Abstract
In the analysis of peripheral blood gene expression, timely processing of samples is essential to ensure that measurements reflect in vivo biology, rather than ex vivo sample processing variables. The effect of processing delays on global gene expression patterns in peripheral blood mononuclear cells (PBMCs) was assessed by isolating and stabilizing PBMC-derived RNA from 3 individuals either immediately after phlebotomy or after a 4 h delay. RNA was labeled using NuGEN Ovation labeling and probed using the Affymetrix HG U133 Plus 2.0 GeneChip®. Comparison of gene expression levels (≥2-fold expression change and P < 0.05) identified 307 probe sets representing genes with increased expression and 46 indicating decreased expression after 4 h. These differentially expressed genes include many that are important to inflammatory, immunologic, and cancer pathways. Among others, CCR2, CCR5, TLR10, CD180, and IL-16 have decreased expression, whereas VEGF, IL8, SOCS2, SOCS3, CD69, and CD83 have increased expression after a 4 h processing delay. The trends in expression patterns associated with delayed processing were also apparent in an independent set of 276 arrays of RNA from human PBMC samples with varying processing times. These data indicate that the time between sample acquisition, initiation of processing, and when the RNA is stabilized should be a prime consideration when designing protocols for translational studies involving PBMC gene expression analysis.
Introduction
Biobanking practices require support from evidence-based biospecimen science to ensure that findings are due to in vivo biological differences rather than ex vivo influences. This is especially important in global gene expression studies such as those using Affymetrix GeneChips® that monitor the expression level of ∼47,000 transcripts from a single sample and can be highly sensitive to preanalytical variability.
Although several studies have examined the effect of preanalytical variables on global gene expression in solid tissues1–3 (among others), much less is known concerning the effect of delayed isolation of peripheral blood mononuclear cells (PBMCs). In one study comparing aliquots of PBMCs processed immediately or after overnight shipping to a single processing center, 2034 out of 6414 genes were found differentially expressed.4 Independently, Affymetrix, Inc., identified extensive expression pattern changes in PBMC samples due to overnight processing delays.5
In this study we identified gene expression changes in PBMCs related to processing delays as short as 4 h. These patterns were also apparent in an independent set of 276 arrays with variable processing times.
Materials and Methods
Subjects
Blood samples from adult volunteers were used to examine the processing time variable directly. In addition, data were available from a large, multicenter, gene expression study related to juvenile idiopathic arthritis (JIA). Patients in the JIA study were followed for up to 2 years and blood samples were collected at up to 10 time points for each patient. Blood was collected after informed consent.
Sample processing
Sample processing has been detailed elsewhere.6–8 Briefly, peripheral blood was collected in acid citrate dextrose (ACD) tubes, and PBMCs were isolated over Ficoll gradient (Ficoll Paque™ Plus; GE Healthcare, Piscataway, NJ) and put into TRIzol® reagent (Invitrogen, Carlsbad, CA) for 5 min at room temperature before slow cooling (−1°C/min), and storage at −80°C. The time between phlebotomy and start of processing is defined as the “processing delay.” The time between phlebotomy and storage at −80°C is defined as “time to freezing” (TTF). RNA was extracted and purified using RNeasy columns (Qiagen, Germantown, MD). cDNA was prepared using NuGEN Ovation kit version 1 (NuGEN Technologies, San Carlos, CA) from 100 ng of RNA and assayed using Affymetrix HG U133 Plus 2.0 GeneChips (Affymetrix, Santa Clara, CA). The timecourse microarray dataset has been deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (NCBI) and is accessible through GEO Series access number GSE21039.
Statistical analysis
Data were preprocessed in GeneSpring GX 7.3.1 using robust multichip analysis.9 Probe sets representing differentially expressed genes were defined as those with ≥2-fold expression difference and P < 0.05. Hierarchical clustering used Pearson correlation for probe sets and distance correlation for samples. To allow comparison to probe sets reported in the literature using an earlier version GeneChip, a conversion was performed using the GeneSpring 7.3 “translate” function.
Results
Short processing delays alter gene expression
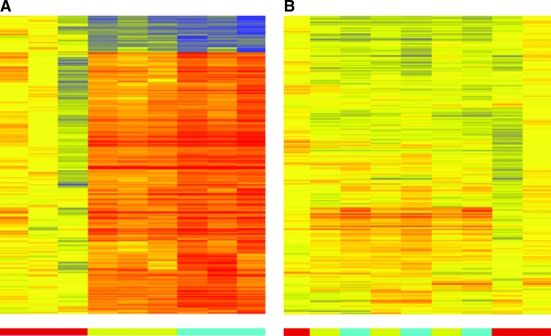
Three tubes of blood were collected from a single venipuncture from each of 3 donors, and PBMCs were isolated after a 0 h (T0), 2 h (T2), or 4 h (T4) delay at room temperature. Comparing GeneChip signal intensity values between T0 and T4 identified 353 probe sets detecting differentially expressed genes (select probe sets in Table 1, and full list in the Appendix. Hierarchical clustering of the samples using these probe sets completely segregated the samples into their respective processing-time category (Fig. 1A).
Table 1.
