Abstract
RNA interference molecules have some advantages as cancer therapeutics, including a proved efficacy on both wild-type (WT) and mutated transcripts and an extremely high sequence-specificity. The most significant hurdle to be overcome if exogenous small interfering RNAs (siRNA) is to be used therapeutically is the specific, effective, nontoxic delivery of siRNA to its intracellular site of action. At present, human applications are confined almost exclusively to targets within the liver, where the delivery systems naturally accumulate, and extra-hepatic targets remain a challenge. Anaplastic lymphoma kinase (ALK) is a receptor tyrosine kinase that has recently been shown to contribute to the cell growth and progression of human neuroblastoma (NB). We investigated its potential as a therapeutic target in NB by generating anti-GD2-targeted nanoparticles that carry ALK-directed siRNA, which are specifically and efficiently delivered to GD2-expressing NB cells. Relative to free ALK-siRNA, anti-GD2-targeted liposomal formulations of ALK-siRNA had low plasma clearance, increased siRNA stability, and improved binding, uptake, silencing and induction of cell death, and specificity for NB cells. In NB xenografts, intravenous (i.v.) injection of the targeted ALK-siRNA liposomes showed gene-specific antitumor activity with no side effects. ALK-selective siRNA entrapped in anti-GD2-targeted nanoparticles is a promising new modality for NB treatment.
Introduction
Overexpression of either mutated or wild-type (WT) anaplastic lymphoma kinase (ALK) tyrosine kinase receptor proteins induces constitutive kinase activity in neuroblastoma (NB).1,2,3,4 On the other hand, the knockdown of ALK expression in cell lines leads to a marked decrease of cell proliferation, which clearly points to ALK as a critical player in NB development. Notably, ALK mutations and amplifications, as well as gene overexpression, were found to have a substantial correlation with poor outcomes in advanced or metastatic NB disease, as compared with localized tumors.1,2,3,4,5,6,7
Herein, we tested the hypothesis that, independent of its mutational status, ALK kinase activity contributes substantially to the malignant growth of human NB. To this end, we designed a liposomal delivery strategy to selectively target NB cells, and to systemically deliver ALK-directed small interfering RNAs (siRNA) as a safe alternative to adeno- or lenti-viral therapy.
Because NB cells are differently sensitive to small molecule inhibitors, depending on the ALK mutation type,3,8 we reasoned that the use of siRNA molecules should confer some advantages in the treatment of NB, such as its proved efficacy on both WT and mutated transcripts, an extremely high sequence-specificity, and a low toxicity. However, despite the considerable potential of RNA interference (RNAi) for treating cancer,9,10 several challenges need to be overcome for exogenous siRNA to be widely used as cancer therapeutics. The most significant hurdle is the specific, effective, nontoxic delivery of siRNA to their intracellular site of action. In recent years, various promising strategies have been developed for systemic siRNA delivery,11,12,13 and some are beginning to progress to nonhuman primate14,15 and to human clinical trials (see: http://www.alnylam.com/Programs-and-Pipeline/Programs/index.php; http://clinicaltrials.gov/ct2/home). To date, human applications are limited almost exclusively to targets within the liver, where the delivery systems naturally accumulate, and delivery of siRNA to extra-hepatic targets remain a considerable challenge.
In an effort to solve these problems, we developed a new tumor-targeted delivery system for siRNA and examined the feasibility of therapeutic targeting of human ALK with siRNA in biologically relevant xenograft models of human NB. Among the specific antigens found on membranes of NB cells, the disialoganglioside GD2 is an attractive target for therapy of tumors of neuroectodermal origin, as it is overexpressed in these tumors relative to nonmalignant tissues.16,17,18 We have previously used GD2-targeted immunoliposomes to deliver the chemotherapeutic agent doxorubicin,19,20 the apoptotic drug fenretinide,21 oncogene-specific antisense oligonucleotides,22,23,24 and immunostimulatory CpG sequences25,26 to NB or melanoma cells, either in vitro or in vivo. Here, we report an anti-GD2-targeted lipidic nanomedicine strategy for the safe, effective delivery of anti-ALK siRNA to NB cells. Administration of the GD2-targeted liposomes showed antitumor activity in NB cells due to specific ALK knockdown, in the absence of toxic side effects.
Results
Anti-GD2-targeted liposomes entrapping siRNA
Plasmid-cationic lipid “lipoplexes” or plasmid-cationic polymer “polyplexes” have a short lifespan in circulation following intravenous (i.v.) injection,27,28 which leads to low tumor uptake. To significantly increase the dose, half-life, and stability of siRNA, which is expected to increase the concentration of siRNA that reaches tumor sites (via the enhanced permeability and retention effect), we used coated cationic liposomes (CCL), which we previously have shown to be efficient in condensing and carrying nucleic acid or antisense oligodeoxynucleotides. CCL have increased half-lives and reduced toxicities compared to free drug, lipoplexes and polyplexes, and result in increased delivery of nucleic acids to target cells, and increased therapeutic effects.22,24,29
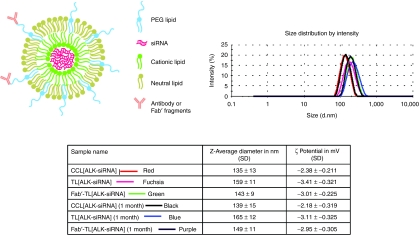
CCL were typically 135–140 (polydispersity 0.085 ± 0.015) nm in diameter. The average diameter of targeted liposomes entrapping siRNA, after coupling of whole anti-GD2 antibodies, named TL[siRNA], or its Fab′ fragment, named Fab′-TL[siRNA], increased by ~20 nm (polydispersity 0.09 ± 0.02) and ~10 nm (polydispersity 0.08 ± 0.02), respectively, with an average coupling efficiency of 53 ± 11 µg whole antibody/µmol phospholipids (PL) and 42 ± 15 µg Fab′ fragment/µmol PL. Particles had a ζ-potential in the range of −2.2 to −3.4 mV. Particles had a ζ-potential in the range of −2.4 to −3.4 mV. The size and surface charge of the nanoparticles were measured as a function of time. Both diameter and ζ-potential measures were identical after 1 month at 4 °C in phosphate-buffered saline, indicating a good stability of our liposomal formulations (Figure 1).
Figure 1.
Cartoon showing the novel technology, the ligand-targeted coated cationic liposomes for small interfering RNA (siRNA) delivery named TL[siRNA] and Fab′-TL[siRNA]. Diameter and ζ-potential of untargeted (CCL[siRNA]) and targeted liposomes entrapping siRNA resulted identical after 1 month at 4 °C in phosphate-buffered saline (PBS). CCL, coated cationic liposomes.
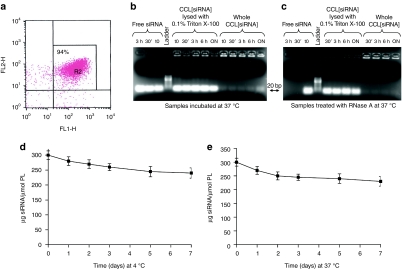
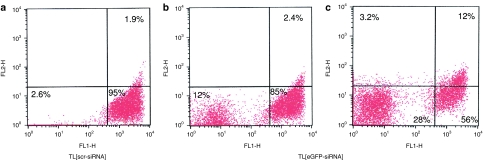
Entrapment of siRNA into liposomes was evaluated by labeling siRNA with 6-carboxyfluorescein (FAM) and liposomes with rhodamine-phosphoethanolamine (PE) and by analyzing them with fluorescence activated cell sorter (FACS). Figure 2a shows that ~94% of siRNA are entrapped into CCL. The loading capacity was ~10 nmol siRNA/µmol of total lipid, which is higher than, or comparable with, loading values obtained by others.30,31
Figure 2.
