Abstract
Time of flight secondary ion mass spectrometry (TOF-SIMS) imaging has been used to investigate the incorporation of phospholipids into the plasma membrane of PC12 cells after incubation with phosphatidylcholine (PC) and phosphatidylethanolamine (PE). The incubations were done at concentrations previously shown to change the rate of exocytosis in model cell lines. The use of TOF-SIMS in combination with an in situ freeze fracture device enables the acquisition of ion images from the plasma membrane in single PC12 cells. By incubating cells with deuterated phospholipids and acquiring ion images at high mass resolution, specific deuterated fragment ions were used to monitor the incorporation of lipids into the plasma membrane. The concentration of incorporated phospholipids relative to the original concentration of PC was thus determined. The observed relative amounts of phospholipid accumulation in the membrane ranges from 0.5 to 2 percent following 19 hours of incubation with PC at 100 to 300 μM and from 1 to 9 percent following incubation with PE at the same concentrations. Phospholipid accumulation is therefore shown to be dependent on the concentration in the surrounding media. In combination with previous exocytosis results, the present data suggests that very small changes in the plasma membrane phospholipid concentration are sufficient to produce significant effects on important cellular processes, such as exocytosis in PC12 cells.
Keywords: TOF-SIMS, deuterated phospholipids, relative quantification, PC12 cells
INTRODUCTION
Communication between neurons primarily occurs through the process of exocytosis. In exocytosis, neurotransmitters are released from small vesicles in the axon into the extracellular matrix by fusion of the vesicles with the plasma membrane.1–3 This delicate process involves both proteins and lipids, and is likely highly regulated by factors still not fully understood. In one study, using dopamine releasing rat pheochromocytoma (PC12) cells and amperometry, the dynamics of exocytosis was shown to change after incubation with phospholipids.4 For example, phosphatidylcholine (PC) was shown to decrease the rate of exocytosis while phosphatidylethanolamine (PE) increased the rate.4 An explanation for this is that the molecular shape of the phospholipids influences the ability of the membrane to rapidly form high curvature regions needed for fusion. Owing to the different sizes of the head groups, PC is more cylindrical making it more stable in low curvature regions and PE is more conical making it more stable in high curvature regions.5 Conceptually, a conical shape would thus assist the formation of high curvature regions, which could explain the increased rate of exocytosis. In another study, chromaffin cells and amperometry was used to investigate the effect of incubation with the inverted cone shaped lysophosphatidylcholine and the cone shaped arachidonic acid. This study concluded that both the steric geometry and the location of the exogenous lipids affected the time course and frequency of exocytosis post incubation.6
In order to better understand the role of lipids in synaptic plasticity, it would be of great value to determine the amount of lipids incorporated into the plasma membrane of the cells during these incubation experiments. In particular, it would be of value to be able to quantify the accumulation of exogenous phospholipids in the plasma membrane of PC12 cells under the same conditions where significant effects on the exocytosis process have been discovered.4 However, it is not trivial to analyze the changes in the lipid composition of the plasma membrane, partly because it is difficult to separate the plasma membrane from the rest of the cell for traditional phospholipid analyses using, e.g., LCMS.7 The incorporation of phospholipids is therefore often compared with the total amount of phospholipids in the cell.8 Another approach is to analyze the incorporation of molecules using fluorescent imaging while the cell is intact. However, these studies require the use of fluorescent tags on the molecule of interest, which might interfere with the normal function of the molecule due to the increased size and possible changes in hydrophobicity.5 Isotopically labeled molecules have previously been used to image molecules in biological structures with dynamic SIMS.9, 10 In addition, the combination of deuterated lipids and Time of Flight Secondary Ion Mass Spectrometry (TOF-SIMS) has been used to quantify lipid transfer in artificial lipid membrane systems.11 In TOF-SIMS only the top molecular layers of a surface are analyzed, which provides molecular information exclusively from the exposed surface of the sample. Quantification using TOF-SIMS is, however, complex since the ionization of molecules on the surface is highly dependent on the sample matrix.12 TOF-SIMS has previously been used to quantify the relative amount of cholesterol incorporated into cells which had been incubated prior to analysis.13 In this study the amount of cholesterol in the incubated cells was compared to the amount in cells that had not been incubated, and the relative increase in cholesterol could be determined. TOF-SIMS has also been used to investigate the relative increase or decrease of specific phospholipids in the native plasma membrane in mating tetrahymena14, 15 and the localization of vitamin E in neuronal membrane.16 In addition, TOFSIMS in combination with different multivariate analysis techniques has been used for quantification of protein films, carbohydrates and polystyrenes.17–20
In this paper, we use deuterated phospholipids to quantify the relative amounts of phospholipids accumulated in the plasma membrane of PC12 cells. After an overnight incubation at different concentrations of PC or PE, the cells are frozen, freeze fractured in situ and analyzed using TOFSIMS. We show that PC12 cells incorporate both PC and PE into the plasma membrane and that the amount of phospholipid accumulation strongly depends on the lipid concentration in the incubation medium.