Selected Genes of Immunological Importance
| |
|
Fold changeb |
|
|---|---|---|---|
| Probe IDa | Gene symbola | 2 h/0 hc | 4 h/0 hd |
| 207794_at | CCR2 | 0.63 | 0.38 |
| 206991_s_at | CCR5 | 0.53 | 0.39 |
| 223751_x_at | TLR10 | 0.64 | 0.45 |
| 216379_x_at | CD24 | 0.55 | 0.45 |
| 206206_at | CD180 | 0.61 | 0.47 |
| 209828_s_at | IL16 | 0.67 | 0.47 |
| 223583_at | TNFAIP8L2 | 0.60 | 0.47 |
| 207907_at | TNFSF14 | 0.62 | 0.49 |
| 227697_at | SOCS3 | 1.71 | 2.14 |
| 218881_s_at | FOSL2 | 1.69 | 2.16 |
| 209795_at | CD69 | 1.60 | 2.23 |
| 230170_at | OSM | 2.26 | 2.29 |
| 218311_at | MAP4K3 | 1.55 | 2.40 |
| 201465_s_at | JUN | 1.33 | 2.41 |
| 203373_at | SOCS2 | 1.53 | 2.50 |
| 202644_s_at | TNFAIP3 | 2.30 | 2.79 |
| 211506_s_at | IL8 | 2.27 | 3.23 |
| 217738_at | PBEF1 | 2.04 | 3.31 |
| 204440_at | CD83 | 2.20 | 3.47 |
| 210512_s_at | VEGF | 1.89 | 3.48 |
Affymetrix U133 Plus 2.0 GeneChip® probe set IDs and annotations.
Fold change as ratio of geometric means.
Expression at 2 h/expression at 0 h.
Expression at 4 h/expression at 0 h.
FIG. 1.
Processing effects of gene expression signatures. (A) Samples clustered according to 353 probe sets identified (P < 0.05 and fold-change ≥2-fold) representing genes differentially expressed between samples processed immediately or after a 2 or 4 h delay. (B) Samples from this study were clustered according to gene expression data related to 2082 probe sets identified by Baechler et al. as differentially expressed after overnight shipment of peripheral blood before RNA extraction.4 Each column represents 1 sample and each row represents 1 probe set. In the heatmap, expression levels are indicated by color (yellow, median; red, increased; blue, decreased). Bar at the bottom indicates class membership: red, immediate processing; yellow, 2 h delay; blue, 4 h delay. See online article at www.liebertonline.com for color figure.
Response to overnight processing delays is different
In a previous report, Baechler et al. identified 2155 probe sets (2082 HG U133 Plus 2.0 equivalents) measuring differentially expressed genes between peripheral bloods processed to PBMCs immediately and those bloods shipped by overnight courier before processing.4 Understanding that there were significant experimental differences between the 2 studies, the current list of 353 probe sets was compared and showed only 62 probe sets (17%) in common. Although there were clusters of genes that appeared different in samples with delayed processing, the 2082 probe sets were unable to distinguish the current samples according to processing time (Fig. 1B).
Validation in a larger cohort
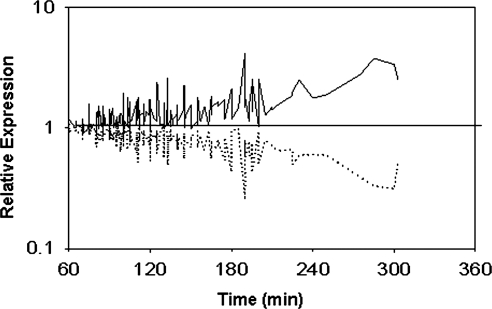
To determine whether the processing time signature would be apparent in samples that exhibit variation in gene expression for other reasons, the expression pattern (shown as the average of the expression levels for observation purposes) was examined in an independent group of 276 arrays (Fig. 2). The PBMC samples were obtained either from healthy controls or from patients with JIA and TTF ranged from 60 to 315 min. Although these samples showed some variation (possibly indicating biological variation between samples), trends of expression changes due to processing time were evident (Fig. 2). The trend was apparent in samples from each clinical site and each disease subtype showing that the processing time effect is independent of either clinical site or disease state (data not shown).
FIG. 2.
Average expression levels of time-sensitive probe sets. In a pilot experiment, 307 probe sets were identified with increased expression and 46 with decreased expression after a 4 h processing delay. Average expression of probe sets with increased (solid line) or decreased (dotted line) expression is shown in 267 independent samples. Samples are arranged according to time to processing.
Discussion
Many studies have reported gene expression differences in PBMCs without providing details of sample processing procedures. This study highlights the importance of considering processing delays since periods as short as 4 h can have significant effects on gene expression. While additional studies with independent samples are necessary to define specific processing gene expression signatures, it is clear that processing delays must be considered in experimental design and data interpretation.
This study was not designed to determine the specific cause of the identified gene expression changes although several explanations can be hypothesized. Phlebotomy itself can have an influence on lab results obtained from blood samples.10 Regarding storage of samples after phlebotomy, a previous study looked at the effect of room temperature storage of serum and plasma.11 Within 4 h after phlebotomy, the environment of the plasma and serum sample changed significantly, which would cause biological stresses. Glucose levels dropped, lactate increased, pH decreased, and hypoxia were identified by a decrease in pO2 and an increase in pCO2. Although this study did not examine the effect of blood processing delays, it can be expected that similar effects would be encountered. Even the simple preanalytical variables of the additive in the collection tube (eg, ACD in this study) or interaction of cells with the surface of the tube itself (a significant change from the vasculature of the donor) for several hours may cause the identified gene expression changes.