Evaluation of entrapment, stability, and release of small interfering RNA (siRNA) liposomal formulation. (a) siRNA entrapment into liposomes analyzed by fluorescence activated cell sorter (FACS) after labeling of siRNA with FAM (FL1-H) and liposomes with rhodamine-PE (FL2-H). (b) Stability over time of siRNA-entrapped into liposomes, CCL[ALK-siRNA] checked on agarose gel. Liposomes were incubated at 37 °C for different times, then, lysed with Triton X-100 and loaded (centre) in parallel with free ALK-siRNA (left side) and whole CCL[ALK-siRNA] (right side). Ladder: 10 basepairs ladder. (c) Stability over time of CCL[ALK-siRNA] pretreated with RNase A at 37 °C before lysis and checked on agarose gel. A clear prevention of siRNA lability was obtained by entrapment into CCL (centre) with weak leakage (right side) compared to free siRNA (left side). (d,e) Release of siRNA from liposomes was measured by dialysis of rhodamine-PE-labeled liposomes, containing FAM-labeled siRNA at 4 °C and 37 °C. ALK, anaplastic lymphoma kinase; CCL, coated cationic liposomes; FAM, 6-carboxyfluorescein; PE, phosphoethanolamine.
To test the lability of siRNA that may result in rapid degradation by serum nucleases, ALK-siRNA liposomal formulations, both untreated and treated with RNase A, were incubated at 37 °C for different times and checked on 2.2% agarose gels. In the absence of pretreatment with RNase A, lysis of liposomes with 0.1% Triton X-100 for 10 minutes at 37 °C showed that ALK-siRNA released from liposomes were intact (Figure 2b). Whole liposomes were retained in the gel wells while only a faint band of ~20 basepairs was detected, indicating a very modest presence of siRNA outside the carrier (Figure 2b). Figure 2c shows the effect of RNase A treatment on free ALK-siRNA and CCL[ALK-siRNA] incubated at 37 °C. Protection of siRNA from degradation was observed when they were entrapped into CCL, as compared to free ALK-siRNA, which were already degraded after 30 minutes of RNase treatment.
The stability of siRNA-entrapped liposomes was measured by dialyzing rhodamine-PE-labeled liposomes, containing FAM-labeled siRNA at 4 °C and 37 °C against 25% human plasma from healthy donors. The dialysis bag was sampled at intervals and the residual fluorescence of the liposomes measured by FACS analysis. In this assay, leakage of siRNA from liposomes was marginal, being not more than 10–20% after 1 week at both 4 °C and 37 °C (Figure 2d,e).
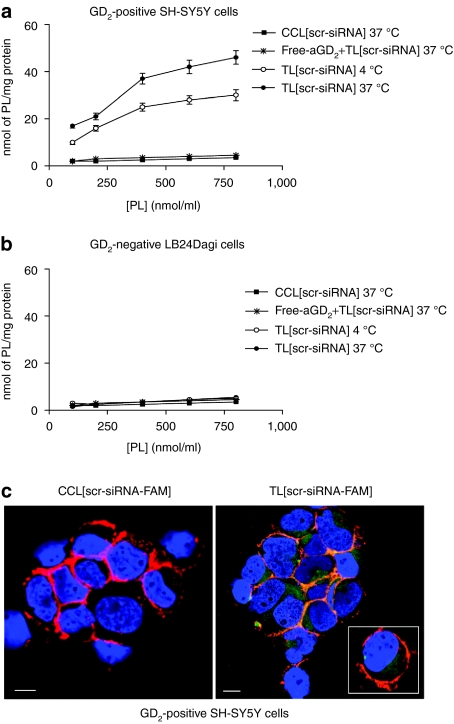
Next, we prepared anti-GD2-targeted liposomes, entrapping various siRNA, i.e., TL[ALK-siRNA], TL[scr-siRNA], TL[GAPDH-siRNA], or TL[eGFP-siRNA]. The binding of TL[scr-siRNA] to GD2-positive (SH-SY5Y) and GD2-negative (LB24Dagi) cells was studied by measuring uptake of PL from liposomes (Figure 3a,b). PL uptake by NB cells increased with increasing PL concentration, showing subsaturation at 400–600 nmol PL/ml at 4 °C; saturation of uptake was less evident at 37 °C. A significant increase in the level of cellular association was observed upon increasing the temperature from a temperature that is nonpermissive for endocytosis (4 °C) to a permissive temperature (37 °C); this provides an indication that the targeted particles are actively internalized by GD2-positive tumor cells. A similar liposome preparation that lacked the anti-GD2 MAb (CCL[scr-siRNA]), showed almost no PL uptake at either 4 °C or 37 °C. In a competition experiment, preincubation of cells for 30 minutes with soluble anti-GD2, blocked PL uptake by cells almost completely, suggesting that TL[scr-siRNA] binding occurred through specific antigen–antibody recognition. Conversely, GD2-negative cells showed very low PL uptake in all the examined cases (Figure 3b). Uptake of siRNA was evaluated by confocal microscopy analysis of the GD2-positive cell line SH-SY5Y, treated with CCL[scr-siRNA-FAM] or TL[scr-siRNA-FAM]. No uptake of CCL[scr-siRNA-FAM] was observed, while TL[scr-siRNA-FAM] (green) were effectively internalized (Figure 3c).
Figure 3.
Binding and uptake of TL[scr-siRNA] to (a) GD2-positive cells (SH-SY5Y) and (b) GD2-negative cells (LB24Dagi). The binding was demonstrated by measuring uptake of [3H]-CHE-labeled phospholipids (PL) from liposome preparations. We observed a significant dose-dependent increase for the extent of cellular association in cells treated with TL[scr-siRNA] at 4 °C (open circle) and 37 °C (closed circle), while the same liposome preparation without anti-GD2 MAb, CCL[scr-siRNA], showed very low PL uptake at 37 °C (closed square). Pretreatment of cells with free anti-GD2 MAb completely abolished PL uptake by cells (*). siRNA uptake in SH-SY5Y cells was evaluated by confocal microscopy analysis (c). While almost no uptake of untargeted liposomes, CCL[scr-siRNA-FAM], was observed, GD2-targeted liposomes, TL[scr-siRNA-FAM], were efficiently internalized. Green: TL[scr-siRNA-FAM] Red: TRITC-labeled aCD56, indicating the cell surface molecule N-CAM; Orange: merged colors resulting from siRNA-liposome binding to the cell surface; Blue: nuclear 4′ 6-diamidino-2-phenylindole (DAPI) staining. White bars = 50 µm. CCL, coated cationic liposomes; CHE, cholesterol; FAM, 6-carboxyfluorescein; siRNA, small interfering RNA.
In general, the results obtained from measurements of mean size, encapsulation efficiency, stability, and selective targeting of TL[siRNA] demonstrate physico-chemical features that meet the requirements of a targeted nonviral particle aiming at systemic gene silencing for cancer treatment.9,27
Pharmacokinetic profiles of anti-GD2-targeted liposomes entrapping siRNA
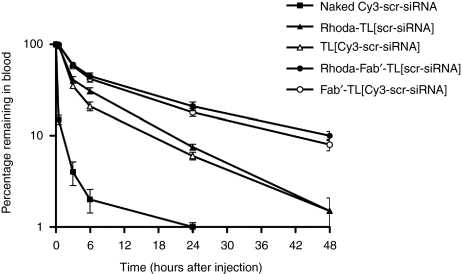
Long circulation times are required for small liposomes to gain access to tumor sites.23 To quantify the fate of both the liposome and the “incorporated drug,” we performed pharmacokinetic (PK) studies as described.20 Specifically, pharmacokinetics of rhoda-PE-TL[scr-siRNA], TL[Cy3-scr-siRNA], rhoda-PE-Fab′-TL[scr-siRNA], Fab′-TL[Cy3-scr-siRNA] and naked Cy3-scr-siRNA were evaluated and expressed as a percentage of the administered dose of either lipid (liposomes) or siRNA remaining in the blood (Figure 4). Both of the targeted liposomal formulations had good stability and long circulation times, with ~10 and 20% of the carriers and the siRNA remaining in the blood at 24 hours after administration of TL[scr-siRNA] and Fab′-TL[scr-siRNA], respectively. The liposomes and the siRNA cleared with similar slow clearance kinetics, indicating that the release rate of siRNA from the liposomes in vivo was slow, however, as expected,20 Fab′-targeted liposomes behave better than liposomal formulations targeted via whole antibody.
