Abstract
Analyses on DNA microarrays depend considerably on spot quality and a low background signal of the glass support. By using betaine as an additive to a spotting solution made of saline sodium citrate, both the binding efficiency of spotted PCR products and the homogeneity of the DNA spots is improved significantly on aminated surfaces such as glass slides coated with the widely used poly-l-lysine or aminosilane. In addition, non-specific background signal is markedly diminished. Concomitantly, during the arraying procedure, the betaine reduces evaporation from the microtitre dish wells, which hold the PCR products. Subsequent blocking of the chip surface with succinic anhydride was improved considerably in the presence of the non-polar, non-aqueous solvent 1,2-dichloroethane and the acylating catalyst N-methylimidazole. This procedure prevents the overall background signal that occurs with the frequently applied aqueous solvent 1-methyl-2-pyrrolidone in borate buffer because of DNA that re-dissolves from spots during the blocking process, only to bind again across the entire glass surface.
INTRODUCTION
DNA microarrays are produced by in situ synthesis of oligonucleotides (1,2) or the immobilisation of pre-fabricated molecules (3). Currently, glass slides are mainly used as support medium because of their favourable optical characteristics. Especially for transcriptional profiling analyses (4,5), PCR products are spotted onto activated glass surfaces coated with poly-l-lysine or aminosilane. Since the efficiency of binding PCR products to glass slides still limits the sensitivity and the dynamic range of such measurements, performance is directly influenced by the amount of DNA that is attached to the surface. Also, DNA spots of high homogeneity are beneficial, since they simplify image analysis and considerably enhance the accuracy of signal detection. One important factor in the spotting process is the chemical properties of the solution in which the DNA is dissolved. With the widely used saline sodium citrate (SSC) buffer, binding efficiency and spot uniformity are often poor. The problems are reduced by supplementing SSC with 50% dimethyl sulfoxide. This reaction buffer has the disadvantage, however, of being both toxic and a solvent for many materials, apart from its only limited effect on spot appearance.
Another critical part of microarray manufacturing is the processing of the glass surface after spotting, during which the remaining, unreacted amino residues of the poly-l-lysine polymer or aminosilane are deactivated. This prevents subsequent binding of DNA, which increases the background signal upon hybridisation of a labelled target. Blocking is usually achieved by reacting the arrays with succinic anhydride in aqueous, borate-buffered 1-methyl-2-pyrrolidinone (NMP), converting the amines into carboxylic moieties (3,5). During this process, however, the spotted DNA comes in contact with the aqueous blocking solution, is partly re-dissolved and spread across the entire slide. To prevent this, we developed a robust processing protocol that makes use of a non-polar, non-aqueous solvent and accelerates the blocking reaction by the addition of a catalyst.
MATERIALS AND METHODS
Probe and target synthesis
For the analysis, non-homologous DNA inserts of ∼500 bp in length were picked at random from a clone library generated by cDNA representational difference analysis (6). They were PCR-amplified in 100 µl reactions with the universal primer d(AGGCAACTGTGCTATCCGAGGGAA), purified by an isopropanol precipitation and resuspended in water. The DNA concentration was determined by measuring the fluorescence signal obtained in the presence of the dye Hoechst-33258. Purity of the fragments was checked by agarose gel electrophoresis. For the generation of complementary hybridisation targets, a Cy5-labelled oligonucleotide primer of identical sequence was used for amplification.
