Abstract
INTRODUCTION:
Williams-Beuren syndrome (WBS; OMIM 194050) is caused by a hemizygous contiguous gene microdeletion at 7q11.23. Supravalvular aortic stenosis, mental retardation, overfriendliness, and ocular and renal abnormalities comprise typical symptoms in WBS. Although fluorescence in situ hybridization is widely used for diagnostic confirmation, microsatellite DNA markers are considered highly informative and easily manageable.
OBJECTIVES:
This study aimed to test the microsatellite markers for the diagnosis of Williams-Beuren syndrome, to determine the size and parental origin of microdeletion, compare the clinical characteristics between patients with different sizes of the deletion and parental origin.
METHODS:
We studied 97 patients with clinical diagnosis of Williams-Beuren syndrome using five microsatellite markers: D7S1870, D7S489, D7S613, D7S2476 and D7S489_A.
RESULTS AND DISCUSSION:
Using five markers together, the result was informative in all patients. The most informative marker was D7S1870 (78.4%), followed by D7S613 (75.3%), D7S489 (70.1%) and D7S2476 (62.9%). The microdeletion was present in 84 (86.6%) patients and absent in 13 (13.4%) patients. Maternal deletions were found in 52.4% of patients and paternal deletions in 47.6% of patients. The observed size of deletions was 1.55 Mb in 76/84 patients (90.5%) and 1.84 Mb in 8/84 patients (9.5%). SVAS as well as ocular and urinary abnormalities were more frequent in the patients with a deletion. There were no clinical differences in relation to either the size or parental origin of the deletion.
CONCLUSION:
Using these five selected microsatellite markers was informative in all patients, thus can be considered an alternative method for molecular diagnosis in Williams-Beuren syndrome.
Keywords: Williams-Beuren syndrome; microsatellite markers, parental origin; size deletion; molecular diagnosis
INTRODUCTION
Williams-Beuren syndrome (WBS; OMIM 194050) is a neurodevelopmental disorder described independently,1,2 as a syndrome involving facial appearance characteristics, supravalvular aortic stenosis (SVAS) and mental retardation. In fact, WBS presents a wide collection of symptoms affecting blood vessels, growth, intelligence, and behavior. Children with this condition have distinctive “elfin” facial features, a hoarse voice associated with growth, mental retardation and an “engaging” personality;3,4 hyperacusis, infantile hypercalcemia, prematurely wrinkled skin and SVAS are also common symptoms.5-8
WBS is generally sporadic with an incidence of 1/13,700–1/25,000 live births with no ethnic or sex preference, although familial cases have been reported with apparent autosomal dominant inheritance.9-11 Despite the consistency of the overall clinical features, the broad spectrum of anomalies and phenotypic variability frequently lead to a significant difference in the number of patients diagnosed.12,13
The most common deletion, found in 90% to 95% of WBS patients, spans a genomic region of ∼1.55 Mb and is the result of mispairing between the centromeric and medial LCR blocks B (Bcen and Bmid).14-16 In 5% to 10% of cases, the breakpoints are within the centromeric and medial LCR blocks A (Acen and Amid), leading to an ∼1.84 Mb deletion.17,18 Atypical (∼0.2 Mb to ∼2.5 Mb) deletions may be the leading cause of the substantial phenotypic variability among WBS patients.
Duplication of the WBS region occurs at half the frequency of deletions with less distinctive and somehow opposite clinical features, such as deficits of social interaction and an autistic-like phenotype.19-20 Autism was also described in an atypically large deletion (2.4 Mb to 3.4 Mb) telomeric to the WBS critical region.21 The presence of inversions in WBS patients has also been reported.22,18
The origin of deletion occurs at the same frequency, and there is no clear-cut clinical association between parental imprinting and phenotype differences, except for hypertension and some aspects of growth, such as height, weight and head circumference.17,13,23,24
Although fluorescence in situ hybridization (FISH) is widely used and considered the gold standard for WBS molecular diagnosis, the use of microsatellite DNA markers has also been widely used and is considered highly informative and easily manageable.15,17,25,26,27 Confirmation of clinical suspicion is essential for clinical monitoring of the patient and genetic counseling of the family.