Figure 4.
Pharmacokinetics of TL[siRNA]. Data are expressed as a percentage of the administered dose of either lipid (liposomes) or siRNA remaining in the blood. Formulations used were: naked Cy3-labeled scrambled-siRNA (Cy3-scr-siRNA), Rhoda-PE-labeled liposomes entrapping scrambled-siRNA and targeted with whole antibody or with the Fab′ fragment (Rhoda-PE-TL[scr-siRNA] and Rhoda-PE-Fab′-TL[scr-siRNA]) or liposomes entrapping scrambled-siRNA-Cy3-labeled (TL[Cy3-scr-siRNA] and Fab′-TL[Cy3-scr-siRNA]). Liposomal formulations demonstrated to have long circulation times and slow clearance rates. PE, phosphoethanolamine; siRNA, small interfering RNA.
Activity of anti-GD2-targeted liposomes entrapping ALK-siRNA in vitro
The proof-of-concept for the efficiency of gene-specific silencing of our siRNA-delivery system were initially evaluated by FACS analysis of SH-SY5Y transducing eGFP, treated or not with TL[eGFP-siRNA]. Figure 5 shows that ~10% of viable cells had eGFP knockdown after 24 hours (Figure 5b) and ~30% after 48 hours (Figure 5c) of treatment with TL[eGFP-siRNA], relative to cells treated with TL[scr-siRNA], which maintained a high eGFP expression up to 48 hours (Figure 5a).
Figure 5.
Efficiency of gene-specific silencing evaluated by fluorescence activated cell sorter (FACS) analysis. SH-SY5Y transducing enhanced green fluorescent protein (eGFP) were treated with aGD2-targeted Rhoda-PE-labeled liposomes entrapping eGFP-directed siRNA, TL[eGFP-siRNA], or TL[scr-siRNA]. While cells treated with TL[scr-siRNA] kept high level eGFP expression at 48 hours from treatment (a), those treated with TL[eGFP-siRNA] after (b) 24 and (c) 48 hours led to a reduced eGFP expression in ~10 and 30% of viable cells, respectively. Expression of eGFP is evaluated on FL1-H, while rhodamine-PE on FL2-H channel. PE, phosphoethanolamine; siRNA, small interfering RNA.
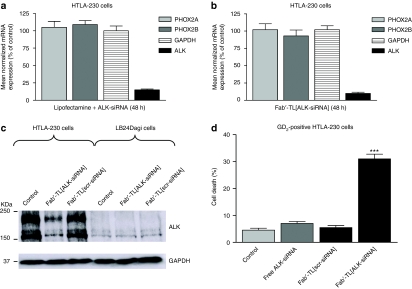
Among the three tested ALK-specific sequences, the most effective siRNA (siRNA-2) was determined by quantitative reverse transcriptase-PCR (qRT-PCR) and chosen for this study (Supplementary Figure S1). To test the feasibility of using the anti-GD2-targeted liposomes to achieve selective silencing in vitro, we transfected GD2+ HTLA-230 cells with lipofectamine RNAi MAX and ALK-siRNA or we incubated cells with Fab′-TL[ALK-siRNA]. The efficiency of ALK-specific silencing was quantitatively evaluated by qRT-PCR at 48 hours post-transfection. Figure 6b demonstrates that the TL formulation achieves a strong, sequence-specific down-modulation of ALK expression, comparable in effect to conventional transfection reagents for RNAi (Figure 6a). Neither expression of GAPDH, nor genes related to NB such as PHOX2A/B,32 was affected by silencing with ALK-siRNA. Western blots confirmed that Fab′-TL[ALK-siRNA] causes a very efficient knockdown of ALK protein, which was highly selective for GD2+ NB cells (Figure 6c). Fab′-TL[ALK-siRNA] also induced a significant increase in cell death in GD2+ cells (Figure 6d), but not in GD2− cells (data not shown). This confirms the high selectivity of Fab′-TL[siRNA] delivery, since the GD2− cell line, LB24Dagi, did not take up anti-GD2-targeted liposomes (Figure 3b).
Figure 6.
siRNA-mediated silencing of ALK and cell death induction in NB cell lines. (a,b) Effects of ALK knockdown evaluated by real-time RT-qPCR 48 hours post-transfection in HTLA-230 cells. A marked ALK knockdown is obtained by in vitro transfection with either ALK-directed siRNA and lipofectamine RNAi MAX (a) or Fab′-TL[ALK-siRNA] (b) demonstrating the high specificity of siRNA sequence, not affecting other tested genes. Relative gene expression analysis is reported on y-axis as expression percentage with respect to the scrambled control. Values are the mean ± 95% confidence intervals (CI) of N = 3 independent experiments performed in duplicate. (c) Western blot showing an extremely selective effects of ALK protein ablation obtained after transfection with Fab′-TL[ALK-siRNA] into GD2-positive HTLA-230 NB cells, but not in GD2-negative cells (LB24Dagi). Both native cell lines (control) and cells treated with Fab′-TL[scr-siRNA] were loaded as negative controls. (d) Fab′-TL[ALK-siRNA] selectively inhibits cell survival in GD2-positive HTLA-230 cells. Cell death after Fab′-TL[ALK-siRNA] treatment are reported on y-axis as percentage with respect to the control and the Fab′-TL[scr-siRNA]-treated cells. Values are the mean ± 95% CI of N = 3 independent assays performed in quadruplicate. ***P < 0.001 versus control. ALK, anaplastic lymphoma kinase; NB, neuroblastoma.
Inhibition of NB growth in vivo by anti-GD2-targeted liposomes entrapping ALK-siRNA
To evaluate the potential of Fab′-TL[ALK-siRNA] for in vivo applications, serum chemistries were investigated in mice administered i.v. with free siRNA or Fab′-TL[siRNA]. Fab′-TL[siRNA] were well-tolerated with no obvious toxicities, i.e., no weight loss or skin rash, no liver and renal toxicities (Supplementary Figure S2).
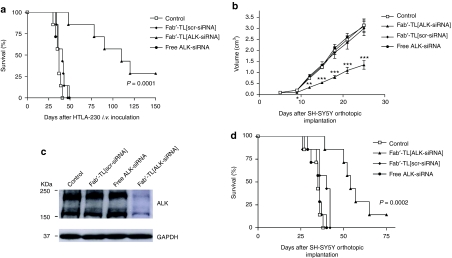
A pseudometastatic model of human NB in mice (i.v. administration of HTLA-230 cells, carrying WT ALK) was treated i.v. with various siRNA formulations. Mice treated with Fab′-TL[ALK-siRNA] liposomes had a significantly increased life span as compared to control mice, or to mice treated with Fab′-TL[scr-siRNA] (Figure 7a). At 150 days after implantation, ~30% of mice treated with Fab′-TL[ALK-siRNA] were still disease-free, while all mice treated with free siRNA or Fab′-TL[scr-siRNA] had died from metastatic disease.
Figure 7.
