Abstract
Vaccinia virus, once widely used for smallpox vaccine, has recently been engineered and used as an oncolytic virus for cancer virotherapy. Their replication has been restricted to tumors by disrupting viral genes and complementing them with products that are found specifically in tumor cells. Here, we show that microRNA (miRNA) regulation also enables tumor-specific viral replication by altering the expression of a targeted viral gene. Since the deletion of viral glycoprotein B5R not only decreases viral pathogenicity but also impairs the oncolytic activity of vaccinia virus, we used miRNA-based gene regulation to suppress B5R expression through let-7a, a miRNA that is downregulated in many tumors. The expression of B5R and the replication of miRNA-regulated vaccinia virus (MRVV) with target sequences complementary to let-7a in the 3′-untranslated region (UTR) of the B5R gene depended on the endogenous expression level of let-7a in the infected cells. Intratumoral administration of MRVV in mice with human cancer xenografts that expressed low levels of let-7a resulted in tumor-specific viral replication and significant tumor regression without side effects, which were observed in the control virus. These results demonstrate that miRNA-based gene regulation is a potentially novel and versatile platform for engineering vaccinia viruses for cancer virotherapy.
Introduction
Oncolytic viruses are promising therapeutic agents for cancer and are currently under preclinical and clinical investigation.1 For example, vaccinia virus is a potential oncolytic virus because it has broad tropism in mammalian cells, a fast replication cycle, and no risk of integration into the host genome.2 The replication cycle of vaccinia viruses only requires about 8 hours and results in cell lysis and release of progeny viruses. Furthermore, there is no risk of the viral genome integrating into the host genome because vaccinia viruses complete their entire life cycle in the cytoplasm, unlike most other DNA viruses. However, since viral toxicity is a potentially serious problem, the virus has been engineered to reduce its pathogenicity while retaining its oncolytic properties.3,4
The attenuated, replicating vaccinia virus strain LC16m8 is an attractive backbone for engineering a novel oncolytic agent because the strain has an extremely low neurovirulence profile.5,6 In addition, LC16m8 has been safely administered to >100,000 infants and adults for smallpox vaccination and induced levels of immunity similar to those of the original Lister strain without serious side effects.5,7,8 LC16m8 was isolated from LC16mO, which is a clone that was isolated from Lister strain through LC16, by repeated passages in primary rabbit kidney cells and selection for their temperature sensitivities.5,6 As a result of attenuation, LC16m8 has a single nucleotide deletion in the open reading frame of the B5R gene.9,10 B5R is a 42 kDa glycoprotein that is involved in virus morphogenesis, trafficking, and dissemination.11,12,13,14,15,16,17,18,19 Previously, the B5R gene was deleted from LC16m8 to develop a more genetically stable strain, LC16m8Δ.20 In this study, we first compared the oncolytic potential of B5R-negative LC16m8Δ with B5R-positive LC16mO in mouse xenograft tumor model to determine the contribution of B5R to the pathogenicity and oncolytic potential of vaccinia virus.
Two strategies have been proposed to reduce the pathogenicity of vaccinia virus in normal cells and selectively target its oncolytic effects to tumor cells. In one strategy, insertional inactivation of vaccinia virus genes encoding thymidine kinase and/or epidermal growth factor-like vaccinia growth factor inhibits pathogenic viral replication in normal cells, while retaining its therapeutic replication in tumor cells that constitutively express thymidine kinase at high levels and have strong activation of the epidermal growth factor receptor pathway.3,4 Thus, the success of this approach requires complementation of the disrupted viral genes by-products that are specifically found in tumor cells. Another strategy, transcriptional targeting, uses tissue-specific promoters to restrict the replication of oncolytic viruses that have been developed from DNA viruses, such as adenovirus and herpes simplex virus, to malignant tissues.21,22 However, this approach is not applicable to vaccinia viruses due to its cytoplasmic life cycle. As a result, an alternative strategy is needed to reduce the pathogenicity of vaccinia virus without impairing its oncolytic activity.
In this study, we used microRNA (miRNA)-based regulation of B5R to specifically target the oncolytic effects of vaccinia virus to tumor cells. Although this strategy has been applied to the development of oncolytic viruses from other DNA23,24 and RNA25,26 viruses, this is first application of the strategy to vaccinia virus. miRNAs are small noncoding RNAs (~22 nucleotides) that repress gene expression by binding to complementary sequences in the 3′-untranslated region (UTR) of messenger RNAs.27,28 These post-transcriptional regulators play important roles in the control of tissue specification, tumorigenesis, and tumor progression. Since many miRNAs are differentially expressed in different tissues29 and tumors,30 they can be used to selectively promote viral replication in tumor cells expressing low levels of miRNA while inhibiting viral replication in normal cells that express higher levels of miRNA. An example of such a miRNA is let-7a, which belongs to the let-7 family of miRNAs that has lower expression in many kinds of cancer cells than in normal cells.31,32,33,34,35,36 We successfully developed a miRNA-regulated vaccinia virus (MRVV) with let-7a miRNA complementary target sequences in the 3′UTR of B5R. This MRVV selectively replicates and induces oncolysis in tumor cells without toxicity in normal cells.
Results
Glycoprotein B5R is associated with viral pathogenicity and oncolytic activity
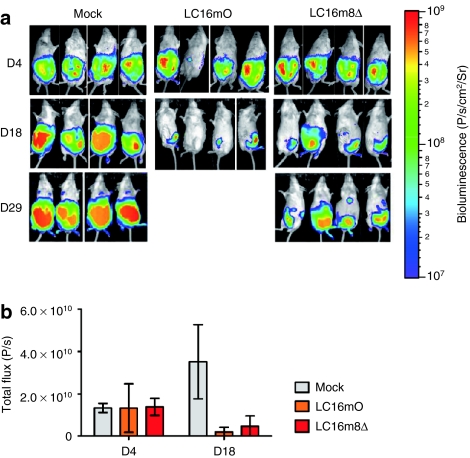
The in vivo oncolytic potentials of B5R-negative LC16m8Δ and B5R-positive LC16mO viruses in xenograft mouse model are compared in Figure 1. Four days after tumor implantation (day 4), the tumor growth in all of the implanted mice was similar. On day 18, tumor growth was significantly inhibited (P < 0.001) in both the LC16mO- and the LC16m8Δ-treated groups compared with the control group (Figure 1a,b). However, there was no significant difference in tumor volume reduction between the LC16mO- and LC16m8Δ-treated groups on this day (Figure 1b). In addition, all of the LC16mO-treated mice died or were sacrificed on days 21–28 because they exhibited symptoms of severe viral toxicity, such as weight loss and pock lesions on their tail, paws, face, and other areas of the body surface (data not shown). Although LC16m8Δ-treated mice did not show these symptoms, tumor regrowth was observed after day 29 (Figure 1a).
Figure 1.
