host. Science, 265, 509–512. [DOI] [PubMed] [Google Scholar]
- 20.Crouzet J., Naudin,L., Orsini,C., Vigne,E., Ferrero,L., Le Roux,A., Benoit,P., Latta,M., Torrent,C., Branellec,D. et al. (1997) Recombinational construction in Escherichia coli of infectious adenoviral genomes. Proc. Natl Acad. Sci. USA, 94, 1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripodi M., Perfumo,S., Ali,R., Amicone,L., Abbott,C. and Cortese,R. (1981) Generation of small mutation in large genomic fragments by homologous recombination: description of the technique and examples of its use. Nucleic Acids Res., 18, 6247–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dastenko K.A. and Wanner,B.L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor M., Peifer,M. and Bender,W. (1989) Construction of large DNA segments in Escherichia coli. Science, 244, 1307–1312. [DOI] [PubMed] [Google Scholar]
- 24.Balows A., Truper,H.G., Dworkin,M., Harder,W. and Schleifer,K.-H. (1992) The Prokaryotes. Springer-Verlag, New York, NY.
- 25.Ray T., Mills,A. and Dyson,P. (1995) Tris-dependent oxidative DNA strand scission during electrophoresis. Electrophoresis, 16, 888–894. [DOI] [PubMed] [Google Scholar]
- 26.Malpartida F., Niemi,J., Navarrete,R. and Hopwood,D.A. (1990) Cloning and expression in a heterologous host of the complete set of genes for biosynthesis of the Streptomyces coelicolor antibiotic undecylprodigiosin. Gene, 93, 91–99. [DOI] [PubMed] [Google Scholar]