Tumor growth inhibition in vivo by Fab′-TL[ALK-siRNA]. Survival curves of NB-bearing mice in response to treatment with liposomal siRNA formulations. nu/nu (n = 7) (a) and SCID/bg (n = 8) (b) mice were treated, two-time a week for 1 week (a) or 3 weeks (b), with Fab′-TL[siRNA] formulations 24 hours after intravenous (i.v.) inoculation of HTLA-230 cells or 7 days after SH-SY5Y cells orthotopic implantation. (a) Treatment with Fab′-TL[ALK-siRNA] showed a significant increased life span compared to control and to Fab′-TL[scr-siRNA]-treated mice (P = 0.0001). (b) Fab′-TL[ALK-siRNA] significantly decrease tumor volume as compared to mice treated with the other formulations: *P < 0.05; **P < 0.01; ***P < 0.001 versus TL[scr-siRNA]-treated mice. (c) Western blot analysis of ALK in the tumor-bearing adrenal gland of treated mice. (d) Fab′-TL[ALK-siRNA] significantly increased life span as compared to mice treated with the other formulations, i.e., P = 0.0002 versus Fab′-TL[scr-siRNA]-treated mice. ALK, anaplastic lymphoma kinase; NB, neuroblastoma; siRNA, small interfering RNA.
Similar results were obtained in SCID/bg mice bearing orthotopic implanted SH-SY5Y cells (carrying F1174L ALK mutation). Indeed, a statistically significant inhibition of tumor volume was already observed after the second injection of Fab′-TL[ALK-siRNA] (day 10), and this improved significantly with further treatments (Figure 7b). One day after the last treatment (day 25), ALK protein was down-modulated in the tumor specimens derived from mice treated with Fab′-TL[ALK-siRNA] (Figure 7c), suggesting that knockdown of ALK signaling contribute strongly to tumor cell kill in our model. Finally, mice receiving Fab′-TL[ALK-siRNA] showed a significant improvement in long term survival compared with all other groups (Figure 7d).
Discussion
High-risk NB patients are currently treated with radiotherapy, intensive chemotherapy, autologous stem-cell rescue, and 13-cis-retinoic acid or monoclonal antibodies targeting NB-specific proteins. The 3-year event-free survival rate is 34%.33,34 Moreover, due to limiting factors linked to intensive induction therapy such as acute and chronic toxicity and the development of secondary neoplasias,35 the overall outcome for high-risk refractory or recurrent NB patients remains very poor. Thus, development of novel antitumor strategies is essential to improve the poor survival rates of high-risk patients.
On the basis of ALK involvement in NB tumor growth and proliferation,1,2,3,4 in this study we tested the therapeutic efficacy of targeting ALK protein in NB cells. To this end, we developed liposomal nanoparticles for the selective delivery of ALK-directed siRNA to NB tumor sites. This new siRNA-based “targeted drug” significantly inhibited tumor growth in two different NB animal models, further suggesting its subsequent clinical application for pediatric NB.
A pseudometastatic model was chosen for our investigation of the therapeutic efficacy of targeted ALK-directed siRNA, since NB cells injected i.v. in mice mimic the metastatic spread observed in advanced stage NB patients.19,36 Circulating NB cells in the blood and micrometastases in the bone marrow at the time of primary surgery of NB patients are strong predictors of relapse and, since they express high levels of the cell surface internalizing receptor disialoganglioside GD2,16,37 they are excellent targets for nanomedicine delivery systems. Furthermore, since bone marrow micrometastases are a direct measurement of the ability of tumor cells to spread systemically, the establishment of a model that closely mimics the clinical situation allows a more realistic evaluation of antitumor therapies. The treatment schedule, beginning at 24 hours after NB cells injection, was deliberately chosen to allow evaluation of the effects of targeted-siRNA during the metastatic cascade, i.e., during the stages in which tumor cells are in intravascular circulation and endothelium-attachment takes place, or when extravasation, stromal invasion, and colonization takes place.19,21
However, an orthotopic model was also used in our studies, as it better reflects the growth of advanced NB in children,38,39 allowing a clinically relevant proof-of-concept and a realistic evaluation of the clinical potential of ALK-targeted siRNA therapies in NB. Interestingly, ALK knockdown in orthotopic, as well as in pseudometastatic NB models, led to significant tumor growth inhibition, independent of ALK mutational status.
RNAi technology is currently one of the most promising classes of nucleic acid molecules for the next generation of pharmaceutical drugs. However, effective delivery of RNAi therapeutics remains a key challenge for the advancement of this technology.9,12,13 The various obstacles for RNAi delivery in vivo include: (i) the lability of siRNA that results in its rapid degradation by serum nucleases, (ii) poor membrane permeability of siRNA, limiting cellular uptake, (iii) quick glomerular filtration of free (i.e., nonencapsulated) siRNA, resulting in rapid clearance and poor tumor uptake, (iv) the need to avoid siRNA-mediated activation of the innate immune response, and (v) the need to achieve efficient intracellular delivery to target cells in vivo avoiding nonspecific distribution. We believe that the development of the herein described targeted nanoparticles, which entrap high levels of siRNA, may be an effective platform technology, when used with other appropriate targeting ligands, for the extra-hepatic application of siRNA therapeutics in a variety of applications.
The present study shows that targeted lipidic nanoparticles as carriers for siRNA result in an extremely high sequence-specificity and an efficient gene knockdown, without “off-target” effects, and, once packaged, siRNA are stable, resistant to degradation by extrinsic RNAses, and only slowly released by nanoparticles. Furthermore, we demonstrated that liposomes with entrapped siRNA can be specifically targeted to proteins on tumor cell surfaces via surface-grafted antibodies (although other ligands could be used), and that specific targeting and internalization of nanocarriers is essential to efficiently kill tumor cells. Targeting of nucleic acids via internalizing carriers will be enabling the delivery and transfection of tumor cells such as NB cells that are difficult to transfect.40,41 Our work also shows a substantial ALK knockdown, which results in a significant inhibition of xenografted tumor growth in vivo, when nanoparticles entrapping the gene-specific siRNA were targeted to NB cells via anti-GD2 monoclonal antibodies.
Some potential limitations of this study should be considered. Targeted nanoparticles appear to lose their advantage when attempts are made to treat established more advanced solid tumors, probably because the “binding site barrier” and focal distribution result in incomplete penetration of targeted-CCL into the tumor.42 Moreover, the use of selective liposomal formulations of ALK-specific siRNA in a clinical setting will most likely be combined with small molecule inhibitors of ALK and/or chemotherapeutic agents, and this issue is the subject of ongoing studies.
In conclusion, we believe that we have demonstrated proof-of-principle for the extra-hepatic delivery of high levels of siRNA via targeted siRNA-containing nanoparticles, This delivery system may extend the scale and diversity of available delivery solutions for all GD2-expressing cancer cells and beyond. Intravenous delivery of NB-targeted nanoparticles, encapsulating ALK-siRNA, to human NB mouse models is able to exert a highly specific ALK knockdown and was well-tolerated and effective. These results may suggest the rational design of selectively targeted anticancer strategies using siRNA-based therapeutics as an adjuvant therapy for advanced stage NB disease, for early micrometastatic lesions, or for treating residual disease following debulking. Although in this study we focused on ALK gene and GD2-positive tumor cells, this is a platform technology that is widely applicable for siRNA delivery. It can be used to design a variety of personalized molecular targeted therapies with a suitable combination of RNAi molecules, choosing selective surface markers depending on the specific molecular signature of each tumor type. In this respect, other neuroectoderma-derived cancers, such as melanoma and osteosarcoma, and additional human tumors expressing abundant amounts of the GD2 antigen, or a variety of other well-characterized internalizing surface antigens, may benefit from this novel and versatile therapeutic strategy.