Comparison of the oncolytic effects of LC16m8Δ and LC16mO in mice bearing intraperitoneal xenografts. (a) BxPC-3 cells stably expressing luciferase (5 × 106 cells) were intraperitoneally injected into female severe combined immunodeficiency mice on day 0. Seven days after tumor implantation, the mice were intraperitoneally injected with a single dose of LC16mO or LC16m8Δ (1 × 107 plaque-forming unit/mouse). (b) In vivo tumor growth was monitored noninvasively by bioluminescence imaging after intraperitoneal administration of -luciferin on days 4, 18, and 29. Quantification of the bioluminescence signals (photons/s) in the imaging data from days 4 and 18 in a. The data are presented as mean ± SD (n = 4).
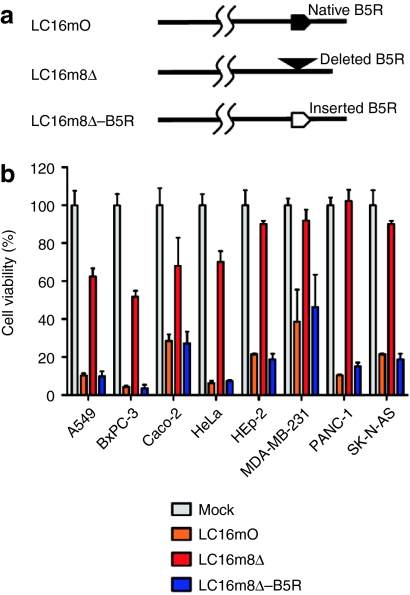
A schematic representation of the restoration of B5R in recombinant vaccinia virus LC16m8Δ-B5R is shown in Figure 2a. LC16m8Δ lysed A549, BxPC-3, Caco-2, and HeLa cells more efficiently than HEp-2, MDA-MB-231, PANC-1, and SK-N-AS cells. Although the cytolytic activity of LC16m8Δ was much lower than that of LC16mO in all tumors, B5R expression fully restored its oncolytic activity (Figure 2b).
Figure 2.
Relationship between B5R expression and oncolytic activity. (a) Schematic representation of the recombinant vaccinia virus LC16m8Δ-B5R. (b) Human cell lines were infected with B5R-positive or B5R-negative viruses at a multiplicity of infection of 0.5. The cell viabilities were determined 120 hours postinfection and are expressed as percentages of the cell survival of mock-infected cultures. The data are presented as mean + SD (n = 3).
B5R expression and the replication of miRNA-regulated vaccinia virus are dependent on endogenous let-7a
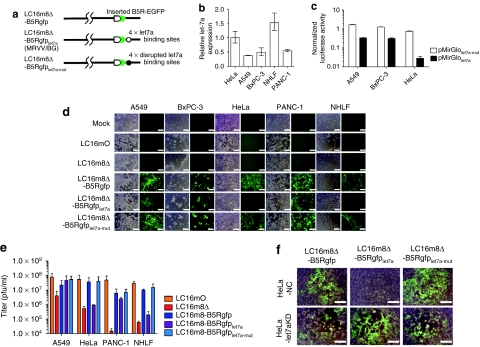
A schematic representation of MRVV with B5R-EGFP (BG) fusion protein (MRVV/BG) is shown in Figure 3a. The expression level of mature let-7a miRNA in normal human lung fibroblasts (NHLF) cells was three to four times higher than that in human lung and pancreatic carcinoma cell lines A549, BxPC-3, and PANC-1. On the other hand, there was less than a twofold difference in let-7a expression between NHLF and cervical carcinoma cell line HeLa (Figure 3b). These expression levels of let-7a correlate with functional activities of let-7a as measured by luciferase reporter assay. As shown in Figure 3c, the presence of four copies of let-7a target sequences in the 3′UTR of firefly luciferase (FLuc) mRNA in HeLa cells resulted in >96% of suppression of FLuc expression compared with the presence of four copies of the disrupted target sequences. In contrast, the suppression of FLuc expression in A549 and BxPC-3 cells was much weaker than that in HeLa cells. Thus, HeLa cells have five to seven times more let-7a activity than A549 and BxPC-3 cells (Figure 3c).
Figure 3.
Construction and characterization of recombinant microRNA (miRNA)-regulated vaccinia virus (MRVV). (a) Schematic representation of the recombinant vaccinia virus genome showing the modified B5R protein fused with enhanced green fluorescent protein at its C-terminus (MRVV/BG). Four copies of let-7a miRNA complementary or disrupted target sequences, flanked by NheI/AgeI restriction sites, were incorporated into the 3′-untranslated region of the B5R gene. (b) Relative expression of mature let-7a miRNA in the indicated cell lines by real-time PCR analysis. The data are the let-7a level normalized with the U6 small nuclear RNA level relative to that in HeLa cells and are represented by the mean ± SD (n = 3). (c) The cell lines expressing different levels of let-7a were transfected with pMirGlolet7a-mut or pMirGlolet7a plasmid containing two expression units encoding firefly luciferase (FLuc) used as the primary reporter to monitor mRNA regulation and Renilla luciferase (RLuc) acting as a transfection control. Dual luciferase assay was performed 24 hours post-transfection. The FLuc activity is normalized to the RLuc activity. The data are presented as mean + SD (n = 3). (d) The cell lines expressing different levels of let-7a were infected with the MRVV/BG at an multiplicity of infection (MOI) of 0.1 and photographed using phase-contrast or fluorescence microscopy of the same field 3 days later. Bar = 200 µm. (e) One-step growth of the MRVV/BG was determined by titration of the viruses that were collected from the infected cells shown in (d). The data are presented as mean + SD (n = 3). (f) HeLa-let7aKD cells (let-7a miRNA knockdown) or HeLa-NC cells (negative control) were infected with the MRVV/BG at an MOI of 0.1 and photographed 3 days later. The combined phase-contrast and fluorescence images are shown. Bar = 200 µm.
During infection of HeLa and NHLF cells, LC16m8Δ-B5Rgfplet7a did not induce a cytopathic effect (CPE) with B5R-enhanced green fluorescent protein (EGFP) expression, whereas LC16mO and the control viruses LC16m8Δ-B5Rgfp (lacking miRNA target sequences) and LC16m8Δ-B5Rgfplet7a-mut (containing the disrupted miRNA target sequences) resulted in a massive CPE after B5R-EGFP expression (Figure 3d). Simultaneously, the replication of LC16m8Δ-B5Rgfplet7a replication in HeLa and NHLF cells, which was equivalent to that of LC16m8Δ, was reduced by two log orders compared with that of LC16mO, LC16m8Δ-B5Rgfp, and LC16m8Δ-B5Rgfplet7a-mut (Figure 3e). In contrast, there were no differences in the CPE and replication in A549, BxPC-3, and PANC-1 cells among LC16m8Δ-B5Rgfp, LC16m8Δ-B5Rgfplet7a, and LC16m8Δ-B5Rgfplet7a-mut (Figure 3d,e). Furthermore, the CPE and replication of LC16m8Δ-B5Rgfp, LC16m8Δ-B5Rgfplet7a, and LC16m8Δ-B5Rgfplet7a-mut in A549, BxPC-3, and PANC-1 cells were comparable with those of LC16mO but were much greater than those of LC16m8Δ (Figure 3d,e). In addition, the presence of miRNA-based gene regulation was confirmed by using HeLa-let7aKD cells where TuD RNA largely suppresses endogenous let-7a activity (Supplementary Figure S1). LC16m8Δ-B5Rgfplet7a induced a CPE after B5R-EGFP expression in HeLa-let7aKD cells but not in HeLa-NC cells, although LC16m8Δ-B5Rgfp and LC16m8Δ-B5Rgfplet7a-mut showed a massive CPE after B5R-EGFP expression in both cell types (Figure 3f). Collectively, these results clearly demonstrate that B5R expression and the replication of LC16m8Δ-B5Rgfplet7a were regulated by endogenous let-7a.