Fabrication of microarrays
Poly-l-lysine-coated glass slides of 75 × 25 mm were prepared as described (3) (http://cmgm.stanford.edu/pbrown/MGuide). Slides with aminosilane surface (CMT-GAPS™) were purchased from Corning (Corning, USA). The DNA spotting solution was adjusted to either 45 mM sodium citrate pH 7.0, 450 mM NaCl (3× SSC) or the same composition supplemented with 1.5 M betaine (N,N,N-trimethylglycine; Sigma, Germany). DNA spotting was done with an SDDC-2 DNA Micro-Arrayer from Engineering Services Inc. (Toronto, Canada). A single SMP3 pin (TeleChem International Inc., Sunnyvale, USA) was used to avoid differences between pins. The DNA samples were printed in quadruplicate at a 200 µm centre-to-centre spacing. Slides were left at room temperature overnight and then heat-treated on a metal block at 80°C for 5 s. The DNA was crosslinked to the support by UV irradiation with a total energy of 60 mJ in a Hoefer UV-crosslinker (Amersham Pharmacia Biotech, Freiburg, Germany). For the blocking process, 1 g succinic anhydride (Fluka, Deisendorf, Germany) was freshly dissolved in 200 ml anhydrous 1,2-dichloroethane (DCE; Fluka). To this solution, 2.5 ml of N-methylimidazol (Fluka) was added and immediately poured into the slide chamber. Incubation was for 1 h, placed on an orbital shaker for slight agitation. Subsequently, the slides were briefly washed in 200 ml of fresh DCE and incubated in boiling water for 2 min for DNA denaturation. After a brief rinse in 95% ethanol, they were left to dry at room temperature. Blocking with succinic anhydride in borate-buffered NMP was carried out according to the protocol published by Eisen and Brown (5).
Hybridisation of labelled samples
For each hybridisation, 0.2 µg of Cy5-labelled and 1.8 µg of unlabelled PCR product were mixed and precipitated with ethanol. The pellet was taken up in 15 µl hybridisation buffer of 50% formamide, 3× SSC, 1% SDS, 5× Denhardt’s reagent and 5% dextran sulfate (7). The sample was denatured at 80°C for 10 min, applied to a microarray and spread evenly by a coverslip of 22 × 22 mm. Hybridisation was carried out for 16 h at 42°C in a humidified hybridisation chamber (TeleChem International Inc.). The slides were washed in 2× SSC, 0.1% SDS for 2 min, then in 1× SSC for 2 min, rinsed briefly in 0.2× SSC and dried by centrifugation at 500 r.p.m. for 5 min. Detection of the fluorescence signals was performed on a ScanArray5000 unit and analysed with the QuantArray1.0 software package (GSI Lumonics, Billerica, USA).
RESULTS
Effectiveness of DNA binding
One critical factor in microarray analyses is the amount of probe material attached to the support that is available for hybridisation. This factor can quickly become limiting to the signal intensities detectable on glass arrays and thus directly influences the sensitivity and dynamic range of measurements. In order to determine how the buffer condition of the spotting solution affects the binding efficiency of the spotted DNA, PCR products of ∼500 bp in length were produced from individual clone inserts, which had been randomly picked from a subtractive human clone library. The DNA was diluted to concentrations of 500, 250, 100, 50 and 25 ng/µl and applied to glass slides in four replica-spots each. Spotting solution without DNA was also deposited. Parallel to 3× SSC and the same buffer supplemented with 1.5 M betaine, the commercial ArrayIt™ micro-spotting solution (TeleChem International Inc.) was tested.