EXPERIMENTAL SECTION
PC12 cell maintenance and incubation
PC12 cells were cultured in supplemented RPMI – 1640 medium as previously described.21 When cells were confluent, they were incubated with fresh medium with an addition of liposomes from deuterated PC or PE at 0, 100, 200 or 300 μM for 19 hours as shown in Figure 1. After incubation the cells were rinsed three times in HEPES (37°C, 10 mM, pH 7.4) while they were still attached to the bottom of the flask. The cells were dislodged from the surface in fresh HEPES using very gentle tapping of the flask to ensure little cell damage. The cells were then centrifuged for 5 min at 1500 rpm, using a Universal 320 centrifuge from Hettich. The supernatant was removed and the cells were resuspended in 8 ml fresh HEPES solution and centrifuged again. The supernatant was removed and the cells were resuspended in 1.5 ml HEPES solution to increase cell density.
Figure 1.
Work flow schematics from left to right showing; the cells being incubated in the culture flask, incorporation of exogenous phospholipids into the plasma membrane of a PC12 cell, the cells being freeze fractured using the freeze fracture device.
Preparation of deuterated phospholipids for incubation
Tail and headgroup deuterated 16:0/16:0 PC (D75PC) and tail group deuterated 16:0/16:0 PE (D62PE), both in chloroform (Avanti Polar Lipids, Inc., USA), were used for cell incubation. The deuterated phospholipids and some additional chloroform were added to a small round bottom flask, using a Hamilton syringe, and the chloroform was removed under 3 hours, using a rotavapor (Büchi Labortechnik AG, Switzerland). Cell medium was added to the lipid film in the round bottom flask and the solution was probe sonicated (5 min at 70 %, Sonics Vibracell, Model 501, Sonics & Materials, Inc, U.S.A) creating phospholipid vesicles in solution. D62PE was further treated using a mini extruder (Avanti Polar Lipids, Inc., USA) at 63°C to increase liposome formation.
Sample preparation and SIMS analysis
A 2 μL droplet of the cell solution was deposited onto the smaller of two Si shards mounted on a freeze fracture device which was held at 37°C, as previously described.22, 23 The device was then closed and fast frozen in liquid propane at −185 °C to prevent formation of large water crystals.24 The lack of large ice crystal formations was confirmed in independent scanning electron microscopy experiments. The frozen freeze fracture device, containing the cell sample, was mounted under liquid nitrogen onto the cold stage sample holder (IONTOF GmbH). It was then taken from the liquid nitrogen and placed immediately in the load lock in contact with a cryo cold finger for introduction into the analysis chamber of the instrument. Inside the main chamber of the TOF-SIMS instrument, the device was opened under vacuum at −110 °C, exposing the fractured cells embedded in ice matrix. The sample temperature during analysis was −105 °C. The scheme for cells from preparation to analysis is shown in Figure 1. This approach allows biological manipulation followed by analysis of frozen and hydrated cells providing images of cells in their native state with conserved molecular distribution.13–15, 22, 25–29
To compare the spectra of deuterated and non-deuterated lipids, and to calculate the secondary ion yield (see below), we examined dried standards of each. Standard samples were prepared by adding a drop of approximately 0.4 mg/mL deuterated lipid, dissolved in chloroform, to a silicon shard. The solvent was evaporated under a stream of nitrogen gas at room temperature before the shard was mounted onto the sample holder and placed into the instrument.
Analysis was performed using a TOF-SIMS IV instrument (IONTOF GmbH, Germany) equipped with a bismuth (Bi) liquid metal cluster primary ion source. Positive ion spectra and images were recorded with the instrument optimized for high mass resolution (bunched mode) with a pulsed current of 0.1 pA using 25 keV Bi3+ primary ions, and using low energy electron flooding for charge neutralization. The lateral resolution was approximately 4 μm and the mass resolution (m/Δm) was 7000. All data used for quantification was recorded under static conditions, with a primary ion dose density below 4 × 1012 ions/cm2, and over analysis areas between 70×70 and 80×80 μm2 with 256×256 pixels.