Transgene insertion into microRNA-regulated vaccinia virus does not affect let-7a miRNA-regulated oncolytic activity
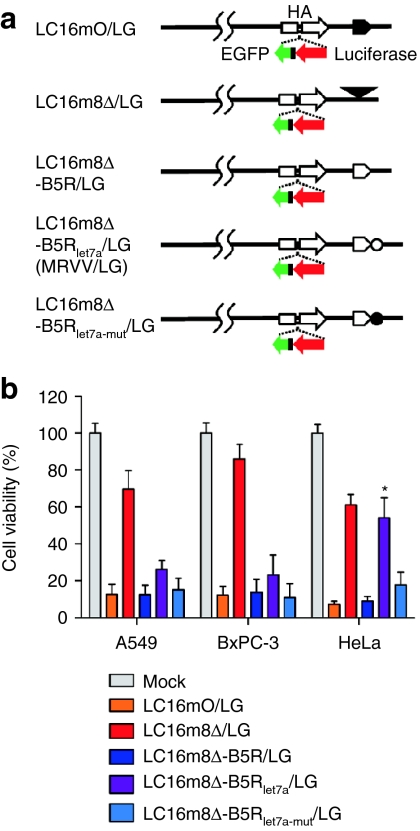
A schematic representation of MRVV with luciferase (L) and EGFP (G) reporters (MRVV/LG) is shown in Figure 4a. Although LC16m8Δ-B5Rlet7a/LG lysed A549 and BxPC-3 cells with low levels of let-7a more efficiently than LC16m8Δ/LG, there was no significant difference in the oncolytic activities of LC16m8Δ-B5Rlet7a/LG and LC16m8Δ/LG against HeLa cells with higher levels of let-7a (Figure 4b). In addition, the oncolytic activity of LC16m8Δ-B5Rlet7a/LG was significantly lower than that of LC16m8Δ-B5Rlet7a-mut/LG in HeLa cells; however, there was no significant difference in their oncolytic activities in A549 and BxPC-3 cells. Finally, LC16mO/LG, LC16m8Δ-B5R/LG, and LC16m8Δ-B5Rlet7a-mut/LG showed a similar oncolytic effect against A549, BxPC-3, and HeLa cells. These results indicated that transgene insertion into the vaccinia virus genome did not affect let-7a miRNA-regulated oncolytic activity.
Figure 4.
Effect of transgene insertion into microRNA (miRNA)-regulated vaccinia virus (MRVV) on its oncolytic activity in vitro. (a) Schematic representation of the recombinant vaccinia virus genome with the expression cassette that encodes both luciferase and enhanced green fluorescent protein reporters miRNA-regulated vaccinia virus (MRVV) luciferase (L) and enhanced green fluorescent protein (G) reporters (MRVV/LG). The symbols are the same as those used in Figures 2a and 3a. (b) The cell viability after infection with MRVV/LG was determined as described in Figure 2b. The data are presented as mean + SD (n = 3). *P < 0.001 for LC16m8Δ-B5Rlet7a/LG versus LC16m8Δ-B5Rlet7a-mut/LG in HeLa cells.
miRNA-based regulation of vaccinia virus inhibits viral replication in normal tissues
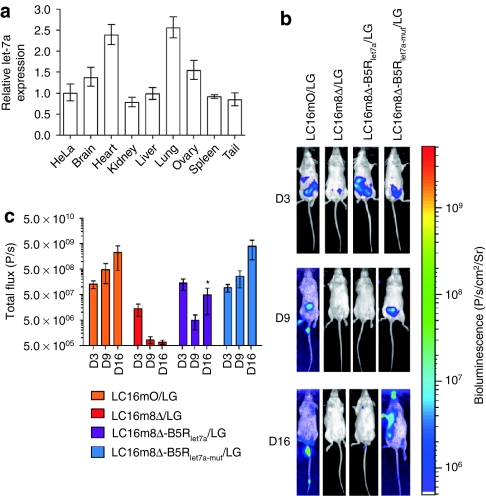
It has been reported that let-7a is highly conserved in humans and mice and is ubiquitous and abundant in normal tissues.27,37,38 Similarly, we confirmed ubiquitous let-7a RNA accumulation in all mouse tissues tested (Figure 5a). Real-time PCR analysis showed that the brain, heart, lung, and ovary have higher expression level of let-7a than HeLa cells do, while the kidney, liver, spleen, and tail have let-7a expression comparable to HeLa cells. Therefore, in vivo let-7a regulation was evaluated by a single intraperitoneal injection of each virus and noninvasive bioluminescence imaging in severe combined immunodeficiency (SCID) mice. Three days after injection of LC16mO/LG, LC16m8Δ/LG, LC16m8Δ-B5Rlet7a/LG, or LC16m8Δ-B5Rlet7a-mut/LG into SCID mice (day 3), the biodistribution of these viruses was concentrated in the abdomen. On days 9 and 16, the LC16mO/LG and LC16m8Δ-B5Rlet7a-mut/LG viruses spread to several areas of their body, including the tail, paws, and face, where pock lesions were observed; however, the LC16m8Δ/LG and LC16m8Δ-B5Rlet7a/LG viruses did not spread much (Figure 5b). In addition, there were significant differences in transgene expression levels and replication between LC16m8Δ-B5Rlet7a/LG (or LC16m8Δ/LG) and LC16m8Δ-B5Rlet7a-mut/LG (or LC16mO/LG) on day 16 but not on days 3 and 9 (Figure 5c). To clear the relationship between B5R expression and the replication of MRVV in normal tissues, LC16m8Δ-B5Rgfp, LC16m8Δ-B5Rgfplet7a, or LC16m8Δ-B5Rgfplet7a-mut was intraperitoneally injected into SCID mice, and the virus-associated B5R-EGFP expression was examined 15 days after injection. As expected, LC16m8Δ-B5Rgfp and LC16m8Δ-B5Rgfplet7a-mut caused pock lesions on the tail, where B5R-EGFP expression was detected. In contrast, no pock lesion and B5R-EGFP expression were observed on the tail of LC16m8Δ-B5Rgfplet7a-injected mice (Supplementary Figure S2). Taken together, these results demonstrate that let-7a miRNA-based regulation inhibits vaccinia virus replication in normal cells by downregulating B5R in cells infected with MRVV.