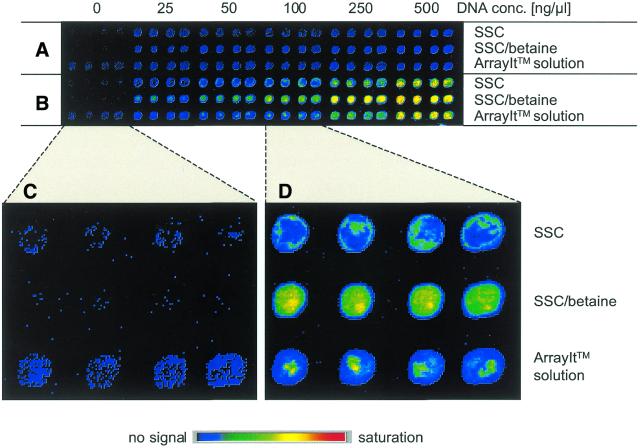
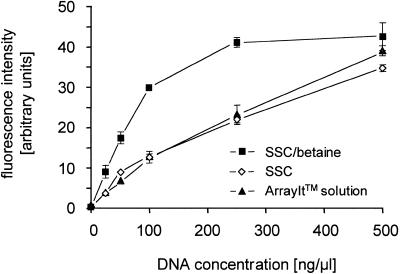
In hybridisations, labelled PCR products were used as target DNA. Figure 1 shows a typical image of fluorescence signal intensities obtained from such experiments. Irrespective of the buffer, hybridisation was specific to the complementary probe molecule. Also, in all cases the signal intensities increased with increasing concentration of the spotted DNA probe solution. However, quantification reveals that, at a DNA concentration in the spotting solution of up to 100 ng/µl, the signal intensities were ∼2.5-fold higher if betaine was present in the spotting buffer (Fig. 2). Correspondingly, the binding capacity of the glass surface is nearly saturated at a DNA concentration of 250 ng/µl, while without betaine this level is reached only at a concentration >500 ng/µl.
Figure 1.
Signal intensities produced upon hybridisation of Cy5-labelled DNA to increasing amounts of spotted PCR product. Spots made with each DNA concentration and buffer system were present in quadruplicate. (A) The background of non-specific binding to a non-complementary sequence. (B) The signals obtained on a fully complementary probe. Enlargements that display in detail (C) the background signal collected in absence of DNA and (D) the homogeneity of signal at spots produced with 100 ng/µl DNA.
Figure 2.
Effect of probe concentration and spotting solution on hybridisation efficiency. The mean signal intensities produced in the experiments shown in Figure 1 are plotted versus the DNA concentration of the spotted DNA. The error bars indicate standard deviation.
Spot homogeneity
Spot homogeneity is dependent on the variation of the DNA concentration across a spot. There are distinct, frequently occurring patterns that can be observed upon hybridisation, such as a higher DNA concentration at the edges (‘doughnut’ effect) or the aggregation of the DNA at few points within a spot. The former effect was seen on slides printed with DNA in pure SSC buffer, while the latter occurred when the ArrayIt™ micro-spotting solution was used (Fig. 1). Supplementing SSC with 1.5 M betaine yielded much more homogenous spots. This effect was evaluated by calculating the variation coefficient of signal intensity across all pixels that represent a spot. At a DNA concentration of 100 ng/µl during spotting (Fig. 1D), for example, the variation coefficient was found to be 7% with the commercial buffer, 14% if SSC was used and only 5% for SSC supplemented with betaine.
Spot-specific background signal
The choice of spotting solution also has a noticeable effect on the background signal produced at the spots in absence of a complementary target DNA. In Figure 1C, typical results are presented where buffer lacking DNA has been spotted. Particular care had been taken to avoid any carry-over of DNA from other samples by extensive washing steps and spotting the buffer probe first before proceeding to samples containing DNA. The signal:noise ratio of each feature was calculated by dividing the mean signal intensity of the four spot areas by the mean of the background signal in between spots. A ratio of 0.7 (±0.2) was found for 3× SSC supplemented with 1.5 M betaine, while much higher ratios of 5.1 (±0.8) and 10.5 (±1.5) were determined for SSC without betaine and the TeleChem ArrayIt™ micro-spotting solution, respectively.