In our previous study,28 we assessed the use of three microsatellite markers (D7S1870, Hei and ELN 17/ exon 18) in the confirmation of the diagnosis of 32 WBS patients. They were informative in 78% of patients and uninformative in 22%. The most informative marker was D7S1870. In the present study, we maintained only the D7S1870 marker and added four other microsatellite markers to determine the size and parental origin of the microdeletion in the 7q11.23 region.
MATERIALS AND METHODS
Subjects
A total of 97 patients with a clinical diagnosis of WBS (61 boys and 36 girls) were ascertained through clinical evaluation by geneticists of the Unit of Clinical Genetics of the Instituto da Criança, Hospital das Clínicas, Faculdade de Medicina, Universidade (ICr-HC-FMUSP) de São Paulo, Brazil.
The clinical diagnosis was made between the ages of 1 and 16 years, with an average of 6 y 2 mo. The mean maternal and paternal ages were 24 y 2 mo and 27 y, respectively.
Methods
The study was approved by the Institutional Review Board, and written consent was obtained from all participants. The molecular study was performed in the Laboratório de Investigação Médica do Instituto da Criança (LIM-36).
Among the 97 patients, DNA from both parents was obtained in 82 cases; in 13 cases, the molecular analysis was performed only with maternal DNA and in two cases, only with paternal DNA.
DNA was isolated from peripheral blood lymphocytes using a salt precipitation technique29 and quantified in a Nano Vue Plus spectrophotometer (GE, New Jersey, USA).
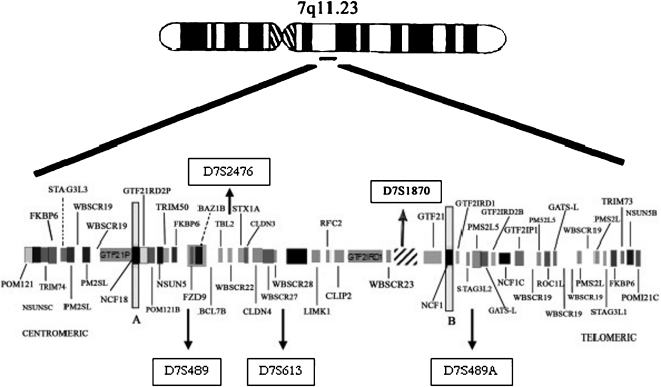
The five microsatellites markers used were D7S1870, D7S489, D7S613 and D7S2476 inside the common 1.55-Mb deletion; D7S489A was used to distinguish deletions of 1.84 Mb (Fig. 1).
Figure 1.
Idiogram of chromosome 7 (7q11.23) illustrating the commonly deleted region and the relative localization of the tested markers.29-32
Polymerase chain reaction
PCR reactions were carried out in a total volume of 25 µL containing 50 ng of genomic DNA, 0.2 mM of dNTP mix, 0.4 µM of the each oligonucleotide, and 0.7 units of EasyTaq DNA Polymerase (LGC, São Paulo, Brazil). Samples were initially denatured for 5 min at 94°C; followed by 35 cycles of 1 min at 94°C, 1 min at the specific annealing temperature of each primer pair and 1 min at 72°C; and a final extension step of 15 min at 72°C. Primer sequences and PCR conditions used in this study are given in table 1. To check PCR amplification, 4 µL of the reaction products were analyzed on 2% agarose gel stained with ethidium bromide and visualized by UV exposure in an AlphaImager HP (Alpha Innotech, Corp. San Leandro, CA).
Table 1.
Primers sequences, amplicon sizes and PCR conditions for the five microsatellites markers used.
| Markers | Primers | Amplicon size | MgCl2 | UniSTS Accesion number * | AT* |
| 7q11.23 | |||||
| D7S1870 | F: TTCACTCAGGAAGTGGC | 108 pb | 2,0 mM | Z51768 | 55°C |
| R: TGGTGATGTGCTTTACTACG | |||||
| D7S489 | F: CTGTTGACTTTCCCACACTC | 140-158 pb | 1,5 mM | Z16646 | 56°C |
| R: GGCAACTCGAGACGTTAGTT | |||||
| D7S613 | F: CAGCCTGGGTAACAAAAGC | 85 pb | 1,4 mM | L16300 | 55°/60°C |
| R: CCTCCCTCCCTAATCCATG | |||||
| D7S2476 | F: GGGCAACATAGCACGATT | 128-160 pb | 2,0 mM | Z53107 | 56°C |
| R: CAGGAGTCAGTTAGATAAGGTCAC | |||||
| D7S489_A | F: GCACCTATGATCACAGCTTCTC | 419 pb | 2,0 mM | BV097124 | 56°C |
| R: ATGACATGAAGGTACTGGCCTT |
*Accession number in the GeneBank; AT – anelling temperature; pb – pairs base.