Figure 5.
Biodistribution and replication of microRNA (miRNA)-regulated vaccinia virus (MRVV) luciferase (L) and enhanced green fluorescent protein (G) reporters (MRVV/LG) in vivo. (a) Relative expression of mature let-7a miRNA in the indicated mouse normal tissues by real-time PCR analysis. The data are the let-7a level normalized with the U6 small nuclear RNA level relative to that in HeLa cells and are represented by the mean ± SD (n = 3). (b) Representative images of the biodistribution of MRVV/LG in severe combined immunodeficiency mice that were intraperitoneally injected with 1 × 107 plaque-forming unit of MRVV/LG (n = 3). The biodistributions were visualized by intraperitoneal injection of -luciferin at 3, 9, and 16 days after viral administration. The vaccinia virus designations are the same as those used in Figure 4a. (c) Quantitation of the bioluminescence signals in photons/s, calculated from the imaging data in (b). The data are presented as mean ± SD (n = 3). *P < 0.001 for LC16m8Δ-B5Rlet7a/LG versus LC16m8Δ-B5Rlet7a-mut/LG on day 16.
miRNA-regulated vaccinia virus reduces viral pathogenicity while maintaining oncolytic activity after tumor-specific replication in mouse tumor models
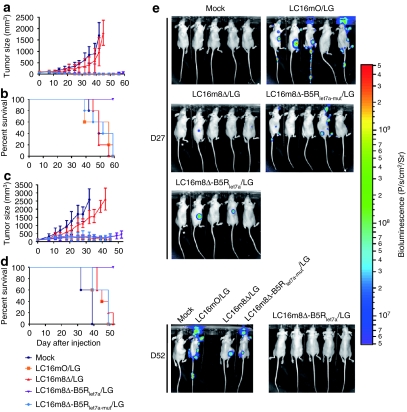
LC16m8Δ-B5Rlet7a/LG induced a significantly stronger antitumor effect than LC16m8Δ/LG in nude mice with subcutaneous BxPC-3 tumors (P < 0.001 on days 39–45; Figure 6a) or A549 tumors (P < 0.001 on days 25–42; Figure 6c) without the severe viral toxicity associated with LC16mO/LG and LC16m8Δ-B5Rlet7a-mut/LG. Furthermore, in the BxPC-3 model, four out of five LC16m8Δ-B5Rlet7a/LG-treated mice showed complete tumor regression without any symptoms of toxicity at the end of the experiment. Similarly, in the A549 model, the LC16m8Δ-B5Rlet7a/LG-treated mice showed reduced tumor growth but not complete tumor regression. Although the degree of tumor regression in the LC16mO/LG- and LC16m8Δ-B5Rlet7a-mut/LG-treated mice was similar to that of the LC16m8Δ-B5Rlet7a/LG-treated mice in both tumor models, all of the mice treated with the former viruses died or were sacrificed on days 39–59 due to severe viral toxicity, such as pock lesions and weight loss. Finally, all of the mock- or LC16m8Δ-treated mice were sacrificed by the end of the experiment due to their great tumor burden. Thus, infection with the LC16m8Δ-B5Rlet7a/LG virus resulted in a significantly longer survival than infection with any of the other viruses in both mouse xenograft models (P < 0.005; Figure 6b,d).
Figure 6.
MicroRNA (miRNA)-regulated vaccinia virus (MRVV) luciferase (L) and enhanced green fluorescent protein (G) reporters (MRVV/LG) reduces viral pathogenicity while maintaining oncolytic activity in vivo. (a,c) Nude mice bearing established subcutaneous (a) BxPC-3 tumors or (c) A549 tumors were treated with intratumoral injections of MRVV/LG [1 × 107 plaque-forming unit (pfu)/injection, 3 × 107 pfu/mouse] on days 0, 3, and 6. The vaccinia virus designations are the same as those used in Figure 4a. The data are presented as mean ± SD (n = 5). (b,d) Survival curves of the mice that are shown in a and c. (e) In vivo biodistribution of MRVV/LG, determined by noninvasive imaging after intraperitoneal injection of -luciferin into the mice that are shown in (a, b) on days 27 and 52.
These results were confirmed by bioluminescence imaging, which showed tumor-specific replication of LC16m8Δ-B5Rlet7a/LG in BxPC-3 xenografts (Figure 6e) and A549 xenografts (Supplementary Figure S3). Three and 10 days after intratumoral injection of LC16mO/LG, LC16m8Δ/LG, LC16m8Δ-B5Rlet7a/LG, or LC16m8Δ-B5Rlet7a-mut/LG (days 3 and 10), the biodistribution of these viruses was concentrated in the tumor in nude mice bearing BxPC-3 (data not shown) or A549 xenografts (Supplementary Figure S3). On day 20, the LC16mO/LG and LC16m8Δ-B5Rlet7a-mut/LG viruses spread to several areas of their body, including the tail, paws, and face, where pock lesions were observed; however, the LC16m8Δ/LG and LC16m8Δ-B5Rlet7a/LG viruses did not spread (Supplementary Figure S3). On day 27, there was no viral replication in three of the tumor-free mice treated with LC16m8Δ-B5Rlet7a/LG, whereas two of the mice with reduced tumor growth still showed tumor-specific viral replication (Figure 6e). On day 52, no viral replication was observed in normal tissues after the tumor had regressed completely in all five mice. In contrast, on day 27, viruses had already spread from tumor to normal tissues even in the tumor-free mice treated with LC16mO/LG and LC16m8Δ-B5Rlet7a-mut/LG. By day 52, all of the surviving mice had widespread viruses in their tail, paws, ears, and face (Figure 6e). Finally, LC16m8Δ/LG replication was tumor-specific; however, it was much slower than that of the other viruses, which resulted in weaker oncolytic activity (Figure 6e and Supplementary Figure S3).
Discussion
The viral glycoprotein B5R plays important roles in packaging intracellular matured virions with membranes derived from the trans-Golgi network or early endosomes to form intracellular enveloped virions.12,16,17 Intracellular enveloped virions are transported along microtubules to the cell periphery15,18 where they adhere to the cell surface as a cell-associated enveloped virions. B5R, along with the A36R and A33R proteins, is also involved in the Src kinase-dependent process of forming actin-containing microvilli and releasing cell-associated enveloped virions from the cell surface to form extracellular enveloped virions (EEVs).13,14 Since EEVs are critical for cell-to-cell and long-range virus spreading, B5R dysfunction markedly reduces the formation of EEVs and results in small viral plaques in vitro and highly attenuated viruses in vivo.10,11,17,19,20
In addition, this study demonstrated that the deletion of B5R weakens its oncolytic activity, as shown by the reduced antitumor efficacy of B5R-negative LC16m8Δ in mouse xenograft tumor models (Figures 1a and 6a,c). These results were confirmed by replication of LC16m8Δ in vivo, which was not only spatially restricted within the injected tumor but also slower replicating than B5R-positive viruses (Figure 6e and Supplementary Figure S3). The EEV is also surrounded by a host cell-derived envelope that contains several host complement control proteins and a few exposed viral proteins.39,40,41 Among these proteins, B5R is the only target for EEV-neutralizing antibodies.39 Nevertheless, high EEV-producing strains of vaccinia virus spread between tumors more efficiently than low EEV-producing strains, even in the presence of EEV-neutralizing antibodies, in a syngeneic mouse tumor model.42 Collectively, these results suggest that strategies that regulate the expression of B5R, such as miRNA, are promising approaches for engineering safe and effective vaccinia viruses for cancer virotherapy.