The amount of ice on the sample surface was carefully controlled by monitoring the signal intensity ratio between the HEPES peak (m/z 239) and several water cluster peaks. Cell data was only acquired when this ratio was kept between 6 and 30% for HEPES compared to the most intense water cluster peak (H307 at m/z 55) and between 10 and 50% of a less intense water cluster peak (H11O5 at m/z 91).
Data treatment
Raw data was collected and analyzed using the IonSpec and IonImage software (ION-TOF GmbH, Germany). The relative amount of phospholipid accumulation in the cell plasma membrane was determined separately for individual cells by reconstruction of spectra from regions of interest that each covers a single cell area. Spectra from the background areas for each analyzed cell were also extracted using regions of interest. The Poisson-corrected intensities of the peaks representing the exogenous and endogenous lipids, respectively, were normalized to the number of pixels in the area of interest before subtracting the background signal from the cell signal. The ratio of the signal intensity from the exogenous lipid to the signal intensity from the endogenous lipid was calculated for each cell and, finally, the relative amount of phospholipid accumulation in the cell plasma membrane was determined using secondary ion yield ratios from analysis of lipid standards, as described in detail below.
RESULTS AND DISCUSSION
Approach for relative quantification in cells with isotopic labeling
In the present work, we use deuterated lipids in the incubation medium to relatively quantify the accumulation of incubated phospholipids in the cell membrane. This has several advantages:
The entire signal from deuterated ions represents phospholipid molecules which have been accumulated in the PC12 cell membrane, i.e. the signal is measured from a zero background and not from a background level representing the endogenous phospholipid concentration.
Since all deuterated ions originate from the deuterated lipid, also small, normally “unspecific”, fragments can be used, providing the opportunity to select ions with high secondary ion yields and non-overlapping peaks.
A specific non-deuterated fragment ion can be used to represent the original lipid concentration in the PC12 cells, providing a direct measurement of the amount of accumulated phospholipid and the endogenous amount of lipid in the same cell membrane.
The headgroup fragment for phosphatidylcholine and sphingomyelin, phosphocholine, has m/z 184.1. For simplicity, as sphingomyelin is present at much lower levels than phosphatidylcholine and as these both are similarly shaped lipids, we treat these two phospholipids together, call this PC and let the fragment represent the endogenous concentration of the phosphocholine headgroup containing lipids. A selected smaller deuterated fragment originating from the fatty acid chain is used to represent the accumulated PC and PE (see below). The same deuterated fragment ion is used when comparing the amounts of accumulated PC and PE partly in order to minimize matrix effects. The reason for providing the amounts of accumulated PE relative to the endogenous concentration of PC is (i) to allow for a direct comparison between the amount of accumulated PC and PE, and (ii) the high yield of the PC peak results in a considerably more reliable measurement and less data scatter as compared to if the characteristic PE head group ions at m/z 124 and m/z 142 were used. The results are calculated based on the assumption that the exogenous lipids are added to the membrane. If they instead are exchanged with phospholipids in the membrane the error will be very small since the accumulated amount of exogenous phospholipid is very small.
Characterization of phospholipids used for incubation and selection of peaks for quantitative analysis
For quantification of lipid accumulation in the cell membrane, the fragment ion used to represent the exogenous phospholipid should ideally have a unique m/z that does not overlap with any endogenous fragments in the mass spectrum. The exogenous molecule must also retain the same chemical properties as the corresponding endogenous species so that it does not limit or change the interactions within the cell; this excludes attaching any functional groups to the exogenous molecule.5 Using deuterated phospholipids, the chance of finding a fragment ion with unique m/z is high and the exchange from hydrogen to deuterium in the exogenous lipid is not likely to produce significant changes in the chemical behavior in the cells.