Proteins derived from the genes listed in Table 1 are important in inflammatory, autoimmune, and cancer pathways. Vascular endothelial growth factor (VEGF) is a prototypical angiogenic factor,12 and anti-VEGF has become the standard treatment for many tumor types.13 CCR2, CCR5, and IL-8 are chemoattractants that activate and recruit immune cells to sites of infection and inflammation. Antagonists to CCR2 have been suggested as therapy for a variety of inflammatory diseases as well as obesity and pulmonary disease.14 The CCR5 (also called CD195) antagonist, Maraviroc, is currently approved for treatment of HIV infection.15 SOCS2 and 3 are key negative regulators of cytokine signaling.16 TLR10 and CD180 (RP105) are involved in toll-like receptor signaling. The fact that these genes have variable expression attributable to processing delays emphasizes the importance of attention to preanalytic variables in sample collection and processing, a fundamental component of biospecimen science.
Various strategies can be employed to address possible changes caused by processing delays. The method we chose to reduce the impact of this variable was to remove samples with extended processing times (>4 h) from analysis.6–8 As an alternative, time-sensitive genes may be removed from consideration.17 Both of these methods have the benefit of simplicity although specific cutoffs are arbitrary. A disadvantage of this approach is that time-sensitive genes might also be involved in pathogenic processes. An interesting, but untested, method would be to delay processing of all samples until a specified time postphlebotomy (eg, 2 h) to reduce this variability. Alternatively, statistical approaches such as linear modeling may be employed using processing time as 1 variable.
To avoid the effect of variable processing time, technologies that stabilize samples instantly have been developed. These methods collect whole blood, including neutrophils that tend to vary in number more than other cellular components, and metabolically active immature red blood cells. Together, these components may overshadow more biologically relevant PBMCs. Additionally, there are issues with the so-called globin effect, where excessive amounts of globin mRNA derived from the reticulocytes present in whole blood interfere with the microarray analysis as seen by decreased present calls.5 It is unclear if this occurs due to inhibition during the labeling or probing steps of the assay. Newer labeling systems may help overcome this issue (as indicated by recovery of detected genes; unpublished data).
It is evident from this study that processing delays affect gene expression patterns obtained from PBMCs in a very short period. It is, therefore, important for this variable to be measured so that its effect can be considered in data interpretation.
Appendix. Three Hundred Fifty-Three Probe Sets Indicating Genes with Differential Expression Between T0 and T4 Samples
| |
|
Fold changeb |
|
|---|---|---|---|
| Probe IDa | Gene symbol | 2 h/0 hc | 4 h/0 hd |
| 204622_x_at | NR4A2 | 5.2877 | 8.9551 |
| 216248_s_at | NR4A2 | 5.4105 | 8.8413 |
| 205239_at | AREG///LOC653193 | 4.6343 | 8.1447 |
| 36711_at | MAFF | 4.7111 | 7.0973 |
| 213933_at | PTGER3 | 3.2406 | 6.4005 |
| 219312_s_at | ZBTB10 | 2.7223 | 5.8898 |
| 202464_s_at | PFKFB3 | 3.266 | 5.8483 |
| 208078_s_at | SNF1LK | 4.1568 | 5.8328 |
| 208937_s_at | ID1 | 3.9189 | 5.8208 |
| 203821_at | HBEGF | 3.1582 | 5.7492 |
| 217739_s_at | PBEF1///RP11-92J19.4 | 2.9129 | 5.6252 |