Materials and Methods
Reagents and chemicals. Hydrogenated soy phosphatidylcholine, cholesterol (CHE), 1,2-distearoyl-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG2000), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] modified with a maleimido group at the distal terminus chain (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000], DSPE-PEG2000-MAL), 1,2-dioleoy-1-3-trimethylammonium-propane (DOTAP), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine Rhodamine B Sulfonyl) (Rhoda-PE) were purchased from Avanti Polar Lipids (Alabaster, AL).
Nuclepore polycarbonate membranes (pore sizes: 0.4, 0.2, and 0.1 µm) were purchased from Avestin (Ottawa, ON, Canada). 2-Iminothiolane (Traut's reagent) was obtained from Sigma (Sigma Chemical, St Louis, MO). Protein A/G column, ImmunoPure IgG elution buffer was purchased from Thermo Scientific-Pierce (Rockford, IL). Bio-Rad Protein Assay Reagent was purchased from Bio-Rad Laboratories (Mississauga, ON, Canada). Sephadex G-50, Sepharose CL-4B and aqueous counting scintillant were purchased from Amersham Pharmacia Biotech (Baie d'Urfe, Qubec, Canada). Cholesteryl-[1,2-3H-(N)]hexadecyl ether ([3H]CHE; 1.48–2.22 TBq/mmol) was purchased from Perkin-Elmer Biosciences (Boston, MA).
The hybridoma cell line 14.G2a, which secretes a murine immunoglobulin G2a specific for the GD2 antigen, was a generous gift of R. A. Reisfeld (The Scripps Institute, La Jolla, CA).
All other chemicals were of analytical grade purity or the highest available purity.
Cell culture. Two human GD2-expressing NB cell lines, namely HTLA-230 (ALK WT) and SH-SY5Y (carrying the F1174L ALK mutation) were used and grown as described.24 The GD2-negative control: LB24Dagi cell line, derived from melanoma cells, kindly provided by Dr D. Castiglia (Istituto Dermopatico dell'Immacolata, Department of Molecular and Cellular Biology, Rome, Italy). SH-SY5Y cells, stably transfected with pcDNA3.1eGFP vector, were kindly provided by Dr G.H. Fey and Dr C. Kellner (University of Erlangen-Nuremberg, Erlangen, Germany). Only cells in the exponential phase of cell growth were used during experiments. All cell lines were tested for mycoplasma contamination, cell proliferation, and morphology evaluation, both after thawing and within four passages in culture.
Transfection of siRNA in NB cell lines. In vitro transfection conditions for gene knockdown by siRNA were optimized using FAM-labeled GAPDH siRNA (SilencerSelect; Ambion Applied Biosystems, Austin, TX) in six-well plates (2 × 105 cells) for HTLA-230 and SH-SY5Y cells, transfected via Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) at different molarities. Transfection efficiencies were evaluated by either fluorescence microscopy or FACS. For each candidate gene, three SilencerSelect predesigned gene-specific siRNA (Ambion Applied Biosystems) were tested in parallel with a scrambled control (Silencer Negative Control #1; Ambion Applied Biosystems), a housekeeping positive control (SilencerSelect GAPDH siRNA; Ambion Applied Biosystems), a blank with the transfection agent only, and the native cells. The most effective siRNA for ALK was selected (siRNA-2 for ALK: ID # s1271: Sense 5′->3′ CAAACCAGUUAAUCCAGAAtt, Antisense 5′->3′ UUCUGGAUUAACUGGUUUGta; Ambion Applied Biosystems) for further experiments and preparation of targeted liposomal siRNA (TL[siRNA], see below). Transfection was carried out at 100 nmol/l siRNA in Dulbecco's modified Eagle's medium without antibiotics in either adherent cells (forward transfection), in serum free Dulbecco's modified Eagle's medium without antibiotics (stopped after 14 hours with complete medium), or freshly harvested cells in suspension (reverse transfection). The efficiency of gene silencing and downstream effect on other genes was evaluated at 24, 48, and 72 hours post-transfection by real-time reverse transcriptase-quantitative PCR and at 72 hours by western blot.
In vitro transfection conditions of NB cells by targeted nanoliposomes (TL) were optimized for the GD2-positive cell line, HTLA-230, and for the GD2-negative cell line, LB24Dagi. Final siRNA concentrations were 750 nmol/l siRNA/750 nmol/l PL, which mimicked the concentration of siRNA-liposomal formulations used for in vivo studies.
Experiments to evaluate knockdown after siRNA treatment of cells (reduction of mRNA) were carried out in six well plates (~2 × 105 cells); protein knockdown was evaluated in 10 mm Petri dishes (~1 × 106 cells). In either case, native cells, as well as cells treated with TL[siRNA], were incubated at 37 °C with moderate shaking for 2 hours in round bottom tubes with small volumes to allow the formation of cell–liposome complexes. Liposomal formulations used were: TL[ALK-siRNA], TL[scr-siRNA] as a negative control, and TL[GAPDH-siRNA] as positive control of transfection (only for RT-qPCR). After 2 hours, cells were seeded onto their respective culture plates. Cells for mRNA evaluation were harvested 48 hours post-treatment, while those for protein evaluation underwent a second treatment at 48 hours and were harvested at 72 hours.
The efficiency of gene silencing and downstream effects on other genes was evaluated at 48 hours post-transfection by real-time RT-qPCR and at 72 hours by western blot.
TL[siRNA] preparation (GD2-targeted CCL entrapping siRNA). Reimer and colleagues showed that plasmid DNA could be taken up into organic solvents when cationic lipid was present, presumably forming inverted micelles in the organic solvent with the plasmid inside the inverted micelle and the acyl chains of the cationic lipid oriented towards the organic phase.43 On the basis of this assumption, we encapsulated siRNA in small and stable CCL, which are prepared and purified following the method of Stuart and colleagues29 slightly modified by us22,24,25 after giving consideration to the number of negative charges of double-stranded RNA molecules (21 mers). Briefly, siRNA were complexed with cationic lipids and coated with neutral lipids as follows: 1,000 µg of siRNA in 500 µl of distilled-deionized water (ddH2O) was mixed with 3.4 µmol of DOTAP in 500 µl of CHCl3 and 1,060 µl of MeOH to form a Bligh–Dyer monophase. After 30 minutes at room temperature, 500 µl of CHCl3 and 500 µl of ddH2O were added. The mixture was vortexed and centrifuged at 800g for 7 minutes at room temperature. The upper aqueous methanol layer phase was removed. Under these conditions, which were developed in pilot studies using 10 µg siRNA and various amounts of DOTAP, 90–96% of the siRNA was recovered in the organic phase. The resulting siRNA-to-lipid, positive-to-negative charge ratio was 1:1. Following the extraction, 8.6 µmol cholesterol and appropriate amounts of hydrogenated soy phosphatidylcholine, DSPE-PEG2000, or DSPE-PEG2000-MAL, to give cholesterol-to-PL mole ratios of 1:2, were added to the organic phase. ddH2O was added to give a final PL concentration of 20 mmol/l. The mixture was sonicated for 5 minutes and the organic phase was removed by rotary evaporation under a stream of N2 until the system reverted to the aqueous phase. The liposomes were sequentially extruded through 400, 200, and 100 nm polycarbonate filters. Liposome size, polydispersity, and ζ-potential were analyzed by dynamic light scattering using the Zeta sizer Nano-S particle sizer at a fixed angle (90°) (Malvern Instruments, Worcestershire, UK). Any free siRNA was separated from liposomes by passing liposomes over a Sephadex G-50 column pre-equilibrated in HEPES-buffered saline (25 mmol/l HEPES, 140 mmol/l NaCl, pH 7.4). Finally, anti-GD2 monoclonal antibody or its Fab′ fragment was coupled to the maleimide terminus of DSPE-PEG2000-MAL as previously described22,44 to give a novel anti-GD2-targeted nanoparticle formulation entrapping siRNA. The amount of siRNA encapsulated in CCLs was evaluated by solubilizing liposomal preparation with 40 mmol/l sodium deoxycholate for 1.5 hours at room temperature followed by spectrophotometer measurement at 260 nm.