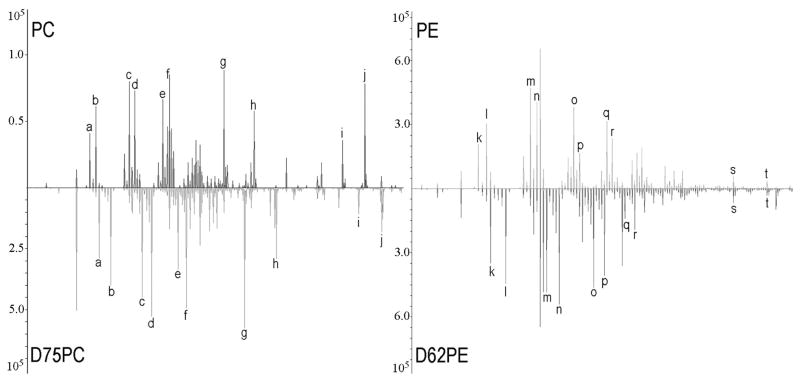
The cells in this study were incubated with either deuterated 16:0/16:0 PC (D75PC, both the tail groups and the headgroup are deuterated) or deuterated 16:0/16:0 PE (D62PE, deuterated tails) prior to analysis. Both D75PC and D62PE have fully deuterated tail groups, providing the possibility to use the same tail group fragment ions to monitor the phospholipid accumulation upon either PC or PE incubation. In Figure 2, the positive ion spectra from standard samples of pure PC, D75PC, PE and D62PE, respectively, are shown in the mass region up to m/z 200. Apart from a shift in mass, caused by the mass difference between hydrogen and deuterium, the fragmentation patterns for the deuterated lipids are very similar to those of native PC and PE. This is highlighted in Fig. 2 by letters that each mark equivalent ions in the spectra from the nondeuterated and deuterated lipids.
Figure 2.
Standard spectra from; PC and D75PC to the left, PE and D62PE to the right. The m/z and fragment ion of the peaks marked with letter are shown in Table 1.
Table 1 contains the m/z value and the empirical formula for each of the fragment ions marked in the spectra. Comparing the spectra for PC and PE, several common fragment ions are observed. One example is the peak at m/z 27 for the non-deuterated lipids and m/z 30 for the deuterated lipids, representing C2H3+ and C2D3+, respectively, which are observed for both PC and PE. In contrast, comparison of the head group ions shows an important difference between PC and PE. The PC head group ion at m/z 184.1 is represented by peaks at m/z 197 through 199 for the deuterated PC (labeled with j in the PC spectra), while the characteristic head group ion of PE at m/z 142 is present also in the spectrum from deuterated PE, since only the tail groups are deuterated in the case of PE.
Table 1.
Letter labels from the peaks in the four spectra in Figure 2 with corresponding m/z and empirical formula for the fragment ion.
| PC spectra | PE spectra | ||||
|---|---|---|---|---|---|
| m/z | Fragment ion | m/z | Fragment ion | ||
| a | 27 | [C2H3]+ | k | 27 | [C2H3]+ |
| 30 | [C2D3]+ | 30 | [C2D3]+ | ||
| b | 29 | [C2H5]+ | l | 29 | [C2H5]+ |
| 34 | [C2D5]+ | 34 | [C2D5]+ | ||
| c | 41 | [C3H5]+ | m | 41 | [C3H5]+ |
| 46 | [C3D5]+ | 46 | [C3D5]+ | ||
| d | 43 | [C3H7]+ | n | 43 | [C3H7]+ |
| 50 | [C3D7]+ | 50 | [C3D7]+ | ||
| e | 55 | [C4H7]+ | o | 55 | [C4H7]+ |
| 62 | [C4D7]+ | 62 | [C4D7]+ | ||
| f | 57 | [C4H9]+ | P | 57 | [C4H9]+ |
| 66 | [C4D9]+ | 66 | [C4D9]+ | ||
| g | 86 | [C5H12N]+ | q | 67 | [C5H7]+ |
| 98 | [C5D12N]+ | 74 | [C5D7]+ | ||
| h | 104 | [C5H14NO]+ | r | 69 | [C5H9]+ |
| 118 | [C5D14NO]+ | 76 | [C5D9]+ | ||
| i | 166 | [C5H13NPO3]+ | s | 124 | [C2H7NPO3]+ |
| 179 | [C5D13NPO3]+ | t | 142 | [C2H9NPO4]+ | |
| J | 184 | [C5H15NPO4]+ | |||
| 197 | [C5D13NPO4H2]+ | ||||
| 198 | [C5Di4NPO4H]+ | ||||
| 199 | [C5D15NPO4]+ | ||||
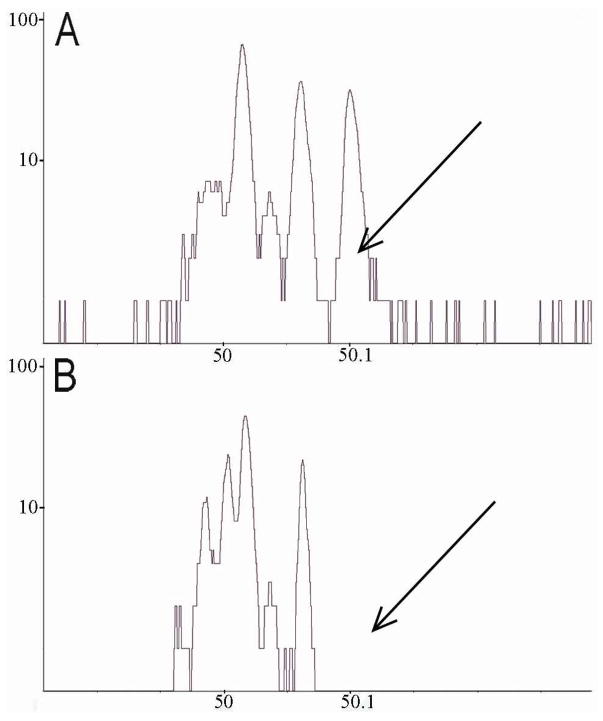
Choice of the fragment ion to use for analysis is critical. In addition to a m/z value well separated from any endogenous peaks in the mass spectrum of a native PC12 cell, the ideal candidate should have a high secondary ion yield to facilitate detection. The biological ice matrix makes it difficult to find a deuterated peak that fills the criteria of being both specific and have high intensity. The peak at m/z 50.1, corresponding to the tail group fragment C3D7+, shows high intensity in the spectra from both D75PC and D62PE. Figure 3 shows the observed peaks in the m/z range from 49.8 to 50.3 in spectra obtained from a non-incubated cell (Fig. 3B) and from a cell, which