| 1554309_at | EIF4G3 | 2.6247 | 5.4238 |
| 209967_s_at | CREM | 3.4671 | 5.2409 |
| 233899_x_at | ZBTB10 | 2.4014 | 5.2317 |
| 228562_at | — | 2.901 | 5.2283 |
| 213524_s_at | G0S2 | 3.772 | 5.1801 |
| 207630_s_at | CREM | 3.5328 | 5.1701 |
| 224454_at | ETNK1 | 2.8282 | 5.1389 |
| 230511_at | CREM | 3.7946 | 5.1284 |
| 204621_s_at | NR4A2 | 3.235 | 5.0914 |
| 214508_x_at | CREM | 3.4216 | 5.049 |
| 222309_at | C6orf62 | 2.4626 | 4.8293 |
| 219228_at | ZNF331 | 2.6557 | 4.6216 |
| 218880_at | FOSL2 | 2.8545 | 4.4736 |
| 222180_at | YES1 | 2.5314 | 4.4144 |
| 240038_at | ELL2 | 2.6019 | 4.3908 |
| 202861_at | PER1 | 2.6954 | 4.3866 |
| 201466_s_at | JUN | 1.781 | 4.2976 |
| 204141_at | TUBB2A | 2.7917 | 4.231 |
| 202859_x_at | IL8 | 2.5225 | 4.0768 |
| 241740_at | CREM | 3.5468 | 3.9794 |
| 227613_at | ZNF331 | 2.4938 | 3.8653 |
| 1552908_at | C1orf150 | 2.6113 | 3.7847 |
| 38037_at | HBEGF | 2.1529 | 3.771 |
| 225262_at | FOSL2 | 2.8867 | 3.7097 |
| 201464_x_at | JUN | 1.9194 | 3.6948 |
| 205548_s_at | BTG3 | 2.2915 | 3.6902 |
| 236495_at | PBEF1 | 1.6075 | 3.6901 |
| 233952_s_at | ZNF295 | 1.9552 | 3.5791 |
| 1562255_at | SYTL3 | 2.335 | 3.5712 |
| 1559975_at | BTG1 | 2.4178 | 3.5277 |
| 210512_s_at | VEGF | 1.8886 | 3.4847 |
| 204440_at | CD83 | 2.1959 | 3.4748 |
| 228062_at | NAP1L5 | 2.3574 | 3.4686 |
| 230133_at | MNAB | 2.5067 | 3.4311 |
| 203543_s_at | KLF9 | 2.1823 | 3.4159 |
| 222044_at | — | 2.2031 | 3.3953 |
| 1554036_at | ZBTB24 | 2.772 | 3.3849 |
| 225884_s_at | ZNF336 | 2.2638 | 3.3824 |
| 204286_s_at | PMAIP1 | 1.8584 | 3.3814 |
| 242712_x_at | LOC653086///LOC653489///LOC653596 | 1.8887 | 3.3689 |
| 1556361_s_at | ANKRD13C | 1.8804 | 3.3257 |
| 1568665_at | RNF103 | 2.2975 | 3.3193 |
| 213134_x_at | BTG3 | 2.2166 | 3.3135 |
| 1557257_at | BCL10 | 2.1335 | 3.3121 |
| 217738_at | PBEF1 | 2.0389 | 3.3118 |
| 1569136_at | MGAT4A | 2.4002 | 3.3043 |
| 231182_at | WASPIP | 2.2977 | 3.2973 |
| 242243_at | TMF1 | 1.5683 | 3.2954 |
| 222815_at | RNF12 | 1.7508 | 3.2557 |
| 211506_s_at | IL8 | 2.2651 | 3.2343 |
| 60084_at | CYLD | 1.862 | 3.2292 |
| 210837_s_at | PDE4D | 1.9635 | 3.2114 |
| 202643_s_at | TNFAIP3 | 2.5643 | 3.2094 |
| 241985_at | JMY | 1.8644 | 3.2086 |
| 1555476_at | IREB2 | 2.5553 | 3.2083 |
| 211458_s_at | GABARAPL1///GABARAPL3 | 2.2323 | 3.1766 |
| 203574_at | NFIL3 | 2.0392 | 3.1158 |
| 219622_at | RAB20 | 1.7122 | 3.0976 |
| 1569263_at | SLC16A3 | 2.0431 | 3.0744 |
| 218319_at | PELI1 | 1.9741 | 3.0721 |
| 208869_s_at | GABARAPL1 | 1.8508 | 3.0551 |
| 242903_at | IFNGR1 | 1.5903 | 3.044 |
| 243664_at | TXNL1 | 1.6132 | 3.0196 |
| 232044_at | RBBP6 | 2.0252 | 3.0159 |
| 225539_at | ZNF295 | 1.8582 | 3.0102 |
| 219094_at | ARMC8 | 1.8875 | 3.001 |
| 226370_at | KLHL15 | 2.0354 | 2.997 |
| 202672_s_at | ATF3 | 1.4894 | 2.9967 |
| 233127_at | ZNF331 | 1.7927 | 2.9944 |
| 221986_s_at | KLHL24 | 1.8226 | 2.966 |
| 1555167_s_at | PBEF1 | 2.0741 | 2.9628 |
| 1554306_at | ITPKB | 2.1793 | 2.9596 |
| 228063_s_at | NAP1L5 | 2.3632 | 2.959 |
| 243857_at | MORF4L2 | 1.3143 | 2.9586 |
| 229718_at | CG018 | 2.0091 | 2.949 |
| 238796_at | YTHDC1 | 1.9338 | 2.9354 |
| 233309_at | TMEM2 | 2.0113 | 2.9252 |
| 203372_s_at | SOCS2 | 2.0505 | 2.9183 |
| 202558_s_at | STCH | 1.7833 | 2.8712 |
| 235592_at | ELL2 | 1.6639 | 2.8409 |
| 1553134_s_at | C9orf72 | 1.7996 | 2.8148 |
| 202932_at | YES1 | 1.6396 | 2.8141 |
| 205214_at | STK17B | 2.1198 | 2.7996 |
| 1555963_x_at | B3GNT7 | 1.9141 | 2.7986 |
| 204285_s_at | PMAIP1 | 1.6905 | 2.795 |