Recently, tumor-targeting approaches using miRNA have been used to develop oncolytic viruses based on adenovirus,23 coxsackievirus A21,26 herpes simplex virus 1,24 and vesicular stomatitis virus.25 The tumor-specific replication of these engineered viruses has decreased their pathogenic effects in normal tissues. For example, the insertion of miRNA target sequences for muscle-specific miRNA into coxsackievirus A21 decreased myositis without compromising antitumor activity.26 Similarly, insertion of miRNA target sequences for hepatocyte-specific miRNA into adenovirus reduced hepatotoxicity.23 Since vaccinia virus exhibits broad host cell tropism, we used tumor-suppressed miRNA rather than tissue-specific miRNA to develop the MRVV. In addition, we selected let-7a miRNA because the let-7 family of miRNAs is highly conserved and abundantly expressed in many types of normal cells.27,38 However, the expression of let-7a is downregulated in tumor cells isolated from patients with breast,31 hepatocellular,32 lung,33,34 melanoma,35 and pancreatic36 carcinomas.
For example, let-7a is reportedly expressed in ~50% of tumor cell lines and tumor tissues from patients with lung or pancreatic cancer at <20% of the expression in normal cells and tissues adjacent to the tumors.33,36 Similarly, the expression of let-7a in the human A549 lung, BxPC-3 pancreatic, and HeLa cervical carcinoma cell lines used in this study was reduced by ~25, 30, and 65% of the expression in NHLF, respectively (Figure 3b). Remarkably, the HeLa cells have five to seven times more let-7a activity than the A549 and BxPC-3 cells despite an approximately twofold difference of let-7a expression between these cells (Figure 3c). Considering that it has been proposed that target suppression depends on a threshold miRNA concentration,43 the concentration of let-7a in HeLa cells may be sufficient to reach the threshold expression level necessary for strong suppression. On the other hand, perfectly complementary target sites for let-7a of MRVV may be subject to regulation by all the other members of the let-7 family that are expressed in HeLa cells as described previously.43 Anyhow, B5R expression and the replication of MRVV/BG were almost completely inhibited by let-7a miRNA-based regulation in not only the NHLF but also HeLa cells (Figure 3d–f). Furthermore, B5R-EGFP expression and the replication of MRVV/BG were also abrogated on the mouse tail that has let-7a expression comparable to HeLa cells (Figure 5a and Supplementary Figure S2). Our findings are consistent with previous reports concerning let-7a miRNA-based regulation of vesicular stomatitis viral25 and polioviral37 replication in HeLa cells and mouse models. In contrast, the expression of let-7a in the A549, BxPC-3, and PANC-1 cells was low enough to induce efficient replication of MRVV/BG in these tumor cells and xenografts, although the residual let-7a activity slightly repressed the B5R expression of MRVV/BG in vitro (Figure 3d) and in vivo (Supplementary Figure S4).
In a rare example, Kelly et al. showed that oncolytic coxsackie virus A21 with inserted muscle-specific miRNA target sequences escaped from the cellular miRNA system by mutation of the target inserts.26 We did not find any indication of escaped mutants from LC16m8Δ-B5Rlet7a/LG, as shown in the bioluminescence images of the BxPC-3 model (Figure 6e). Furthermore, sequence analysis did not show any mutations in the target inserts of LC16m8Δ-B5Rlet7a/LG or LC16m8Δ-B5Rgfplet7a during cell culture passages. As reported previously,20 B5R+ revertants spontaneously emerged from LC16m8 by frameshift mutation resulting from a single nucleotide insertion at site just upstream of the deletion site in the open reading frame of the B5R gene. Since the MRVV has four copies of miRNA complementary target sequences for let-7a in the 3′UTR (and not the open reading frame) of the B5R gene, it is unlikely that a slight mutation would result in a significantly different phenotype, even if a mutation in these target inserts occurred during viral replication.
On the other hand, the spread of LC16m8Δ-B5Rlet7a/LG was much less than that of the unregulated LC16mO/LG or the control LC16m8Δ-B5Rlet7a-mut/LG in SCID mice and therefore did not cause any pock lesions in normal mouse tissues where let-7a is abundant (Figure 5a–c). However, quantitation of the bioluminescence signal from the luciferase-expressing vaccinia revealed that the LC16m8Δ-B5Rlet7a/LG signal was still higher by 1–2 log orders than the B5R-deleted LC16m8Δ/LG signal (Figure 5c). These results suggest that miRNA-mediated inhibition of B5R expression may be overcome by miRNA saturation, which has been observed by Kelly et al. previously.44 The possibility is also supported by another data that no B5R-EGFP expression was observed in HeLa cells infected with LC16m8Δ-B5Rgfplet7a at an multiplicity of infection (MOI) of 0.1 (Figure 3d); however, 100-fold higher input multiplicities of LC16m8Δ-B5Rgfplet7a allowed B5R-EGFP expression in HeLa cells (data not shown). Thus, more attention should be paid to miRNA saturation rather than to mutation of miRNA target inserts in designing vaccinia viruses for future use. In this regard, incorporation of different miRNA target sequences of more than one miRNA species might be one strategy to address the question of miRNA saturation.
In conclusion, we developed a highly attenuated MRVV with let-7a miRNA complementary target sequences in the 3′UTR of the B5R gene. This MRVV could selectively replicate and induce oncolysis in tumor cells without affecting normal cells, depending on the miRNA expression level. More generally, this study shows that control of viral replication and oncolytic activity by miRNA-based gene regulation provides a potentially novel and versatile platform for engineering vaccinia viruses for cancer virotherapy.
Materials and Methods
Plasmid construction. The construction of all plasmids used in this study is described in the Supplementary Materials and Methods.
Cell culture. Human carcinoma cell lines [lung A549 (Ham's F12K); pancreatic BxPC-3, PANC-1 and neuroblastoma SK-N-AS (RPMI-1640); colorectal Caco-2, epidermoid HEp-2 (E-MEM); cervical HeLa and breast MDA-MB-231 (D-MEM)] and rabbit kidney-derived RK13 cells (E-MEM) were obtained from the American Type Culture Collection (Manassas, VA) and grown in their respective mediums (Wako, Osaka, Japan) with 10% fetal bovine serum (Hyclone, Waltham, MA) at 37 °C in a humidified atmosphere with 5% CO2. NHLF cells was purchased from TaKaRa Biomedicals (Otsu, Japan) and cultured according to the manufacturer's protocol. HeLa-let7aKD or HeLa-NC cells were generated by infecting HeLa cells with lentivirus expressing tough decoy (TuD) RNA against let-7a or a negative control, respectively, with the human 7SK RNA polymerase III promoter,45 as described previously.46 Single HeLa-let7aKD or HeLa-NC cell isolates were expanded and selected in media containing 5 µg/ml puromycin (Sigma, St Louis, MO).