In some preparations, cholesteryl-[1,2-3H-(N)]-hexadecyl ether ([3H]-CHE) is added as a nonexchangeable, nonmetabolizable lipid tracer. In some experiments, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine Rhodamine B Sulfonyl) (rhoda-PE) is used at 1 mol% of total PLs as a fluorimetric lipid tracer.
Cellular binding and uptake studies in vitro. Cells were incubated for 1 hour at 4 °C and 37 °C (nonpermissive and permissive temperature, respectively, for receptor internalization) with [3H]-CHE-labeled targeted liposomes at concentrations from 0.1 to 0.8 µmol PL/ml. After extensive washing, cells were trypsinized and lysed with 1 N NaOH for evaluation of protein and for β-counting. Competition for binding to tumor cells between free Ab and liposomes-coupled specific mAb (anti-GD2) was determined by addition of 50-fold excess of free mAb for 30 minutes before adding [3H]-CHE-labeled TL.22,24
The uptake and internalization of liposome-encapsulated FAM-labeled scrambled-siRNA into GD2-expressing cells was detected by fluorescence confocal microscope. Cells were trypsinized, counted, and incubated at 37 °C for various times, with either untargeted or GD2-targeted liposomes. After washing and cytospin centrifugation, the samples were fixed and stained with a monoclonal antibody specific for the cellular adhesion molecule N-CAM (a-CD56 Zymed; Invitrogen) to reveal plasma membrane localization. Binding of the primary antibody was detected with tetramethylrhodamine isothiocyanate (PE)–conjugated rabbit anti-mouse (Sigma) and cell nuclei were stained with 4′ 6-diamidino-2-phenylindole. The cellular distribution of FAM (green), PE (red) and 4′ 6-diamidino-2-phenylindole (blue) fluorescence was analyzed with a laser scanning spectral confocal microscope TCS SP2 AOBS (Leica, Heidelberg, Germany).
siRNA stability and release. To test the lability of siRNA resulting from degradation by serum nucleases, ALK-siRNA liposomal formulations (TL[ALK-siRNA]) were incubated at 37 °C for different times, with or without RNase A (2 µg/ml), washed, lysed with 0.1% Triton X-100 and then were run on a 2.2% agarose (Invitrogen) gel.
Entrapment in Rhoda-PE-labeled liposomes of scr-siRNA and FAM-scr-siRNA (10% of total) (Silencer Negative Control #1, Ambion Applied Biosystems) was assessed and analyzed using a FACS (FACS Calibur; Beckton Dickinson, San Josè, CA) equipped with a xenon lamp. FAM and rhodamine fluorescence intensities were recorded through the reading channel FL1-H and FL2-H, respectively.
Leakage of entrapped siRNA from liposomes labeled with [3H]-cholesterol was measured by dialyzing the TL[siRNA] against either 25% human plasma from healthy donors or complete medium at 4 °C and 37 °C, using tubes with a molecular mass cutoff of 100,000 daltons. Samples were taken at intervals for measurements of radioactivity and siRNA.
Primer design and gene expression analysis by real-time RT-qPCR. Total RNA was extracted from cell lines of interest by RNeasy mini and micro kit (QIAGEN, Hilden, Germany). Analysis of mRNA expression levels of the target genes and the positive control (GAPDH) were carried out by a two-step real-time RT-qPCR, using a random priming-based reverse-transcription (High Capacity cDNA Reverse Transcription Kit; Ambion Applied Biosystems) and SYBR Green I binding dye (Platinum SYBR Green qPCR SuperMix-UDG; Invitrogen), according to the manufacturers' conditions. Primers were designed using Primer 3 software (http://fokker.wi.mit.edu/primer3/input.htm).45 Primer amplification efficiencies, compared to a standard curve, were assayed in triplicates of each cDNA sample (12.5 ng), amplified in the iCycler (Bio-Rad Laboratories, Hercules, CA) following an initial denaturation at 95 °C for 2 minutes, then 50 cycles at 95 °C for 15 seconds, and 60 °C for 30 seconds. Melting curves were calculated between 55 °C and 95 °C, with an increment of 0.5 °C every 15 seconds. PCRs were repeated at least twice.
Three human genes (POLR2A, POLR3F; NDUFB3) were chosen as endogenous controls, and data normalization was performed using BestKeeper software (http://www.gene-quantification.de/bestkeeper.html). For relative quantification of gene expression in native cell lines, we prepared an equimolar pool of RNAs from eight normal tissues of different embryonic origin (adrenal gland, bladder, breast, brain, colon, lung, placenta, and prostate), FirstChoice Human Normal Tissue Total RNA (Ambion Applied Biosystems), was employed as reference sample. In transfected cells, gene expression level was compared to appropriate negative controls (i.e., native cells, transfectant agent only, scrambled transfected cells).
The comparative Ct method was adopted for relative quantification of gene expression in native and transfected cell lines and data were analyzed by qGene software (http://www.gene-quantification.de/download.html#qgene),46 corrected for amplification efficiencies.
Western blot analysis. Inhibition of gene expression was checked at the protein level by western blotting from whole cell lysates of cultured cells or in tumor specimens of orthotopic NB-bearing mice, using specific antibodies, as previously reported.23,24,47 For immunoblotting, proteins were separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis and transferred to nitrocellulose, incubated with the specific antibody. Immune complexes were detected with sheep-anti-mouse Ig antibody conjugated to horseradish peroxidase (Amersham), and visualized by enhanced chemiluminescence reagent (Amersham) according to the manufacturer's instructions.
The membranes were incubated with monoclonal antibodies against ALK (1:1,000, clone C26G7; Cell Signaling Technology, Danvers, MA) and the housekeeping GAPDH (1:1,000, clone 14C10; Cell Signaling Technology).
Cell viability assay following TL[ALK-siRNA] treatment. The in vitro cytotoxicity of free siRNA or Fab′-TL[ALK-siRNA] was evaluated, as previously described.48 Assays were carried out in quadruplicate, at the conditions and concentrations used for in vitro transfection of siRNA, adapted for 96 well plates. Treatments were scheduled at time 0, 48, and 96 hours. At 120 hours cells were harvested, washed with complete medium, and incubated with trypan blue (0.04%, Sigma) for 1 minute at 37 °C and results were evaluated.
Animal models. All animals were purchased from Harlan Laboratories (Harlan Italy, S.Pietro al Natisone, Udine, Italy) and housed under pathogen-free conditions. Experiments on animals were reviewed and approved by the licensing and ethical committee of the National Cancer Research Institute, Genoa, Italy, and by the Italian Ministry of Health. All experiments in vivo were performed with 7–8 mice per group and repeated twice. Body weight and general physical status of the animals were recorded daily, and mice were killed by cervical dislocation after the administration of xilezine (Xilor 2%; Bio98, Milan, Italy), when they showed signs of poor health, such as abdominal dilatation, dehydration, or paraplegia.