| 1553861_at | TCP11L2 | 1.8632 | 2.7942 |
| 231990_at | USP15 | 1.945 | 2.7884 |
| 202644_s_at | TNFAIP3 | 2.3037 | 2.7877 |
| 211924_s_at | PLAUR | 1.6585 | 2.7822 |
| 226608_at | LOC388272 | 1.8214 | 2.7687 |
| 213281_at | JUN | 1.3926 | 2.7683 |
| 242176_at | MEF2A | 1.8762 | 2.7647 |
| 221563_at | DUSP10 | 1.4504 | 2.7587 |
| 202499_s_at | SLC2A3 | 1.9353 | 2.7522 |
| 1556239_a_at | HERPUD2 | 2.0926 | 2.7461 |
| 205281_s_at | PIGA | 1.7789 | 2.738 |
| 228846_at | MXD1 | 1.838 | 2.7347 |
| 209545_s_at | RIPK2 | 1.763 | 2.7331 |
| 210845_s_at | PLAUR | 1.7088 | 2.7267 |
| 228181_at | SLC30A1 | 1.4527 | 2.7179 |
| 1554037_a_at | ZBTB24 | 2.2592 | 2.7114 |
| 225955_at | MED25///METRNL///LOC653506 | 2.1526 | 2.6876 |
| 222624_s_at | ZNF639 | 2.203 | 2.6807 |
| 214696_at | MGC14376 | 2.0627 | 2.6761 |
| 226275_at | MXD1 | 1.7789 | 2.6754 |
| 209803_s_at | PHLDA2 | 1.3385 | 2.6439 |
| 218486_at | KLF11 | 1.8841 | 2.641 |
| 224352_s_at | CFL2 | 1.8999 | 2.6398 |
| 202843_at | DNAJB9 | 1.8769 | 2.6257 |
| 227309_at | YOD1 | 1.8195 | 2.6186 |
| 222142_at | CYLD | 1.5623 | 2.6162 |
| 227718_at | PURB | 1.6592 | 2.6145 |
| 1555274_a_at | SELI | 1.6311 | 2.6088 |
| 225557_at | AXUD1 | 2.1334 | 2.5998 |
| 213758_at | COX4I1 | 2.1092 | 2.5914 |
| 203751_x_at | JUND | 1.9964 | 2.5908 |
| 202988_s_at | RGS1 | 1.8381 | 2.5897 |
| 1555281_x_at | ARMC8 | 1.9612 | 2.5884 |
| 210836_x_at | PDE4D | 1.9332 | 2.5716 |
| 204014_at | DUSP4 | 1.7219 | 2.5612 |
| 225950_at | SAMD8 | 1.7527 | 2.555 |
| 228962_at | — | 2.4711 | 2.552 |
| 211137_s_at | ATP2C1 | 1.66 | 2.5507 |
| 209457_at | DUSP5 | 2.352 | 2.5505 |
| 209020_at | C20orf111 | 1.869 | 2.5422 |
| 230134_s_at | MNAB | 1.9244 | 2.5405 |
| 202241_at | TRIB1 | 1.5014 | 2.539 |
| 230802_at | — | 1.8712 | 2.5229 |
| 207361_at | HBP1 | 1.972 | 2.5225 |
| 202393_s_at | KLF10 | 1.7447 | 2.5212 |
| 1555962_at | B3GNT7 | 2.053 | 2.5179 |
| 218009_s_at | PRC1 | 1.7305 | 2.5112 |
| 203373_at | SOCS2 | 1.5295 | 2.5006 |
| 222088_s_at | SLC2A3 | 2.0713 | 2.4999 |
| 202498_s_at | SLC2A3 | 1.9717 | 2.499 |
| 1553133_at | C9orf72 | 1.7513 | 2.4989 |
| 234993_at | ABHD13 | 1.8074 | 2.4978 |
| 238389_s_at | — | 1.6874 | 2.4961 |
| 204958_at | PLK3 | 2.0159 | 2.4923 |
| 224739_at | PIM3 | 1.8682 | 2.4803 |
| 1557285_at | LOC653193 | 1.513 | 2.4758 |
| 211302_s_at | PDE4B | 2.606 | 2.4689 |
| 242669_at | UFM1 | 1.557 | 2.4684 |
| 224657_at | ERRFI1 | 2.1992 | 2.4629 |
| 223584_s_at | KBTBD2 | 1.7845 | 2.4509 |
| 232576_at | — | 1.9155 | 2.4416 |
| 239143_x_at | RNF138 | 1.652 | 2.4353 |
| 1552644_a_at | PHC3 | 1.8204 | 2.4195 |
| 235780_at | PRKACB | 1.7088 | 2.4114 |
| 201465_s_at | JUN | 1.3337 | 2.4088 |
| 206374_at | DUSP8 | 1.7425 | 2.4062 |
| 218311_at | MAP4K3 | 1.5526 | 2.4006 |
| 226811_at | FAM46C | 2.115 | 2.3995 |
| 1554549_a_at | WDR20 | 1.9178 | 2.3993 |
| 211423_s_at | SC5DL | 1.8531 | 2.3989 |
| 1557459_at | SNF1LK2 | 1.5047 | 2.395 |
| 216834_at | RGS1 | 1.7235 | 2.3864 |
| 213805_at | ABHD5 | 1.4863 | 2.3721 |
| 201195_s_at | SLC7A5 | 1.6834 | 2.372 |
| 37028_at | PPP1R15A | 1.4221 | 2.3542 |
| 203542_s_at | KLF9 | 1.4991 | 2.3531 |
| 212665_at | TIPARP | 1.8848 | 2.346 |
| 243463_s_at | RIT1 | 1.3348 | 2.3438 |
| 229899_s_at | HSUP1 | 1.8462 | 2.3411 |
| 215501_s_at | DUSP10 | 1.484 | 2.3342 |
| 224797_at | ARRDC3 | 1.8174 | 2.3328 |
| 201745_at | PTK9 | 1.8115 | 2.3255 |
| 224453_s_at | ETNK1 | 1.4933 | 2.3238 |
| 215889_at | SKIL | 1.3378 | 2.3207 |
| 230170_at | OSM | 2.265 | 2.2923 |
| 226345_at | — | 1.67 | 2.2914 |