has been incubated with deuterated PC (Fig. 3A). This clearly shows that there is no overlap between the exogenous peak at m/z 50.1 and any endogenous peak from these cells as marked by the arrows. It is important to note that to avoid any effects from the matrix, all cells are embedded in the same HEPES matrix.12 The deuterated fragment ion at m/z 50.1 can thus be used to detect phospholipid accumulation in the cell plasma membrane with a high secondary ion yield and from a zero background for both D75PC and D62PE. No other deuterated peaks filled the criteria and C3D7+ is therefore used for quantification purposes in the present study.
Figure 3.
A) The deuterated fragment peak at m/z 50.1 in an incubated cell. B) The lack of the deuterated peak at m/z 50.1 in a native cell. Peak or lack of is indicated by an arrow.
To quantify the relative amount of deuterated phospholipids incorporated into the plasma membrane of a PC12 cell using the deuterated fragment ion at m/z 50.1, an endogenous fragment ion must also be selected to represent the endogenous PC concentration in the cell. One approach might be to use the non-deuterated form of the same fragment, i.e. the C3H7+ fragment ion at m/z 43, as the endogenous fragment. However, since this fragment ion can originate from many different compounds in the plasma membrane, it is not a suitable choice to represent the concentration of endogenous PC. The evident endogenous fragment for this purpose is instead the head group fragment ion of PC at m/z 184.1, corresponding to [C5H15NPO4]+. This head group fragment is readily formed during SIMS analysis. Further, PC is highly abundant in the PC12 cell, approximately 39%,30 and is present in the plasma membrane at a high concentration.
The deuterated PC used for incubation does not add to the intensity of the native peak at m/z 184.1 since D75PC has a fully deuterated head group and its spectrum does not contain overlapping peaks at m/z 184.1.
Using the peaks at m/z 50.1 and m/z 184.1 for relative quantification of the phospholipid accumulation in the cell plasma membrane makes it necessary to determine relative sensitivity factors that transforms the measured signal intensity ratios into concentration ratios. By analyzing standard samples for D75PC, D62PE and PC, the secondary ion yields (Y) are determined for the relevant ions in the pure lipids, according to Equation 1.31 The result shows that Y for m/z 184.1 in PC and m/z 50.1 in D75PC are similar, but that Y for m/z 50.1 in D62PE is about 1.5 times higher, as shown in Table 2. The relative concentrations are then calculated from the measured intensity ratios by division with the corresponding Y ratios (see below).
Table 2.
Secondary ion yield of fragments from PC, D75PC and D62PE standards used for calculation. n = 8.
| m/z | Fragment ion | SI Yield | Stdev | |
|---|---|---|---|---|
| PC | 184.1 | C5H15NPO4 | 0.013 | 5.2% |
| D75PC | 50.1 | C3D7+ | 0.012 | 6.2% |
| D62PE | 50.1 | C3D7+ | 0.018 | 4.0% |
| (1) |
A disadvantage of using relative sensitivity factors from lipid standards is that it does not take into account possible matrix effects due to the different chemical environment in cells as compared to in the lipid standards. An alternative method, which better would compensate for such matrix effects, would be to use the same fragment ion for the exogenous and endogenous lipids, i.e., the m/z 197, 198, 198 peaks for deuterated PC and m/z 184 for the non-deuterated PC. However, this was not possible in the present case owing to strong overlaps between the deuterated ion peaks at m/z 197, 198, 199 and peaks from endogenous ions in the cells.