Polyacrylamide gel
The PCR amplification product was mixed in an equal volume of denaturing solution (99% formamide, 0.01% bromophenol blue; 0.01% xylenecyanol and 10 mM EDTA), denatured for 10 min at 95°C and separated by electrophoresis in a 7% denaturing polyacrilamide gel with urea at a final concentration of 7.5 M. Electrophoresis was performed in a Mini-Proteans III Cell apparatus (Bio-Rad®). The electrophoresis conditions were 105 V, 25 mA, and 2 W for two hours for the markers D7S1870, D7S489, D7S613 and D7S2476 and 230 V, 60 mA, and 5 W for four hours for marker D7S489A. The gels were fixed in 10% ethanol, silver stained and stored in 10% acetic acid until they could be photographed and interpreted.
The patient genotypes were compared with their parents. Deletions were diagnosed as maternal when the proband presented with gel-bands representing the allele marker inherited only from the father. When, by chance, both parents have the same alleles, the monoallelic inheritance of the corresponding microsatellite marker by the proband indicated an uninformative result.
We first used a two-step algorithm to identify the most common 1.55-Mb deletion. We then tested the D7S489A marker either to identify the larger 1.84-Mb deletion (in those patients in which a deletion of at least one marker was detected in the first step) or to confirm the lack of a deletion.
Statistical Analysis
Pairwise comparisons between clinical features of WBS and the presence of deletion, clinical features and deletion size and clinical features and parental origin of deletion were tested for significance using two-tailed Fisheŕs exact test. A 2x2 contingency table was used to compare clinical features. P analysis was performed in SPSS 13.0 software and considered statistically significant when p ≤ 0.05.
RESULTS
Using five markers (D7S1870, D7S489, D7S613, D7S2476 and D7S489A) were informative in all patients. The most informative marker was D7S1870 (78.4%), followed by D7S613 (75.3%), D7S489 (70.1%) and D7S2476 (62.9%) (Table 2).
Table 2.
Results of the analysis of microdeletions by microsatellite markers.
| D7S1870 | % | D7S613 | % | D7S489 | % | D7S2476 | % | |
| Positive | 65/97 | 67,1 | 66/97 | 68,1 | 58/97 | 59,8 | 56/97 | 57,7 |
| Negative | 11/97 | 11,3 | 7/97 | 7,2 | 10/97 | 10,3 | 5/97 | 5,2 |
| Uninformative | 21/97 | 21,6 | 24/97 | 24,7 | 29/97 | 29,9 | 36/97 | 37,1 |
| Informative | 76/97 | 78,4 | 73/97 | 75,3 | 68/97 | 70,1 | 61/97 | 62,9 |
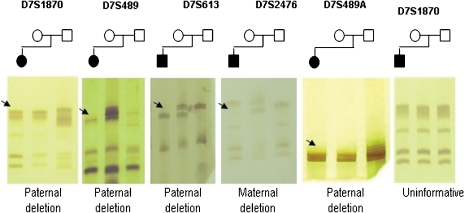
The microdeletion was present in 84/97 (86.6%) patients and absent in 13 (13.4%) patients. A prototypic acrylamide gel showing some of the results is shown in Figure 2.
Figure 2.
Genotyping of the five microsatellites markers in WBS families. DNA fragments of those affected are always the first column of each gel followed by DNA from the mother and the last column the DNA of the father. Black arrows indicate allelic loss.
All deletions were de novo, and none of the available parents had any clinical manifestation of WBS. It was also possible to determine the presence of a deletion for the 15 patients for whom DNA was available from only one of the parents.