Suppression of overall background
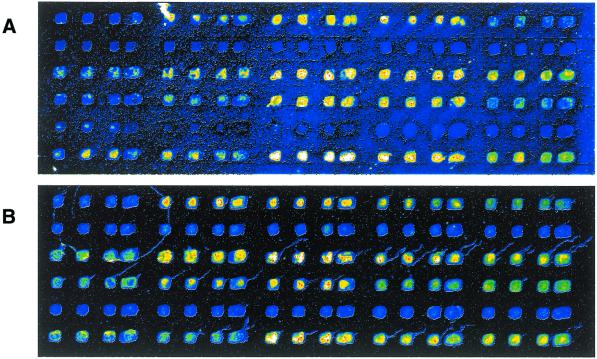
The protocol of slide post-processing with succinic anhydride was introduced by Schena et al. (3) and is widely used for the blocking of aminated surfaces by acylating the unreacted primary amines. In this process, succinic anhydride is first dissolved in NMP before sodium borate buffer pH 8 is added; the final concentrations are 164 mM succinic anhydride, 96% (v/v) NMP and 4% (v/v) aqueous sodium borate buffer. We suspected that an incubation in this solution re-dissolves part of the DNA deposited on the glass surface, which could then spread across the slide, causing additional background. In an effort to avoid this effect, we substituted the non-polar, non-aqueous solvent DCE for NMP. The concentration of succinic anhydride was decreased to 50 mM. Also, no aqueous buffer was added to the solution. Instead, the acylating catalyst N-methylimidazol was added for acceleration of the process. We did comparisons of slides produced and processed in parallel but acylated by either the NMP method or our DCE protocol. With the latter blocking reaction, an overall significantly reduced background is achieved (Fig. 3). Since using the DCE-based process as our routine blocking procedure, we have not encountered any background problems that could be attributed to the blocking, whereas before, when using the NMP method, we experienced all commonly known problems, such as inverted signal phenomena or a higher background around DNA spots.
Figure 3.
Comparison of blocking reactions. Two microarray slides were produced simultaneously before being subjected to the blocking procedures. Acylation was performed using (A) 164 mM succinic anhydride in borate-buffered NMP or (B) 50 mM succinic anhydride and 150 mM N-methylimidazol in DCE. The slides were hybridised in parallel with a Cy5-labelled, complementary PCR product, washed briefly and scanned under identical conditions. The slight DNA ‘tails’ seen in (B) are caused by target DNA left after the brief washing. Such features occur on both types of slides, as could be determined by radioactive hybridisations (not shown), but are submerged in the background signal of (A).
DISCUSSION
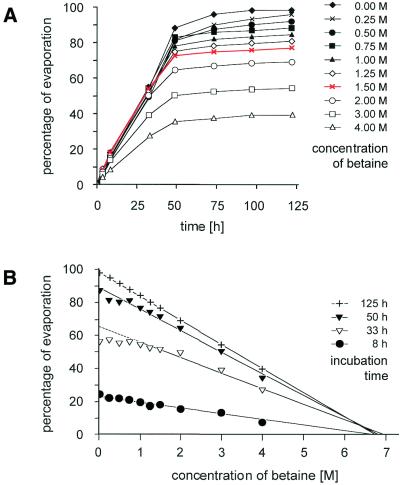
The results described above suggest that binding of DNA to poly-l-lysine slides in the presence of betaine is a different reaction from spotting DNA in SSC alone. Betaine is a naturally occurring substance that serves as an osmoprotectant factor in bacteria (8). It is known to alter DNA and protein stability and to reduce the difference in stability of A:T and G:C base pairs (9,10). Because of these effects, it has been introduced as an additive in sequencing reactions and different PCR strategies (11,12). In physical terms, betaine increases the viscosity of a solution and reduces the rate of evaporation, dependent on its concentration (Fig. 4). However, surface tension is less affected (data not shown). These characteristics are likely to account for its effects as an additive to spotting solution. The increased binding efficiency and spot homogeneity is most likely due to the reduced evaporation rate. Only very small volumes in the nanolitre range are deposited on the microarray surface during spotting. Because of the swift evaporation of such small volumes, the electrostatic binding of DNA on the positively charged surface must occur within a very short period. Also, the quickly receding liquid film dictates where binding takes place. If spots stay humid for longer, however, the DNA is more likely to bind at equal rates across the entire surface. As a side effect, betaine reduces the evaporation of the DNA samples in the microtiter plates during the microarray manufacturing process. Varying the parameters, we found that a concentration of 1.5 M betaine had the overall best effect on the quality of DNA microarrays.