Relative quantification of lipid incorporation in incubated cells
All cells were analyzed in the frozen hydrated state at −105 °C in order to keep the chemical distribution as close to native as possible. All cells that were used for quantification were embedded in ice with the plasma membrane exposed after freeze fracture. This was determined using the signal intensities from potassium (K+) (m/z 39.0) and PC (m/z 184.1), as shown previously.28 High potassium levels indicate that the cell has been fractured through the cytoplasm and high PC levels indicate that the fracture occurred above the plasma membrane, i.e., with the plasma membrane exposed on the sample surface. By calculating the ratio of the K+ signal over the PC signal for each cell, the plane of fracture could be determined. Only the cells with a very low ratio of K+/PC, ranging from 0.05 to 0.23, indicating that the plasma membrane was exposed, have been used in the study.
Figure 4 shows ion images of a single freeze fractured PC12 cell, incubated with 100 μM D62PE. The image in Figure 4A shows the signal intensity from the fragment ion at m/z 50.1 (C3D7+), originating from D62PE, while the signal from the endogenous PC fragment ion at m/z 184.1, is shown in Figure 4B. Even though the intensity of the C3D7+ ion is low, it is clearly localized to the cell area, as displayed in the m/z 184.1 image. In order to enable subtraction of the lipid signal from the background area, the images were divided into two regions of interest. These regions are marked in Figure 4C, where the red striped region corresponds to the cell area and the green striped region corresponds to the background.
Figure 4.
Ion images of a PC12 cell after incubation with D62PE at 100 μM. A) The fragment ion at m/z 50.1 (C3D7+), originating from D62PE, total counts are 104, B) the endogenous PC fragment ion at m/z 184.1, total counts are 9163, C) same image as in B) with region of interest shown using red and green outlines. The red ring circles the cell area, red stripes, and the area inside the green lines (green stripes) corresponds to the background. There is a region between the red and green lines that is not marked as either cell or background to clearly distinguish between the two. The color bars show 0 to 1 and 0 to 5 counts, for A and B, respectively, and the scale bars in A and B is 10 μm.
To calculate the incorporation of exogenous phospholipids in PC12 cells, the Poisson-corrected intensities of the two selected fragment ions (m/z 50.1 and m/z 184.1) were normalized to the number of pixels in the region of interest, as marked in the image in Figure 4C, making it possible to subtract the background signal from the cell signal. The signal intensities were then normalized to the secondary ion yields (Y) determined for the pure lipid sample (Table 2) as shown in Equation 2, where c is the normalized signal intensity (counts) for each fragment over each region. The relative amount of deuterated phospholipid incorporated into the plasma membrane of the PC12 cell can then be calculated from the signal intensity ratio of m/z 50.1 and m/z 184.1 as shown in equation 2.
| (2) |
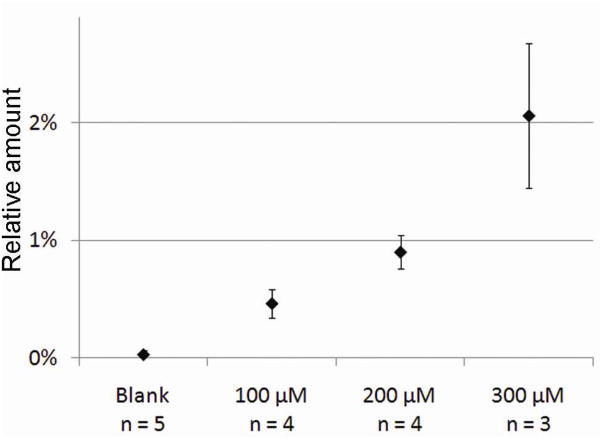
Three to five cells were analyzed at each lipid incubation concentration to determine the accumulation of deuterated phospholipid in the plasma membrane of the PC12 cells after incubation at 0, 100, 200 and 300 μM phospholipid concentration. The obtained relative amounts of accumulated phospholipid are plotted against the exogenous phospholipid concentration in the incubation medium in Figures 5 and 6. As expected, the relative amount of phospholipids accumulated into the plasma membrane increased with an increasing concentration of phospholipids in the incubation media. For PC (Figure 5), the incorporated amount of exogenous D75PC ranges from 0.5 % at 100 μM to 2.1% at 300 μM. Since PC makes up about 39% of the phospholipids in PC12 cells,30 the observed change of 2.1% is less than 1% of the total phospholipid pool in the cell.
Figure 5.