There was no difference in clinical features found in patients with and without a deletion except SVAS as well as ocular and urinary abnormalities, more frequent with statistically significant association with a deletion. (Table 3) Other cardiovascular diseases, such as pulmonary artery stenosis, mitral valve prolapse and ventricular septum defects, were similar between the two groups. Ocular abnormalities observed were strabismus, stellate irides, punctuate opacities in the lens, glaucoma and myopia. Urinary abnormalities present included enuresis, diverticulosis, asymmetric kidneys, narrowing of the renal artery and renal microlithiasis.
Table 3.
Clinical fidings in patients with and without deletion.
| Clinical Findings | With deletion | Without deletion | p |
| Frequency % | Frequency % | ||
| Typical facies | 100.0 | 100.0 | – |
| Mental retardation | 100.0 | 100.0 | – |
| Friendly personality | 92.5 | 91.7 | – |
| Hyperacusis | 81.9 | 58.3 | 0.120 |
| Height (<5%) | 51.2 | 23.1 | 0.059 |
| Microcephaly | 24.7 | 23.1 | – |
| Cardiovascular disease | 77.5 | 61.5 | 0.297 |
| SVAS | 58.1 | 12.5 | 0.022 |
| Ocular abnormalities | 65.2 | 20.0 | 0.012 |
| Urinary abnormalities | 48.1 | 16.7 | 0.041 |
The observed size of deletions was 1.55 Mb in 76/84 patients (90.5%) and 1.84 Mb in 8/84 patients (9.5%). The parental origin of deletion was maternal in 44/84 patients (52.4%), and paternal deletions in 40/84 patients (47.6%). There was no clinical difference in relation to either the size of deletion or the parental origin of deletion.
Twenty-three patients were previously studied by FISH, and 21 proved to have a deletion (FISH positive); whereas 2 patients were FISH negative. Microsatellite marker analysis of these cases revealed microdeletions in all patients, including the two FISH negative patients.
DISCUSSION
The five microsatellite markers (D7S1870, D7S489, D7S613, D7S2476, and D7S489A) used in our study were informative in all patients.
The D7S1870 microsatellite marker showed the highest power of detection, identifying 78.4% of the cases alone and confirming the results found by previous studies.17,18,26,28,34-37
Another two markers (D7S613, D7S489) were also able to diagnose over 70% of our patients, while the D7S2476 marker showed the poorest results (62.9%). These results were similar to those found in the literature.17,25,26,37
The two best markers (D7S1870 and D7S613) in our study were informative in 97.6% to the detect deletion when used together. When the third marker, D7S489, was included informative detection increased to almost 99%. To increase the detection closer to 100%, additional useful markers will need to be chosen.27,23,35-37
The microsatellite marker D7S489A was effective in the analysis of the size of the deletion and in the confirmation of the absence of fragments in the region critical for WBS. The 1.55-Mb deletion was found in 76/84 (90.5%) patients, and the 1.84-Mb deletion found in 8/84 patients (9.5%); these observed proportions are similar to those found in studies in the literature.18,21
Clinical findings in our patients were similar in the groups with a deletion and without a deletion, except for SVAS and ocular and urinary abnormalities. There was also no relationship between the size of the deletion and observed clinical features. These results are similar to those found in literature,23,33 where the clinical features found in WBS patients were very similar with both the 1.55-Mb and 1.84-Mb deletions.
In the 13 cases without a deletion, there is the possibility of a deletion smaller than 1.0 Mb that is not detectable by microsatellite markers; thus, this diagnostic should not be ruled out.38,39
There was no significant difference between the frequencies of maternal and paternal deletions (52.4% and 47.6% respectively). The literature 18,23,28,36,37,40-42 is concordant with our findings, except for one study,25 where the maternal deletion was more frequent (77.8%) than the paternal deletion (22.2%).
Despite the frequency of the paternal and maternal deletions being very close, some authors have suggested a slight tendency for deletion due to the effect of maternal genomic imprinting, when all data are combined.25,36,42
The phenotypic aspects between the groups of the patients with paternally (n = 44/84) and maternally derived (n = 40/84) deletions were similar. Some reports in the literature, however, suggest a parent-of-origin effect on microcephaly and growth retardation in WBS.17,36
The FISH technique is currently considered the “gold standard” for molecular diagnosis of WBS, but partial microdeletions can lead to false negative results depending on the sequence and composition of the probe used.25,30,38 In the present study, the results obtained by FISH were compared with microsatellite marker analysis. The results were concordant, except for two negative FISH.