| 236223_s_at | RIT1 | 1.1963 | 2.2896 |
| 234907_x_at | POLB | 1.6498 | 2.2841 |
| 200731_s_at | PTP4A1 | 1.811 | 2.2823 |
| 202497_x_at | SLC2A3 | 2 | 2.2822 |
| 216236_s_at | SLC2A3 | 2.0204 | 2.2787 |
| 214326_x_at | JUND | 1.697 | 2.2744 |
| 202014_at | PPP1R15A | 1.4726 | 2.2677 |
| 204908_s_at | BCL3 | 1.824 | 2.2656 |
| 206877_at | MXD1 | 1.9677 | 2.2589 |
| 201751_at | JOSD1 | 1.8399 | 2.2569 |
| 214007_s_at | PTK9 | 2.2865 | 2.2558 |
| 209383_at | DDIT3 | 1.4827 | 2.2509 |
| 219624_at | BAG4 | 1.4673 | 2.2485 |
| 1560739_a_at | UBE3C | 1.6187 | 2.2419 |
| 225283_at | ARRDC4 | 1.5707 | 2.2404 |
| 209694_at | PTS | 1.7733 | 2.2386 |
| 225954_s_at | MIDN | 1.4918 | 2.2373 |
| 209795_at | CD69 | 1.5999 | 2.2349 |
| 238035_at | SP3 | 1.7086 | 2.2321 |
| 225699_at | C7orf40 | 1.691 | 2.231 |
| 206919_at | ELK4 | 1.38 | 2.2309 |
| 209211_at | KLF5 | 1.6051 | 2.2256 |
| 208868_s_at | GABARAPL1 | 1.7702 | 2.2253 |
| 223746_at | STK4 | 1.3786 | 2.2227 |
| 204299_at | FUSIP1///LOC642558 | 1.4299 | 2.2204 |
| 202657_s_at | SERTAD2 | 1.5966 | 2.2196 |
| 244103_at | C1orf55 | 1.4718 | 2.2185 |
| 243371_at | — | 1.2476 | 2.217 |
| 219382_at | SERTAD3 | 1.5925 | 2.2096 |
| 1559582_at | RHOQ | 1.634 | 2.2063 |
| 227979_at | RBM4B | 1.6985 | 2.2013 |
| 227521_at | FBXO33 | 2.0717 | 2.2007 |
| 205241_at | SCO2 | 1.6203 | 2.1977 |
| 205409_at | FOSL2 | 1.9516 | 2.1974 |
| 201170_s_at | BHLHB2 | 1.5799 | 2.1973 |
| 230380_at | THAP2 | 1.646 | 2.1963 |
| 226732_at | RBM33 | 1.6297 | 2.1953 |
| 36829_at | PER1 | 1.5855 | 2.1948 |
| 220330_s_at | SAMSN1 | 1.4587 | 2.1929 |
| 1555279_at | ARMC8 | 1.5154 | 2.1899 |
| 1559121_s_at | ARIH2 | 1.7287 | 2.1812 |
| 227680_at | ZNF326 | 1.556 | 2.1762 |
| 200733_s_at | PTP4A1 | 1.481 | 2.1759 |
| 209185_s_at | IRS2 | 1.8139 | 2.1745 |
| 241018_at | TMEM59 | 1.7747 | 2.1732 |
| 213538_at | SON | 1.7143 | 2.1713 |
| 201502_s_at | NFKBIA | 1.8469 | 2.1713 |
| 1556750_at | LOC153577 | 1.4175 | 2.1679 |
| 1554469_at | BTBD15 | 1.355 | 2.1672 |
| 212373_at | FEM1B | 1.7173 | 2.1652 |
| 203659_s_at | RFP2 | 1.6565 | 2.1647 |
| 223527_s_at | CDADC1 | 1.5857 | 2.1636 |
| 229955_at | FBXO3 | 1.553 | 2.1617 |
| 218881_s_at | FOSL2 | 1.6893 | 2.1598 |
| 226650_at | ZFAND2A | 1.7917 | 2.1594 |
| 230304_at | — | 1.6304 | 2.1564 |
| 202083_s_at | SEC14L1 | 1.3439 | 2.1561 |
| 204244_s_at | DBF4 | 1.5495 | 2.1544 |
| 200730_s_at | PTP4A1 | 1.6072 | 2.1488 |
| 221727_at | — | 1.6197 | 2.1482 |
| 235670_at | STX11 | 1.3933 | 2.1472 |
| 221919_at | — | 1.5308 | 2.1456 |
| 209300_s_at | NECAP1 | 1.6498 | 2.1448 |
| 218940_at | C14orf138 | 1.6503 | 2.1436 |
| 1554089_s_at | SBDS///SBDSP | 1.6481 | 2.1423 |
| 230748_at | SLC16A6 | 1.6112 | 2.1411 |
| 222669_s_at | SBDS | 1.6738 | 2.14 |
| 216015_s_at | CIAS1 | 1.7615 | 2.1398 |
| 227697_at | SOCS3 | 1.7134 | 2.1357 |
| 1554571_at | APBB1IP | 1.5946 | 2.1335 |
| 231863_at | ING3 | 1.582 | 2.1324 |
| 238719_at | — | 1.471 | 2.1315 |
| 204491_at | PDE4D | 1.8871 | 2.1279 |
| 218379_at | RBM7 | 1.5762 | 2.1261 |
| 202284_s_at | CDKN1A | 1.6065 | 2.1256 |
| 220306_at | FAM46C | 1.8422 | 2.125 |
| 226830_x_at | LOC440309 | 1.5305 | 2.1241 |
| 212240_s_at | PIK3R1 | 1.756 | 2.1235 |
| 214060_at | AMY1A///SSBP1 | 1.8757 | 2.1225 |
| 234055_s_at | ZNF336 | 1.5654 | 2.1211 |
| 1556911_at | ALMS1 | 1.1536 | 2.1195 |
| 241385_at | LARP7 | 1.4494 | 2.119 |
| 228693_at | CCDC50 | 1.4426 | 2.1181 |
| 238488_at | IPO11 | 1.2186 | 2.1138 |
| 226970_at | FBXO33 | 1.6576 | 2.1127 |
| 222808_at | GLT28D1 | 1.4785 | 2.1123 |