Virus construction. To construct viruses with B5R, namely, LC16m8Δ-B5R and LC16m8Δ-B5Rgfp, RK13 cells were infected with B5R-deleted LC16m8Δ viruses20 at a MOI of 0.02, and then transfected with pB5R or pTN-B5Rgfp. After harvesting the progeny viruses 2–5 days later, LC16m8Δ-B5R and LC16m8Δ-B5Rgfp were selected on the basis of larger plaque size and/or enhanced EGFP expression, by three serial plaque purifications. Finally, the insertion of B5R was verified by sequencing the modified region.
Similarly, miRNA-regulated viruses, namely, LC16m8Δ-B5Rlet7a, LC16m8Δ-B5Rlet7a-mut, LC16m8Δ-B5Rgfplet7a, and LC16m8Δ-B5Rgfplet7a-mut were constructed by infecting RK13 cells with LC16m8Δ viruses, as described above, and then transfecting them with pTN-B5Rlet7a × 4, pTN-B5Rlet7a-mut × 4, pTN-B5Rgfplet7a × 4, or pTN-B5Rgfplet7a-mut × 4, respectively.
Likewise, the viruses expressing luciferase and EGFP, namely, LC16mO/LG, LC16m8Δ/LG, LC16m8Δ-B5R/LG, LC16m8Δ-B5Rlet7a/LG, and LC16m8Δ-B5Rlet7a-mut/LG, were constructed by infecting RK13 cells with LC16mO, LC16m8Δ, LC16m8Δ-B5R, LC16m8Δ-B5Rlet7a, or LC16m8Δ-B5Rlet7a-mut viruses, respectively, as described above, and then transfecting them with pSFJvnc110-LucIRESgfp. All viruses were propagated and titrated in RK13 cells and stored at −80 °C.
Quantification of let-7a miRNA. First, total RNA was isolated from A549, BxPC-3, HeLa, PANC-1, and NHLF cells and also normal brain, heart, kidney, liver, lung, ovary, spleen, and tail of 6-week-old female athymic nude mice (Charles River Laboratories, Yokohama, Japan) using the mirVana microRNA isolation kit (Ambion, Carlsbad, CA). Then, expression of mature let-7a miRNA and the endogenous control were quantified by real-time PCR using the TaqMan microRNA assay kit (Applied Biosystems, Carlsbad, CA) for has-let-7a miRNA and U6 small nuclear RNA (snRNA), respectively. The relative expression of let-7a was calculated by using the comparative threshold method (Applied Biosystems User Bulletin No. 2).
Luciferase reporter assay. Cells in 96-well optical-bottom white plates (Nunc, Rochester, NY) were transfected with 0.1 µg of pMirGlolet7a or pMirGlolet7a-mut plasmid containing two expression units that encode Renilla luciferase (RLuc) acting as a transfection control and FLuc with four copies of let-7a target sequences or the disrupted sequences in the 3′UTR respectively, using Fugene HD (Roche, Basel, Switzerland). At 24 hours after transfection, the cells were analyzed for luciferase activities using the Dual-Glo Luciferase Assay System (Promega, Madison, WI).
Viral infection. Each cell line was infected with a vaccinia virus at an MOI of 0.1 or 0.5 plaque-forming unit (pfu)/cell, respectively, in Opti-MEM medium (Invitrogen, Carlsbad, CA) for 1 hour at 37 °C in 24-well or 96-well plates. Seventy-two hours after infection, the cells in the 24-well plate were photographed under phase-contrast or fluorescence microscopy. Subsequently, the infected cells were harvested into 1 ml of growth medium and sonicated to release the replicated viruses for titration in RK13 cells. One hundred twenty hours after infection, the viability of the cells in the 96-well plate was determined by using the CellTiter 96 AQueous cell proliferation assay kit (Promega).
In vivo experiments. The protocols for the following animal experiments were approved by the Animal Experiment Committee of the Institute of Medical Science, University of Tokyo, Japan.
In the first in vivo experiment, BxPC-3 cells stably expressing luciferase (5 × 106 cells in 100 µl of phosphate-buffered saline, pH 7.4) were intraperitoneally injected into 6-week-old female SCID mice (Charles River Laboratories) on day 0. Seven days later, the mice were administered a single intraperitoneal injection of LC16mO or LC16m8Δ (1 × 107 pfu in 100 µl of Opti-MEM per mouse). Control animals (mock therapy) were injected with 100 µl of Opti-MEM without any virus. To monitor in vivo tumor growth noninvasively, 150 µl of -luciferin (15 mg/ml) was administered to the treated mice on days 4, 18, and 29. The mice were anesthetized with isoflurane before imaging the tumors with the IVIS 100 bioluminescence imaging system (Xenogen, Hopkinton, MA). The bioluminescence signals were quantified according to the manufacturer's protocol.
In the second in vivo experiment, 6-week-old female SCID mice were intraperitoneally injected with a single dose of each vaccinia virus expressing luciferase (1 × 107 pfu in 100 µl of Opti-MEM per mouse) on day 0. To monitor the in vivo viral growth, -luciferin was injected into the mice on days 3, 9, or 16, and then they were examined by bioluminescence imaging, as described above.
In the third in vivo experiment, subcutaneous tumors were established by injecting A549 or BxPC-3 cells (5 × 106 cells in 100 µl of phosphate-buffered saline, pH 7.4) into the right flank of 6-week-old female athymic nude mice (Charles River Laboratories). When the tumors reached 5–8 mm in diameter, the mice received three intratumoral injections of each vaccinia virus (1 × 107 pfu in 100 µl of Opti-MEM per mouse) on days 0, 3, and 6. Control animals (mock therapy) were injected with 100 µl of Opti-MEM without any virus. The mice were euthanized at the end of the experiment or when any of the following occurred: tumor burden exceeded 2,500 mm3, tumor ulceration occurred, or symptoms of severe viral toxicity, such as pock lesions on body surfaces and weight loss of >30%, manifested. The diameter of tumors was measured three times per week, and the volume of a tumor was calculated according to the formula: volume = 0.5 × length × width2. The virus biodistribution was determined by injecting -luciferin into the mice on day 27 or 52, followed by bioluminescence imaging, as described above.
Statistical analysis. The differences in cytolytic activity, in vivo viral replication, and tumor burden between treatment groups were analyzed for statistical significance by one-way or two-way ANOVA and the Bonferroni test when ANOVA showed overall significance. P values <0.05 were considered to be statistically significant. Survival curves were constructed using the Kaplan–Meier method. Survival times were statistically analyzed by using the log-rank test. Data were analyzed using GraphPad Prism Ver 5 (GraphPad Software).