Other techniques in molecular biology are also being used for the molecular diagnosis of WBS, such as quantitative real-time PCR, multiplex ligation dependent probe amplification (MLPA) and array comparative genomic hybridization (aCGH), however, many other factors should be considered in relation to the cost-benefit and practicality.43-45 In developed country, aCGH is currently the method of choice for genome-wide screening for chromosome, but in the developing country this technic is not in a disposal due to an expensive cost.46,47
CONCLUSION
The diagnosis of WBS based on clinical assessments may be difficult because of the great variability of its manifestations. Laboratory tests to detect the microdeletion in 7q11.23 are essential to confirm the clinical diagnosis for WBS. Although fluorescence in situ hybridization (FISH) is widely used, microsatellite DNA markers are considered highly informative and easily manageable for diagnostic confirmation.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Ulysses Dória-Filho (Nucleo de Apoio Metodológico Instituto da Criança - FMUSP) for his help with the statistical analysis. This work was supported by FAPESP 2008/55391-6.
REFERENCES
- 1.Williams JC, Barratt-Boyes BG, Lowe JB. Supravalvular aortic stenosis. Circulation. 1961;24:1311–18. doi: 10.1161/01.cir.24.6.1311. [DOI] [PubMed] [Google Scholar]
- 2.Beuren AJ, Apitz J, Harmjanz D. Supravalvular aortic stenosis in association with mental retardation and a certain facial appearance. Circulation. 1962;26:1235–40. doi: 10.1161/01.cir.26.6.1235. [DOI] [PubMed] [Google Scholar]
- 3.Preus M. The Williams syndrome: objective definition and diagnosis. Clin Genet. 1984;25:422–28. doi: 10.1111/j.1399-0004.1984.tb02011.x. 10.1111/j.1399-0004.1984.tb02011.x [DOI] [PubMed] [Google Scholar]
- 4.Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. 10.1038/ng0993-11 [DOI] [PubMed] [Google Scholar]
- 5.Martin ND, Snodgrass GJ, Cohen RD. Idiopathic infantile hypercalcemia: a continuing enigma. Arch Dis Child. 1984;59:605–613. doi: 10.1136/adc.59.7.605. 10.1136/adc.59.7.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burn J. Williams syndrome. J Med Genet. 1986;23:389–95. doi: 10.1136/jmg.23.5.389. 10.1136/jmg.23.5.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: physical characteristics. J Pediatr. 1988;113:318–26. doi: 10.1016/s0022-3476(88)80272-5. 10.1016/S0022-3476(88)80272-5 [DOI] [PubMed] [Google Scholar]
- 8.Udwin O. A survey of adults with Williams syndrome and idiopathic infantile hypercalcaemia. Dev Med Child Neurol. 1990;32:129–41. doi: 10.1111/j.1469-8749.1990.tb16912.x. 10.1111/j.1469-8749.1990.tb16912.x [DOI] [PubMed] [Google Scholar]
- 9.Morris CA, Thomas IT, Greenberg F. Williams syndrome: autosomal dominant inheritance. Am J Med Genet. 1993;47:478–81. doi: 10.1002/ajmg.1320470409. 10.1002/ajmg.1320470409 [DOI] [PubMed] [Google Scholar]
- 10.Sadler LS, Robinson LK, Verdaasdonk KR, Gingell R. The Williams syndrome: evidence for possible autosomal dominant inheritance. Am J Med Genet. 1993;47:468–70. doi: 10.1002/ajmg.1320470406. 10.1002/ajmg.1320470406 [DOI] [PubMed] [Google Scholar]
- 11.Strømme P, Bjørnstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17:269–71. doi: 10.1177/088307380201700406. 10.1177/088307380201700406 [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Rangel E, Maurice M, McGillivray B, Friedman JM. Williams syndrome in adults. Am J Med Genet. 1992;44:720–29. doi: 10.1002/ajmg.1320440605. 10.1002/ajmg.1320440605 [DOI] [PubMed] [Google Scholar]