Figure 4.
Effect of betaine on evaporation. Spotting solution (1 ml) was supplemented with different concentrations of betaine in a 1.5 ml Eppendorf tube, which was incubated with an open lid at 30°C. (A) The percentage of evaporation is presented. Note that by the increase in betaine concentration at the liquid surface the evaporation eventually stops. From this data, it can be extrapolated that a concentration of 6.8 M betaine prevents further evaporation (B).
The substitution of DCE for the NMP solution during the blocking of the glass surface subsequent to DNA spotting and the addition of an acylation catalyst improved the background considerably. In most published reports, NMP has been used as solvent of the acylating succinic anhydride. In this procedure, sodium borate buffer is added to keep the pH at 8, thus enhancing deprotonation of the charged amines. Deprotonation leaves a free pair of electrons on the amine, which can undergo a nucleophilic attack on the carbon of succinic anhydride. We chose DCE because of its non-polar, non-aqueous nature, while nevertheless being a solvent of succinic anhydride and N-methylimidazol. N-methylimidazol is a tertiary amine, which is used as a standard acylation catalyst in organic chemistry. Since it not only acts as a coupling activator but also has a basic character, there is no need for the use of other buffer components.
In combination, the modifications in the compositions of the spotting solution and the blocking reagent led to a significant improvement in the quality of microarrays, affecting sensitivity and accuracy of measurements, thereby moving such analyses another step toward more quantitative performance.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Achim Stephan for the preparation of slides and Philipp Angenendt for technical help. This work was funded by the German Ministry of Education and Research (BMBF).
References
- 1.Maskos U. and Southern,E.M. (1992) Oligonucleotide hybridisations on glass supports: a novel linker for oligonucleotide synthesis and hybridisation properties of oligonucleotides synthesised in situ. Nucleic Acids Res., 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fodor S.P.A., Rava,R.P., Huang,X.C., Pease,A.C., Holmes,C.P. and Adams,C.L. (1993) Multiplexed biochemical assays with biological chips. Nature, 364, 555–556. [DOI] [PubMed] [Google Scholar]
- 3.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 4.The Chipping Forecast (1999) Nature Genet., 21 (suppl.), 1–60. [Google Scholar]
- 5.Eisen M.B. and Brown,P.O. (1999) DNA-arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 6.Hubank M. and Schatz,D.G. (1994) Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res., 22, 5640–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welford S.M., Gregg,J., Chen,E., Garrison,D., Sorensen,P.H., Denny,C.T. and Nelson,S.F. (1998) Detection of differentially expressed genes in primary tumor tissues using representational differences analysis coupled to microarray hybridisation. Nucleic Acids Res., 26, 3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csonka L.N., Ikeda,T.P., Fletcher,S.A. and Kustu,S. (1994) The accumulation of glutamate is necessary for optimal growth of Salmonella typhimurium in media of high osmolality but not induction of the proU operon. J. Bacteriol ., 176, 6324–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoro M.M., Liu,Y., Khan,S.M., Hou,L.X. and Bolen,D.W. (1992) Increased thermal stability of proteins in the presence of naturally occurring osmolytes. Biochemistry, 16, 5278–5283. [DOI] [PubMed] [Google Scholar]
- 10.Rees W.A., Yager,T.D., Korte,J. and Hippel,P.H. (1993) Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry, 32, 137–144. [DOI] [PubMed] [Google Scholar]
- 11.Mytelka D.S. and Chamberlin,M.J. (1996). Analysis and suppression of DNA polymerase pauses associated with a trinucleotide consensus. Nucleic Acids Res., 24, 2774–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henke W., Herdel,K., Jung,K., Schnorr,D. and Loening,S.A. (1997) Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res., 25, 3957–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]