Measured amounts of incorporated D75PC relative to the amount of endogenous PC following 19 hours of incubation at different D75PC concentrations. All points are significantly different (students t-test p<0.05) and the error bars are standard deviation.
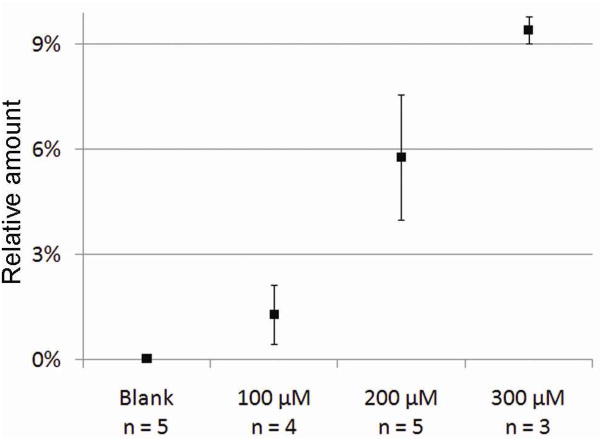
Figure 6.
Measured amounts of incorporated D62PE relative to the amount of endogenous PC following 19 hours of incubation at different concentrations. All points are significantly different (students t-test p<0.05) and the error bars are standard deviation.
The measured amounts of D62PE incorporated into the cell membrane relative to the amount of endogenous PC are shown in Figure 6. The relative amount ranges from 1.3 % for incubation at 100 μM to 9.4% at 300 μM. By assuming that PE makes up about 22% of the phospholipids in a PC12 cell,30 and that this is constant in all PC12 cells, the amount of exogenous PE incorporated into the cell after 300 μM incubation corresponds to 16.7% of the total PE pool and 3.7% of the total phospholipid pool in the cell.
The results in this study demonstrate the capability of TOF-SIMS in combination with the use of a freeze fracture device and deuterated lipids to determine the relative amount of phospholipid accumulation in the cell plasma membrane with high sensitivity (<1%) and single cell resolution. The key aspect of these measurements is that the deuterated lipid provides unique secondary ions that can be used for the detection of the incorporated lipid. In this work, we used a relatively small fragment ion originating from the fatty acid chain (C3D7+) to represent the incorporated lipid, partly due to its high secondary yield resulting in a high sensitivity for the measurement of phospholipid accumulation. However, since the C3D7+ ion originates from a small part of the phospholipid molecule, its detection does not necessarily reflect the presence of an intact phospholipid molecule, although it seems unlikely that the deuterated lipid is decomposed or degraded inside the cell. Another aspect of the present study is that we only measured the phospholipid accumulated into the plasma membrane of the cells and no information is obtained about the phospholipid accumulation into the internal part of the cell. As organic depth profiling using SIMS32, 33 becomes more developed, it may be possible to measure lipid levels in different cell compartments. Here we have been specifically interested in measuring the relative amount of phospholipid in the plasma membrane following incubation.
Conclusions
The results of this work clearly show that PC12 cells incorporate PC and PE into their plasma membrane from the surrounding medium. Interestingly, however, our results show that even at 100 μM incubation concentration, which may be considered extremely high, the change in lipid concentration of the cell membrane is quite small, ranging from 0.5% for PC to 1.3% for PE. This is an important result as our measurements were done under conditions similar to earlier work where PC12 cells were shown to change exocytosis rate following incubation.4, 6 Thus, the fact that a significant change in exocytosis is observed following identical incubation with lipids to those here strongly suggests that only a small change in the relative amount of lipid in the cell membrane is needed to cause a significant effect on the cell function, in this case exocytosis.
Acknowledgments
This work was supported by grants from the National Institutes of Health (2R01EB002016-16 (A.G.E.), the Swedish Research Council (A.G.E.) and the Swedish Governmental Agency for Innovation Systems (VINNOVA) (P.S.). A.G.E. acknowledges support from an ERC Advanced grant and a center grant from the Knut and Alice Wallenberg Foundation. The authors thank Ulf Lanekoff for illustrations in Figure 1.