- 13.Ashkenas J. Williams syndrome starts making sense. Am J Hum Genet. 1996;59:756–61. [PMC free article] [PubMed] [Google Scholar]
- 14.Meng X, Lu X, Li Z, Green ED, Massa H, Trask BJ, et al. Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes. Hum Genet. 1998;103:590–99. doi: 10.1007/s004390050874. 10.1007/s004390050874 [DOI] [PubMed] [Google Scholar]
- 15.Peoples R, Franke Y, Wang YK, Pérez-Jurado L, Paperna T, Cisco M, et al. A physical map, including a BAC/PAC clone contig, of the Williams-Beuren syndrome deletion region at 7q11.23. Am J Hum Genet. 2000;66:47–68. doi: 10.1086/302722. 10.1086/302722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merla G, Brunetti-Pierri N, Micale L, Reymond A. Copy number variants at Williams-Beuren syndrome 7q11.23 region. Hum Genet. 2010;128:3–26. doi: 10.1007/s00439-010-0827-2. 10.1007/s00439-010-0827-2 [DOI] [PubMed] [Google Scholar]
- 17.Pérez Jurado LA, Peoples R, Kaplan P, Hamel BC, Francke U. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet. 1996;59:781–92. [PMC free article] [PubMed] [Google Scholar]
- 18.Bayés M, Magano LF, Rivera N, Flores R, Pérez Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–51. doi: 10.1086/376565. 10.1086/376565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, Fong CT, Salamone J, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–41. doi: 10.1097/gim.0b013e3180986192. 10.1097/GIM.0b013e3180986192 [DOI] [PubMed] [Google Scholar]
- 20.Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, et al. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet. 2009;52:94–100. doi: 10.1016/j.ejmg.2009.02.006. 10.1016/j.ejmg.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 21.Edelmann L, Prosnitz A, Pardo S, Bhatt J, Cohen N, Lauriat T, et al. An atypical deletion of the Williams-Beuren syndrome interval implicates genes associated with defective visuospatial processing and autism. J Med Genet. 2007;44:136–43. doi: 10.1136/jmg.2006.044537. 10.1136/jmg.2006.044537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–25. doi: 10.1038/ng753. 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang MS, Schinzel A, Kotzot D, Balmer D, Casey R, Chodirker BN, et al. Molecular and clinical correlation study of Williams-Beuren syndrome: No evidence of molecular factors in the deletion region or imprinting affecting clinical outcome. Am J Med Genet. 1999;86:34–43. 10.1002/(SICI)1096-8628(19990903)86:1<34::AID-AJMG7>3.0.CO;2-4 [PubMed] [Google Scholar]
- 24.Collette JC, Chen XN, Mills DL, Galaburda AM, Reiss AL, Bellugi U, et al. William's syndrome: gene expression is related to parental origin and regional coordinate control. J Hum Genet. 2009;54:193–98. doi: 10.1038/jhg.2009.5. 10.1038/jhg.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson WP, Waslynka J, Bernasconi F, Wang M, Clark S, Kotzot D, et al. Delineation of 7q11.2 deletions associated with Williams-Beuren syndrome and mapping of a repetitive sequence to within and to either side of the common deletion. Genomics. 1996;34:17–23. doi: 10.1006/geno.1996.0237. 10.1006/geno.1996.0237 [DOI] [PubMed] [Google Scholar]
- 26.Hockenhull EL, Carette MJ, Metcalfe K, Donnai D, Read AP, Tassabehji M. A complete physical contig and partial transcript map of the Williams syndrome critical region. Genomics. 1999;58:138–45. doi: 10.1006/geno.1999.5815. 10.1006/geno.1999.5815 [DOI] [PubMed] [Google Scholar]
- 27.Fomin AB, Pastorino AC, Kim CA, Pereira CA, Carneiro-Sampaio M, Abe-Jacob CM. DiGeorge Syndrome: a not so rare disease. Clinics. 2010;65:865–69. doi: 10.1590/S1807-59322010000900009. 10.1590/S1807-59322010000900009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sbruzzi IC, Pereira AC, Vasconcelos B, Honjo RS, Krieger JE, Kim CA. Williams-Beuren Syndrome: Diagnosis by Polymorphic Markers. Genet Test Mol Biomarkers. 2010;14:209–14. doi: 10.1089/gtmb.2009.0120. 10.1089/gtmb.2009.0120 [DOI] [PubMed] [Google Scholar]