SUPPLEMENTARY MATERIAL Figure S1. Inhibitory effects of TuD RNA on endogenous let-7a activity. Cells in 96-well optical-bottom white plates (Nunc) were transfected with 0.1μg of pMirGlolet7a or pMirGlolet7a-mut plasmid containing two expression units that encode Renilla luciferase (RLuc) acting as a transfection control and firefly luciferase (FLuc) with four copies of let-7a target sequences or the disrupted sequences in the 3′UTR respectively, using Fugene HD (Roche). At 24 h after transfection, the cells were analyzed for luciferase activities using the Dual-Glo Luciferase Assay System (Promega). The FLuc activity is normalized to the RLuc activity. The data are presented as mean + SD (n = 3). Figure S2. B5R expression of miRNA-regulated vaccinia virus in normal tissues. 6-week-old female SCID mice were intraperitoneally injected with a single dose of each vaccinia virus expressing B5R-EGFP fusion protein (3 × 107 pfu in 100 μL of Opti-MEM per mouse) on day 0 (n=3). Control animals (mock therapy) were injected with 100 μL of Opti-MEM without any virus. On day 15, the virus-associated B5R-EGFP expression was examined on the tail using macro fluorescence imaging system (MVX10; Olympus). Figure S3. Representative images of the biodistribution of MRVV/LG, determined by noninvasive imaging after intraperitoneal injection of -luciferin into the mice that are shown in (Figure 6c,d) on days 3, 10, and 20. Figure S4. B5R expression of miRNA-regulated vaccinia virus in subcutaneous mouse xenografts that expressed low levels of let-7a. Subcutaneous tumors were established by injecting BxPC-3 cells (5 × 106 cells in 100 μL of phosphate-buffered saline, pH 7.4) into the right flank of 6-week-old female athymic nude mice. When the tumors reached 5–8 mm in diameter, the mice received a single injection of each vaccinia virus (3 × 107 pfu in 100 μL of Opti-MEM per mouse) on day 0 (n=3). Control animals (mock therapy) were injected with 100 μL of Opti-MEM without any virus. On day 11, the virus-associated B5R-EGFP expression was examined in the tumor using macro fluorescence imaging system (MVX10; Olympus). Materials and Methods.
Acknowledgments
This work was supported by the Precursory Research for Embryonic Science and Technology (PRESTO) program in RNA and Biofunctions from the Japan Science and Technology Agency and partly supported by a Grant-in-Aid for Young Scientists (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.N.).
Supplementary Material
Inhibitory effects of TuD RNA on endogenous let-7a activity. Cells in 96-well optical-bottom white plates (Nunc) were transfected with 0.1μg of pMirGlolet7a or pMirGlolet7a-mut plasmid containing two expression units that encode Renilla luciferase (RLuc) acting as a transfection control and firefly luciferase (FLuc) with four copies of let-7a target sequences or the disrupted sequences in the 3′UTR respectively, using Fugene HD (Roche). At 24 h after transfection, the cells were analyzed for luciferase activities using the Dual-Glo Luciferase Assay System (Promega). The FLuc activity is normalized to the RLuc activity. The data are presented as mean + SD (n = 3).
B5R expression of miRNA-regulated vaccinia virus in normal tissues. 6-week-old female SCID mice were intraperitoneally injected with a single dose of each vaccinia virus expressing B5R-EGFP fusion protein (3 × 107 pfu in 100 μL of Opti-MEM per mouse) on day 0 (n=3). Control animals (mock therapy) were injected with 100 μL of Opti-MEM without any virus. On day 15, the virus-associated B5R-EGFP expression was examined on the tail using macro fluorescence imaging system (MVX10; Olympus).
Representative images of the biodistribution of MRVV/LG, determined by noninvasive imaging after intraperitoneal injection of -luciferin into the mice that are shown in (Figure 6c,d) on days 3, 10, and 20.
B5R expression of miRNA-regulated vaccinia virus in subcutaneous mouse xenografts that expressed low levels of let-7a. Subcutaneous tumors were established by injecting BxPC-3 cells (5 × 106 cells in 100 μL of phosphate-buffered saline, pH 7.4) into the right flank of 6-week-old female athymic nude mice. When the tumors reached 5–8 mm in diameter, the mice received a single injection of each vaccinia virus (3 × 107 pfu in 100 μL of Opti-MEM per mouse) on day 0 (n=3). Control animals (mock therapy) were injected with 100 μL of Opti-MEM without any virus. On day 11, the virus-associated B5R-EGFP expression was examined in the tumor using macro fluorescence imaging system (MVX10; Olympus).
REFERENCES
- Parato KA, Senger D, Forsyth PA., and, Bell JC. Recent progress in the battle between oncolytic viruses and tumours. Nat Rev Cancer. 2005;5:965–976. doi: 10.1038/nrc1750. [DOI] [PubMed] [Google Scholar]
- Moss B. Knipe, DM, Howley PM. Fields Virology. Lippincott: Philadelphia, PA; 2001. Poxviridae: The viruses and their replication; pp. pp 2849–2883. [Google Scholar]
- Park BH, Hwang T, Liu TC, Sze DY, Kim JS, Kwon HC.et al. (2008Use of a targeted oncolytic poxvirus, JX-594, in patients with refractory primary or metastatic liver cancer: a phase I trial Lancet Oncol 9533–542. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yu YA, Wang E, Chen N, Danner RL, Munson PJ.et al. (2007Eradication of solid human breast tumors in nude mice with an intravenously injected light-emitting oncolytic vaccinia virus Cancer Res 6710038–10046. [DOI] [PubMed] [Google Scholar]
- Hashizume K, Yoshikawa H, Morita M., and, Suzuki K. Quinnan GV. Elsevier Science: Amsterdam; 1985. Properties of attenuated mutant of vaccinia virus, LC16m8, derived from Lister strain; Vaccinia Virus as Vectors for Vaccine Antigens; pp. pp 421–428. [Google Scholar]
- Kenner J, Cameron F, Empig C, Jobes DV., and, Gurwith M. LC16m8: an attenuated smallpox vaccine. Vaccine. 2006;24:7009–7022. doi: 10.1016/j.vaccine.2006.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Fujii T, Kanatani Y, Saijo M, Morikawa S, Yokote H.et al. (2009Clinical and immunological response to attenuated tissue-cultured smallpox vaccine LC16m8 JAMA 3011025–1033. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Kimura M., and, Hirayama M. Report of the National Smallpox Vaccination Research Committee: Study of side effects, complications and their treatments. Clin Virol. 1975;3:269–278. [Google Scholar]
- Morikawa S, Sakiyama T, Hasegawa H, Saijo M, Maeda A, Kurane I.et al. (2005An attenuated LC16m8 smallpox vaccine: analysis of full-genome sequence and induction of immune protection J Virol 7911873–11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi-Nishimaki F, Funahashi S, Miki K, Hashizume S., and, Sugimoto M. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Engelstad M., and, Smith GL. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- Hollinshead M, Rodger G, Van Eijl H, Law M, Hollinshead R, Vaux DJ.et al. (2001Vaccinia virus utilizes microtubules for movement to the cell surface J Cell Biol 154389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, Ward BM, Weisberg AS., and, Moss B. Mutations in the vaccinia virus A33R and B5R envelope proteins that enhance release of extracellular virions and eliminate formation of actin-containing microvilli without preventing tyrosine phosphorylation of the A36R protein. J Virol. 2003;77:12266–12275. doi: 10.1128/JVI.77.22.12266-12275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Scaplehorn N., and, Way M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- Rietdorf J, Ploubidou A, Reckmann I, Holmström A, Frischknecht F, Zettl M.et al. (2001Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus Nat Cell Biol 3992–1000. [DOI] [PubMed] [Google Scholar]
- Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G.et al. (1994Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network J Virol 68130–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL, Vanderplasschen A., and, Law M. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol. 2002;83 Pt 12:2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- Ward BM., and, Moss B. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J Virol. 2001;75:4802–4813. doi: 10.1128/JVI.75.10.4802-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Isaacs SN., and, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro M, Tashiro M., and, Shida H. Genetically stable and fully effective smallpox vaccine strain constructed from highly attenuated vaccinia LC16m8. Proc Natl Acad Sci USA. 2005;102:4152–4157. doi: 10.1073/pnas.0406671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruppu D., and, Tanabe KK. Viral oncolysis by herpes simplex virus and other viruses. Cancer Biol Ther. 2005;4:524–531. doi: 10.4161/cbt.4.5.1820. [DOI] [PubMed] [Google Scholar]
- Mathis JM, Stoff-Khalili MA., and, Curiel DT. Oncolytic adenoviruses - selective retargeting to tumor cells. Oncogene. 2005;24:7775–7791. doi: 10.1038/sj.onc.1209044. [DOI] [PubMed] [Google Scholar]
- Cawood R, Chen HH, Carroll F, Bazan-Peregrino M, van Rooijen N., and, Seymour LW. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 2009;5:e1000440. doi: 10.1371/journal.ppat.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Rennie PS., and, Jia WW. MicroRNA regulation of oncolytic herpes simplex virus-1 for selective killing of prostate cancer cells. Clin Cancer Res. 2009;15:5126–5135. doi: 10.1158/1078-0432.CCR-09-0051. [DOI] [PubMed] [Google Scholar]
- Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H., and, Bell JC. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Hadac EM, Greiner S., and, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W., and, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R., and, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W., and, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G.et al. (2007Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer Cancer Res 6711612–11620. [DOI] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG.et al. (2007Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma Cancer Res 676092–6099. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H.et al. (2004Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival Cancer Res 643753–3756. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A.et al. (2005RAS is regulated by the let-7 microRNA family Cell 120635–647. [DOI] [PubMed] [Google Scholar]
- Müller DW., and, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J.et al. (2009let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression Hum Gene Ther 20831–844. [DOI] [PubMed] [Google Scholar]
- Barnes D, Kunitomi M, Vignuzzi M, Saksela K., and, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B.et al. (2000Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA Nature 40886–89. [DOI] [PubMed] [Google Scholar]
- Bell E, Shamim M, Whitbeck JC, Sfyroera G, Lambris JD., and, Isaacs SN. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology. 2004;325:425–431. doi: 10.1016/j.virol.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Pütz MM, Midgley CM, Law M., and, Smith GL. Quantification of antibody responses against multiple antigens of the two infectious forms of Vaccinia virus provides a benchmark for smallpox vaccination. Nat Med. 2006;12:1310–1315. doi: 10.1038/nm1457. [DOI] [PubMed] [Google Scholar]
- Vanderplasschen A, Mathew E, Hollinshead M, Sim RB., and, Smith GL. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc Natl Acad Sci USA. 1998;95:7544–7549. doi: 10.1073/pnas.95.13.7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn DH, Wang Y, Liang W, Contag CH., and, Thorne SH. Enhancing poxvirus oncolytic effects through increased spread and immune evasion. Cancer Res. 2008;68:2071–2075. doi: 10.1158/0008-5472.CAN-07-6515. [DOI] [PubMed] [Google Scholar]
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A.et al. (2007Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state Nat Biotechnol 251457–1467. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Hadac EM, Cullen BR., and, Russell SJ. MicroRNA antagonism of the picornaviral life cycle: alternative mechanisms of interference. PLoS Pathog. 2010;6:e1000820. doi: 10.1371/journal.ppat.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna F, Santel A, Hinz M, Fechtner M, Durieux B, Fisch G.et al. (2003Inducible shRNA expression for application in a prostate cancer mouse model Nucleic Acids Res 31e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Ozaki Y., and, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inhibitory effects of TuD RNA on endogenous let-7a activity. Cells in 96-well optical-bottom white plates (Nunc) were transfected with 0.1μg of pMirGlolet7a or pMirGlolet7a-mut plasmid containing two expression units that encode Renilla luciferase (RLuc) acting as a transfection control and firefly luciferase (FLuc) with four copies of let-7a target sequences or the disrupted sequences in the 3′UTR respectively, using Fugene HD (Roche). At 24 h after transfection, the cells were analyzed for luciferase activities using the Dual-Glo Luciferase Assay System (Promega). The FLuc activity is normalized to the RLuc activity. The data are presented as mean + SD (n = 3).
B5R expression of miRNA-regulated vaccinia virus in normal tissues. 6-week-old female SCID mice were intraperitoneally injected with a single dose of each vaccinia virus expressing B5R-EGFP fusion protein (3 × 107 pfu in 100 μL of Opti-MEM per mouse) on day 0 (n=3). Control animals (mock therapy) were injected with 100 μL of Opti-MEM without any virus. On day 15, the virus-associated B5R-EGFP expression was examined on the tail using macro fluorescence imaging system (MVX10; Olympus).
Representative images of the biodistribution of MRVV/LG, determined by noninvasive imaging after intraperitoneal injection of -luciferin into the mice that are shown in (Figure 6c,d) on days 3, 10, and 20.
B5R expression of miRNA-regulated vaccinia virus in subcutaneous mouse xenografts that expressed low levels of let-7a. Subcutaneous tumors were established by injecting BxPC-3 cells (5 × 106 cells in 100 μL of phosphate-buffered saline, pH 7.4) into the right flank of 6-week-old female athymic nude mice. When the tumors reached 5–8 mm in diameter, the mice received a single injection of each vaccinia virus (3 × 107 pfu in 100 μL of Opti-MEM per mouse) on day 0 (n=3). Control animals (mock therapy) were injected with 100 μL of Opti-MEM without any virus. On day 11, the virus-associated B5R-EGFP expression was examined in the tumor using macro fluorescence imaging system (MVX10; Olympus).