| 244219_at | WTAP | 1.7084 | 2.1063 |
| 200989_at | HIF1A | 1.4106 | 2.1018 |
| 211998_at | H3F3B | 1.8898 | 2.1008 |
| 218708_at | NXT1 | 1.6595 | 2.0998 |
| 242975_s_at | GNAS | 1.2272 | 2.0946 |
| 226206_at | MAFK | 1.7976 | 2.0904 |
| 41577_at | PPP1R16B | 1.5225 | 2.0887 |
| 1554929_at | KIAA0999 | 1.5522 | 2.0872 |
| 212666_at | SMURF1 | 1.3985 | 2.0866 |
| 228468_at | MASTL | 1.3022 | 2.085 |
| 218810_at | ZC3H12A | 1.7462 | 2.082 |
| 222018_at | NACA///NACAP1///LOC389240 | 1.6582 | 2.0819 |
| 213537_at | HLA-DPA1 | 1.6066 | 2.0813 |
| 224663_s_at | CFL2 | 2.0207 | 2.081 |
| 226390_at | STARD4 | 1.5828 | 2.0764 |
| 227577_at | EXOC8 | 1.6527 | 2.0717 |
| 200664_s_at | DNAJB1 | 1.8485 | 2.0689 |
| 204799_at | ZBED4 | 1.6269 | 2.0675 |
| 238633_at | EPC1 | 1.6999 | 2.0655 |
| 228180_at | SMU1 | 1.6351 | 2.0634 |
| 201169_s_at | BHLHB2 | 1.7168 | 2.0625 |
| 209345_s_at | PI4KII | 1.5147 | 2.0621 |
| 239494_at | LOC646725///LOC649431 | 1.6368 | 2.0608 |
| 238455_at | — | 1.6264 | 2.0598 |
| 201341_at | ENC1 | 1.9072 | 2.0533 |
| 204370_at | HEAB | 1.7027 | 2.0516 |
| 236196_at | — | 1.5068 | 2.0429 |
| 200666_s_at | DNAJB1 | 1.6874 | 2.0397 |
| 212374_at | FEM1B | 1.6998 | 2.0381 |
| 220239_at | KLHL7 | 1.5973 | 2.0372 |
| 221768_at | SFPQ | 1.5509 | 2.0359 |
| 223598_at | RAD23B | 1.6276 | 2.0343 |
| 203708_at | PDE4B | 2.0592 | 2.0342 |
| 225951_s_at | LOC440309///LOC649908 | 1.5941 | 2.0342 |
| 221763_at | JMJD1C | 1.3942 | 2.0265 |
| 242255_at | WDR37 | 1.2723 | 2.0258 |
| 205681_at | BCL2A1 | 1.3674 | 2.0221 |
| 1569864_at | SERAC1 | 1.3365 | 2.0106 |
| 1570394_at | XRN1 | 1.4304 | 2.0053 |
| 207513_s_at | ZNF189 | 0.5791 | 0.4999 |
| 212407_at | KIAA0859 | 0.6241 | 0.4969 |
| 218805_at | GIMAP5 | 0.6386 | 0.4966 |
| 223404_s_at | C1orf25 | 0.5826 | 0.4961 |
| 208893_s_at | DUSP6 | 0.5503 | 0.4955 |
| 1552316_a_at | GIMAP1 | 0.5609 | 0.4949 |
| 218979_at | RMI1 | 0.5514 | 0.4947 |
| 207907_at | TNFSF14 | 0.6168 | 0.4946 |
| 208891_at | DUSP6 | 0.4888 | 0.4937 |
| 238907_at | LOC284323 | 0.6243 | 0.4922 |
| 230226_s_at | JARID1A | 0.6606 | 0.4913 |
| 220235_s_at | C1orf103 | 0.5597 | 0.488 |
| 228920_at | ZNF260 | 0.5932 | 0.4869 |
| 231576_at | ETNK1 | 0.7761 | 0.4838 |
| 221081_s_at | DENND2D | 0.6306 | 0.4826 |
| 219777_at | GIMAP6 | 0.5575 | 0.48 |
| 223583_at | TNFAIP8L2 | 0.5994 | 0.4747 |
| 209828_s_at | IL16 | 0.6659 | 0.4737 |
| 206206_at | CD180 | 0.6085 | 0.4686 |
| 226230_at | SMEK2 | 0.618 | 0.4657 |
| 228190_at | CTR9 | 0.5411 | 0.4623 |
| 226481_at | VPRBP | 0.5429 | 0.4613 |
| 235085_at | DKFZp761P0423 | 0.5794 | 0.4582 |
| 226977_at | LOC492311 | 0.6134 | 0.4544 |
| 216379_x_at | CD24 | 0.5512 | 0.4533 |
| 223751_x_at | TLR10 | 0.6385 | 0.4482 |
| 220992_s_at | C1orf25 | 0.5581 | 0.4437 |
| 224953_at | YIPF5 | 0.5 | 0.4376 |
| 219243_at | GIMAP4 | 0.5749 | 0.4371 |
| 218242_s_at | SUV420H1 | 0.5972 | 0.4344 |
| 227335_at | DIDO1 | 0.5739 | 0.4299 |
| 229367_s_at | GIMAP6 | 0.5808 | 0.427 |
| 226423_at | PAQR8 | 0.5985 | 0.4188 |
| 200799_at | HSPA1A | 0.617 | 0.4116 |
| 206978_at | CCR2 | 0.5425 | 0.408 |
| 227626_at | PAQR8 | 0.5885 | 0.4058 |
| 240646_at | GIMAP8 | 0.5375 | 0.4037 |
| 206991_s_at | CCR5///LOC653725 | 0.5257 | 0.3864 |
| 226041_at | NAPE-PLD | 0.5368 | 0.3817 |
| 207794_at | CCR2 | 0.6309 | 0.3811 |
| 205898_at | CX3CR1 | 0.5622 | 0.3734 |
| 222566_at | SUV420H1 | 0.4814 | 0.3719 |
| 233461_x_at | ZNF226 | 0.5794 | 0.3675 |
| 200800_s_at | HSPA1A///HSPA1B | 0.6225 | 0.3664 |
| 235306_at | GIMAP8 | 0.4554 | 0.3547 |