References
- 1.Chernomordik LV, Kozlov MM. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R, Lang T, Sudhof TC. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 3.Churchward MA, Coorssen JR. Biochem J. 2009;423:1–14. doi: 10.1042/BJ20090969. [DOI] [PubMed] [Google Scholar]
- 4.Uchiyama Y, Maxson MM, Sawada T, Nakano A, Ewing AG. Brain Res. 2007;1151:46–54. doi: 10.1016/j.brainres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Meer G. Embo J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amatore C, Arbault S, Bouret Y, Guille M, Lemaitre F, Verchier Y. ChemBioChem. 2006;7:1998–2003. doi: 10.1002/cbic.200600194. [DOI] [PubMed] [Google Scholar]
- 7.Shevchenko A, Simons K. Nat Rev Mol Cell Bio. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 8.Kainu V, Hermansson M, Somerharju PJ. Lipid Res. 2010;51:3533–3541. doi: 10.1194/jlr.D009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, Moo Park K, Ito S, Schwartz M, Benichou G, Slodzian G. J Biol. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Science. 2006:313. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 11.Kunze A, Sjovall P, Kasemo B, Svedberg S. J Am Chem Soc. 2009;131:2450–2451. doi: 10.1021/ja809608n. [DOI] [PubMed] [Google Scholar]
- 12.Belu AM, Graham DJ, Castner DG. Biomaterials. 2003;24:3635–3653. doi: 10.1016/s0142-9612(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 13.Ostrowski SG, Kurczy ME, Roddy TP, Winograd N, Ewing AG. Anal Chem. 2007;79:3554–3560. doi: 10.1021/ac061825f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurczy ME, Piehowski PD, Van Bell CT, Heien ML, Winograd N, Ewing AGP. Natl Acad Sci USA. 2010;107:2751–2756. doi: 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. J Am Chem Soc. 2005;127:12152–12153. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 17.Wagner MS, Shen M, Horbett TA, Castner DG. J Biomed Mater Res A. 2003;64A:1–11. doi: 10.1002/jbm.a.10263. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari S, Ratner BD. Surf Interface Anal. 2000;29:837–844. [Google Scholar]
- 19.Sjovall P, Lausmaa J, Johansson BL, Andersson M. Anal Chem. 2004;76:1857–1864. doi: 10.1021/ac035457g. [DOI] [PubMed] [Google Scholar]
- 20.Vanden Eynde X, Bertrand P. Appl Surf Sci. 1999;141:1–20. [Google Scholar]
- 21.Kozminski KD, Gutman DA, Davila V, Sulzer D, Ewing AG. Anal Chem. 1998;70:3123–3130. doi: 10.1021/ac980129f. [DOI] [PubMed] [Google Scholar]
- 22.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, Sjovall P, Ewing AG. Anal Chem. 2010;82:6652–6659. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanekoff I, Kurczy ME, Adams KL, Malm J, Karlsson R, Sjovall P, Ewing AG. Surf Interface Anal. 2010 [Google Scholar]
- 24.Severs NJ, Shotton D. Rapid Freezing, Freeze Fracture, and Deep Etching. Wiley-Liss; New York: 1995. [Google Scholar]
- 25.Colliver TL, Brummel CL, Pacholski ML, Swanek FD, Ewing AG, Winograd N. Anal Chem. 1997;69:2225–2231. doi: 10.1021/ac9701748. [DOI] [PubMed] [Google Scholar]
- 26.Piehowski PD, Kurczy ME, Willingham D, Parry S, Heien ML, Winograd N, Ewing AG. Langmuir. 2008;24:7906–7911. doi: 10.1021/la800292e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roddy TP, Cannon DM, Meserole CA, Winograd N, Ewing AG. Anal Chem. 2002;74:4011–4019. doi: 10.1021/ac0255734. [DOI] [PubMed] [Google Scholar]
- 28.Roddy TP, Cannon DM, Ostrowski SG, Winograd N, Ewing AG. Anal Chem. 2002;74:4020–4026. doi: 10.1021/ac025574w. [DOI] [PubMed] [Google Scholar]
- 29.Cliff B, Lockyer N, Jungnickel H, Stephens G, Vickerman JC. Rapid Commun Mass Sp. 2003;17:2163–2167. doi: 10.1002/rcm.1169. [DOI] [PubMed] [Google Scholar]
- 30.Ariga T, Macala LJ, Saito M, Margolis RK, Greene LA, Margolis RU, Yu RK. Biochemistry. 1988;27:52–58. doi: 10.1021/bi00401a010. [DOI] [PubMed] [Google Scholar]
- 31.Kotter F, Benninghoven A. Appl Surf Sci. 1998;133:47–57. [Google Scholar]
- 32.Fletcher JS, Lockyer NP, Vickerman JC. Mass Spectrom Rev. 2010;30:142–174. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- 33.Nygren H, Hagenhoff B, Malmberg P, Nilsson M, Richter K. Microsc Res Technique. 2007;70:969–974. doi: 10.1002/jemt.20502. [DOI] [PubMed] [Google Scholar]