For the pseudometastatic animal model, 4-week-old female nu/nu mice were injected i.v. in the tail vein with 3.5 × 106 HTLA-230 tumor cells, as described.19,21 In the orthotopic animal model, 5-week-old female SCID/bg mice were anesthetized with ketamine (Imalgene 1000; Merial Italia, Milan, Italy), subjected to laparotomy, and injected with 1.5 × 106 SH-SY5Y cells in the capsule of the left adrenal gland, as described.38,49 No mice died as a result of this treatment. Mice were monitored at least twice weekly for evidence of tumor development and were killed as above. Tumor expansion over time, as well as the response to treatment were measured by calipers, as described.23
Pharmacokinetic experiments. Athymic (nu/nu) mice were injected via the tail vein with a single dose of: Rhoda-PE-TL[scr-siRNA], Rhoda-PE-Fab′-TL[scr-siRNA], Cy3-labeled scr-siRNA entrapped into TL and into Fab′-TL, (0.8 µmol PL/mouse), and naked Cy3-labeled scr-siRNA (Cy3-scr-siRNA). At selected time points (2, 12, 24, and 48 hours) postinjection, mice (three/group) were anaesthetized, blood was collected through the retro-orbital sinus from each mouse into a tube containing heparin (2 IU/ml; Parke-Davis, Milan, Italy) and the mice were killed. Blood was centrifuged (at 500g for 10 minutes at 4 °C), and 100 µl of the isolated plasma was analyzed for the Rhoda-PE and Cy3 labels using a fluorescence plate reader (INFINITE M2000 Monochromator Instrument; TECAN, Grodig/Salzburg, Austria). Cy3 and Rhoda-PE excitation/emission were 547/563 nm and 557/571, respectively. Pharmacokinetic data are expressed as the percentage of the injected dose remaining in blood at the various time points postinjection.
In vivo therapeutic studies. For the pseudometastatic model, obtained by inoculation of HTLA-230 cells, mice were treated i.v. 24 hours postinoculation of tumor cells with free ALK-siRNA, Fab′-TL[ALK-siRNA], and Fab′-TL[scr-siRNA] (1.25 mg/kg siRNA) twice a week for a total of 1 week. For the orthotopic model, obtained by implantation of SH-SY5Y cells, tumors were allowed to grow from the implanted cells for 7 days, before treatments started, as above, but for a total of six times. Control mice received weekly injections of HEPES-buffered saline. Experiments were performed twice, and mice were monitored routinely for weight loss and were killed when signs of poor health became evident. Survival times were used as the main criterion for determination of treatment efficacy in the pseudometastatic model, while tumor expansion over time, as well as the response to treatment were measured by calipers in the orthotopic one, as described.23,38 In this latter model, the day after the last treatment (25 day), three mice per group were anaesthetized with xylezine, killed, and tumors were recovered for western blot analyses.
Systemic toxicity. To detect liver and renal toxicity, athymic mice were treated every other day for six total doses with 1.5 mg/kg of siRNA, free or encapsulated into Fab′-TL, and blood samples evaluated 3 hours after the first injection (acute), and 1 day after the end of treatment (chronic) using commercial kits (Fisher Scientific, Pitsburgh, PA) according to the manufacturer's instructions.
Statistical methods. Results are expressed as mean ± 95% confidence intervals. The statistical significance of differential findings between experimental groups and controls was determined by ANOVA, with the Tukey's multiple comparison test, using Graph-Pad Prism 3.0 software (GraphPad Software, San Diego, CA). The significance of the differences between experimental groups (n = 7–8 mice/group) in the survival experiments was determined by Kaplan–Meier curves and the χ2 log-rank test (Graph-Pad Prism 3.0). Findings were considered significant if P values were <0.05.
SUPPLEMENTARY MATERIAL Figure S1. Efficiency and specificity of gene knockdown over time evaluated in SH-SY5Y by RT-qPCR. Figure S2. Liver and renal toxicity studies.
Acknowledgments
We thank Dr Georg Fey and Dr Christian Kellner for providing us with eGFP-transfected SH-SY5Y cell line. We thank Vito Pistoia for critical reading of the manuscript; Pamela Beccherini and Roberto Taverniti for technical help. The work was supported by Associazione Italiana Ricerca Cancro (AIRC grant IG 5505), Italian Ministry of Health (Programma integrato per la ricerca oncologica), and by the European Commission FP7 program grant “INFLA-CARE” (EC contract number 223151; http://inflacare.imbb.forth.gr/) to M.P. and grant ERC-2009-StG (Proposal number 242965-“Lunely”) to R.C. D.D.P. is a recipient of a FIRC fellowship. The authors do not have financial conflict of interest.
Supplementary Material
Efficiency and specificity of gene knockdown over time evaluated in SH-SY5Y by RT-qPCR.
Liver and renal toxicity studies.
REFERENCES
- Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF.et al. (2008Identification of ALK as a major familial neuroblastoma predisposition gene Nature 455930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M.et al. (2008Oncogenic mutations of ALK kinase in neuroblastoma Nature 455971–974. [DOI] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Fröhling S, Luther W., 2nd, , Zhang J.et al. (2008Activating mutations in ALK provide a therapeutic target in neuroblastoma Nature 455975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V.et al. (2008Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma Nature 455967–970. [DOI] [PubMed] [Google Scholar]
- Carén H, Abel F, Kogner P., and, Martinsson T. High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem J. 2008;416:153–159. doi: 10.1042/bj20081834. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Vernersson E, Grabbe C., and, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passoni L, Longo L, Collini P, Coluccia AM, Bozzi F, Podda M.et al. (2009Mutation-independent anaplastic lymphoma kinase overexpression in poor prognosis neuroblastoma patients Cancer Res 697338–7346. [DOI] [PubMed] [Google Scholar]
- Webb TR, Slavish J, George RE, Look AT, Xue L, Jiang Q.et al. (2009Anaplastic lymphoma kinase: role in cancer pathogenesis and small-molecule inhibitor development for therapy Expert Rev Anticancer Ther 9331–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Van Buren G, Dallas NA, Xia L, Wang X, Yang AD.et al. (2008Therapeutic targeting of neuropilin-2 on colorectal carcinoma cells implanted in the murine liver J Natl Cancer Inst 100109–120. [DOI] [PubMed] [Google Scholar]
- MacDiarmid JA, Amaro-Mugridge NB, Madrid-Weiss J, Sedliarou I, Wetzel S, Kochar K.et al. (2009Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug Nat Biotechnol 27643–651. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Bar-Eli M., and, Sood AK. The dicey role of Dicer: implications for RNAi therapy. Cancer Res. 2010;70:2571–2574. doi: 10.1158/0008-5472.CAN-09-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E.et al. (2010Sustained small interfering RNA delivery by mesoporous silicon particles Cancer Res 703687–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V.et al. (2010Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study Lancet 3751896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK.et al. (2010Rational design of cationic lipids for siRNA delivery Nat Biotechnol 28172–176. [DOI] [PubMed] [Google Scholar]
- Mujoo K, Cheresh DA, Yang HM., and, Reisfeld RA. Disialoganglioside GD2 on human neuroblastoma cells: target antigen for monoclonal antibody-mediated cytolysis and suppression of tumor growth. Cancer Res. 1987;47:1098–1104. [PubMed] [Google Scholar]