| 230337_at | SOS1 | 0.3715 | 0.344 |
Affymetrix U133 Plus 2.0 GeneChip® probe set IDs and annotations.
Fold change as ratio of geometric means.
Expression at 2 h/expression at 0 h.
Expression at 4 h/expression at 0 h.
Acknowledgments
This work was supported by the NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (Grants P01AR048929, P30AR047363, and P60AR047784), the Cincinnati Children's Hospital Research Foundation, and the Ohio Valley Chapter of the Arthritis Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Dash A. Maine IP. Varambally S, et al. Changes in differential gene expression because of warm ischemia time of radical prostatectomy specimens. Am J Pathol. 2002;161:1743–1748. doi: 10.1016/S0002-9440(10)64451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J. Qi R. Quackenbush J, et al. Effects of ischemia on gene expression. J Surg Res. 2001;99:222–227. doi: 10.1006/jsre.2001.6195. [DOI] [PubMed] [Google Scholar]
- 3.Spruessel A. Steimann G. Jung M, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 2004;36:1030–1037. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 4.Baechler EC. Batliwalla FM. Karypis G, et al. Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes Immun. 2004;5:347–353. doi: 10.1038/sj.gene.6364098. [DOI] [PubMed] [Google Scholar]
- 5.An Analysis of Blood Processing Methods to Prepare Samples for GeneChip Expression Profiles (Technical Note) http://media.affymetrix.com/support/technical/technotes/blood_technote.pdf. 2003. http://media.affymetrix.com/support/technical/technotes/blood_technote.pdf
- 6.Barnes MG. Grom AA. Thompson SD, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:2102–2112. doi: 10.1002/art.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin TA. Barnes MG. Ilowite NT, et al. Gene expression signatures in polyarticular juvenile idiopathic arthritis demonstrate disease heterogeneity and offer a molecular classification of disease subsets. Arthritis Rheum. 2009;60:2113–2123. doi: 10.1002/art.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fall N. Barnes M. Thornton S, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56:3793–3804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 9.Irizarry RA. Bolstad BM. Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey IR. Thurlow VR. Is suboptimal phlebotomy technique impacting on potassium results for primary care? Ann Clin Biochem. 2008;45(Pt 3):266–269. doi: 10.1258/acb.2007.007123. [DOI] [PubMed] [Google Scholar]
- 11.Boyanton BL., Jr. Blick KE. Stability studies of twenty-four analytes in human plasma and serum. Clin Chem. 2002;48:2242–2247. [PubMed] [Google Scholar]
- 12.Shweiki D. Itin A. Soffer D, et al. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 13.Bose D. Meric-Bernstam F. Hofstetter W, et al. Vascular endothelial growth factor targeted therapy in the perioperative setting: implications for patient care. Lancet Oncol. 2010;11:373–382. doi: 10.1016/S1470-2045(09)70341-9. [DOI] [PubMed] [Google Scholar]
- 14.Xia M. Sui Z. Recent developments in CCR2 antagonists. Expert Opin Ther Pat. 2009;19:295–303. doi: 10.1517/13543770902755129. [DOI] [PubMed] [Google Scholar]
- 15.Emmelkamp JM. Rockstroh JK. CCR5 antagonists: comparison of efficacy, side effects, pharmacokinetics and interactions—review of the literature. Eur J Med Res. 2007;12:409–417. [PubMed] [Google Scholar]
- 16.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 17.Baechler EC. Batliwalla FM. Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]