- Schulz G, Cheresh DA, Varki NM, Yu A, Staffileno LK., and, Reisfeld RA. Detection of ganglioside GD2 in tumor tissues and sera of neuroblastoma patients. Cancer Res. 1984;44 12 Pt 1:5914–5920. [PubMed] [Google Scholar]
- Cheresh DA, Harper JR, Schulz G., and, Reisfeld RA. Localization of the gangliosides GD2 and GD3 in adhesion plaques and on the surface of human melanoma cells. Proc Natl Acad Sci USA. 1984;81:5767–5771. doi: 10.1073/pnas.81.18.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino F, Brignole C, Marimpietri D, Sapra P, Moase EH, Allen TM.et al. (2003Doxorubicin-loaded Fab′ fragments of anti-disialoganglioside immunoliposomes selectively inhibit the growth and dissemination of human neuroblastoma in nude mice Cancer Res 6386–92. [PubMed] [Google Scholar]
- Pastorino F, Brignole C, Di Paolo D, Nico B, Pezzolo A, Marimpietri D.et al. (2006Targeting liposomal chemotherapy via both tumor cell-specific and tumor vasculature-specific ligands potentiates therapeutic efficacy Cancer Res 6610073–10082. [DOI] [PubMed] [Google Scholar]
- Raffaghello L, Pagnan G, Pastorino F, Cosimo E, Brignole C, Marimpietri D.et al. (2003In vitro and in vivo antitumor activity of liposomal Fenretinide targeted to human neuroblastoma Int J Cancer 104559–567. [DOI] [PubMed] [Google Scholar]
- Pastorino F, Stuart D, Ponzoni M., and, Allen TM. Targeted delivery of antisense oligonucleotides in cancer. J Control Release. 2001;74:69–75. doi: 10.1016/s0168-3659(01)00312-1. [DOI] [PubMed] [Google Scholar]
- Pastorino F, Brignole C, Marimpietri D, Pagnan G, Morando A, Ribatti D.et al. (2003Targeted liposomal c-myc antisense oligodeoxynucleotides induce apoptosis and inhibit tumor growth and metastases in human melanoma models Clin Cancer Res 94595–4605. [PubMed] [Google Scholar]
- Pagnan G, Stuart DD, Pastorino F, Raffaghello L, Montaldo PG, Allen TM.et al. (2000Delivery of c-myb antisense oligodeoxynucleotides to human neuroblastoma cells via disialoganglioside GD2-targeted immunoliposomes: antitumor effects J Natl Cancer Inst 92253–261. [DOI] [PubMed] [Google Scholar]
- Brignole C, Pastorino F, Marimpietri D, Pagnan G, Pistorio A, Allen TM.et al. (2004Immune cell-mediated antitumor activities of GD2-targeted liposomal c-myb antisense oligonucleotides containing CpG motifs J Natl Cancer Inst 961171–1180. [DOI] [PubMed] [Google Scholar]
- Brignole C, Marimpietri D, Pastorino F, Di Paolo D, Pagnan G, Loi M.et al. (2009Anti-IL-10R antibody improves the therapeutic efficacy of targeted liposomal oligonucleotides J Control Release 138122–127. [DOI] [PubMed] [Google Scholar]
- Allen TM., and, Cullis PR. Drug delivery systems: entering the mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- Fenske DB., and, Cullis PR. Liposomal nanomedicines. Expert Opin Drug Deliv. 2008;5:25–44. doi: 10.1517/17425247.5.1.25. [DOI] [PubMed] [Google Scholar]
- Stuart DD, Kao GY., and, Allen TM. A novel, long-circulating, and functional liposomal formulation of antisense oligodeoxynucleotides targeted against MDR1. Cancer Gene Ther. 2000;7:466–475. doi: 10.1038/sj.cgt.7700145. [DOI] [PubMed] [Google Scholar]
- Santos AO, da Silva LC, Bimbo LM, de Lima MC, Simões S., and, Moreira JN. Design of peptide-targeted liposomes containing nucleic acids. Biochim Biophys Acta. 2010;1798:433–441. doi: 10.1016/j.bbamem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN.et al. (2006RNAi-mediated gene silencing in non-human primates Nature 441111–114. [DOI] [PubMed] [Google Scholar]
- Longo L, Panza E, Schena F, Seri M, Devoto M, Romeo G.et al. (2007Genetic predisposition to familial neuroblastoma: identification of two novel genomic regions at 2p and 12p Hum Hered 63205–211. [DOI] [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R., and, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Matthay KK, Quach A, Huberty J, Franc BL, Hawkins RA, Jackson H.et al. (2009Iodine-131–metaiodobenzylguanidine double infusion with autologous stem-cell rescue for neuroblastoma: a new approaches to neuroblastoma therapy phase I study J Clin Oncol 271020–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesundheit B, Moser A, Or R., and, Klement G. Successful antiangiogenic therapy for neuroblastoma with thalidomide. J Clin Oncol. 2007;25:5321–5324. doi: 10.1200/JCO.2007.12.3331. [DOI] [PubMed] [Google Scholar]
- Raffaghello L, Marimpietri D, Pagnan G, Pastorino F, Cosimo E, Brignole C.et al. (2003Anti-GD2 monoclonal antibody immunotherapy: a promising strategy in the prevention of neuroblastoma relapse Cancer Lett 197205–209. [DOI] [PubMed] [Google Scholar]
- Handgretinger R, Baader P, Dopfer R, Klingebiel T, Reuland P, Treuner J.et al. (1992A phase I study of neuroblastoma with the anti-ganglioside GD2 antibody 14.G2a Cancer Immunol Immunother 35199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino F, Brignole C, Marimpietri D, Cilli M, Gambini C, Ribatti D.et al. (2003Vascular damage and anti-angiogenic effects of tumor vessel-targeted liposomal chemotherapy Cancer Res 637400–7409. [PubMed] [Google Scholar]
- Pastorino F, Di Paolo D, Loi M, Becherini P, Caffa I, Zorzoli A.et al. (2009Recent advances in targeted anti-vasculature therapy: the neuroblastoma model Curr Drug Targets 101021–1027. [DOI] [PubMed] [Google Scholar]
- Giles RV, Spiller DG, Grzybowski J, Clark RE, Nicklin P., and, Tidd DM. Selecting optimal oligonucleotide composition for maximal antisense effect following streptolysin O-mediated delivery into human leukaemia cells. Nucleic Acids Res. 1998;26:1567–1575. doi: 10.1093/nar/26.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrias MV, Guarnaccia F., and, Ponzoni M. Bioavailability of antisense oligonucleotides in neuroblastoma cells: comparison of efficacy among different types of molecules. J Neurooncol. 1997;31:171–180. doi: 10.1023/a:1005726623591. [DOI] [PubMed] [Google Scholar]
- Yuan F, Leunig M, Huang SK, Berk DA, Papahadjopoulos D., and, Jain RK. Microvascular permeability and interstitial penetration of sterically stabilized (stealth) liposomes in a human tumor xenograft. Cancer Res. 1994;54:3352–3356. [PubMed] [Google Scholar]
- Reimer DL, Zhang Y, Kong S, Wheeler JJ, Graham RW., and, Bally MB. Formation of novel hydrophobic complexes between cationic lipids and plasmid DNA. Biochemistry. 1995;34:12877–12883. doi: 10.1021/bi00039a050. [DOI] [PubMed] [Google Scholar]
- Pagnan G, Montaldo PG, Pastorino F, Raffaghello L, Kirchmeier M, Allen TM.et al. (1999GD2-mediated melanoma cell targeting and cytotoxicity of liposome-entrapped fenretinide Int J Cancer 81268–274. [DOI] [PubMed] [Google Scholar]
- Rozen S., and, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR., and, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques. 2002;32:1372–4, 1376, 1378. [PubMed] [Google Scholar]
- Piva R, Chiarle R, Manazza AD, Taulli R, Simmons W, Ambrogio C.et al. (2006Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas Blood 107689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignole C, Marimpietri D, Pastorino F, Nico B, Di Paolo D, Cioni M.et al. (2006Effect of bortezomib on human neuroblastoma cell growth, apoptosis, and angiogenesis J Natl Cancer Inst 981142–1157. [DOI] [PubMed] [Google Scholar]
- Pastorino F, Di Paolo D, Piccardi F, Nico B, Ribatti D, Daga A.et al. (2008Enhanced antitumor efficacy of clinical-grade vasculature-targeted liposomal doxorubicin Clin Cancer Res 147320–7329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Efficiency and specificity of gene knockdown over time evaluated in SH-SY5Y by RT-qPCR.
Liver and renal toxicity studies.