- 29.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heller R, Rauch A, Lüttgen S, Schröder B, Winterpacht A. Partial deletion of the critical 1.5 Mb interval in Williams-Beuren syndrome. J Med Genet. 2003;40:e99. doi: 10.1136/jmg.40.8.e99. 10.1136/jmg.40.8.e99",-1,"xxx/8.e99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonell A, Del Campo M, Flores R, Campuzano V, Perez-Jurado LA. Williams syndrome: its clinical aspects and molecular bases. Rev Neurol. 2006;1:S69–75. [PubMed] [Google Scholar]
- 32.Schubert C. The genomic basis of the Williams-Beuren syndrome. Cell Mol Life Sci. 2009;66:1178–97. doi: 10.1007/s00018-008-8401-y. 10.1007/s00018-008-8401-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pober BR. Williams-Beuren syndrome. N Engl J Med. 2010;362:239–52. doi: 10.1056/NEJMra0903074. 10.1056/NEJMra0903074 [DOI] [PubMed] [Google Scholar]
- 34.Gilbert-Dussardier B, Bonneau D, Gigarel N, et al. A novel microsatellite DNA marker at locus D7S1870 detects hemizygosity in 75% of patients with Williams syndrome. Am J Hum Genet. 1995;56:542–44. [PMC free article] [PubMed] [Google Scholar]
- 35.Urbán Z, Helms C, Fekete G, Le Merrer M, Bonnet D, Philip N, et al. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am J Hum Genet. 1996;59:958–62. [PMC free article] [PubMed] [Google Scholar]
- 36.Brøndum-Nielsen K, Beck B, Gyftodimou J, Hørlyk H, Liljenberg U, Petersen MB, et al. Investigation of deletions at 7q11.23 in 44 patients referred for Williams-Beuren syndrome, using FISH and four DNA polymorphisms. Hum Genet. 1997;99:56–61. doi: 10.1007/s004390050311. [DOI] [PubMed] [Google Scholar]
- 37.Wu YQ, Sutton VR, Nickerson E, Lupski JR, Potocki L, Korenberg JR, et al. Delineation of the common critical region in Williams syndrome and clinical correlation of growth, heart defects, ethnicity, and parental origin. Am J Med Genet. 1998;78:82–89. doi: 10.1002/(sici)1096-8628(19980616)78:1<82::aid-ajmg17>3.0.co;2-k. 10.1002/(SICI)1096-8628(19980616)78:1<82::AID-AJMG17>3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- 38.Botta A, Novelli G, Mari A, Novelli A, Sabani M, Korenberg J, et al. Detection of an atypical 7q11.23 deletion in Williams syndrome patients which does not include the STX1A and FZD3 genes. J Med Genet. 1999;36:478–80. [PMC free article] [PubMed] [Google Scholar]
- 39.Gagliardi C, Bonaglia MC, Selicorni A, Borgatti R, Giorda R. Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J Med Genet. 2003;40:526–30. doi: 10.1136/jmg.40.7.526. 10.1136/jmg.40.7.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Campo M, Antonell A, Magano LF, Muñoz FJ, Flores R, Bayés M, et al. Hemizygosity at the NCF1 gene in patients with Williams-Beuren syndrome decreases their risk of hypertension. Am J Hum Genet. 2006;78:533–42. doi: 10.1086/501073. 10.1086/501073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milà M, Carrió A, Sánchez A, Gómez D, Jiménez D, Estivill X, et al. Caracterización clínica y genética de 80 pacientes con sospecha clínica de síndrome de Williams-Beuren. Med Clin (Barc) 1999;113:46–49. [PubMed] [Google Scholar]
- 42.Thomas NS, Durkie M, Potts G, Sandford R, Van Zyl B, Youings S, et al. Parental and chromosomal origins of microdeletion and duplication syndromes involving 7q11.23, 15q11-q13 and 22q11. Eur J Hum Genet. 2006;14:831–37. doi: 10.1038/sj.ejhg.5201617. 10.1038/sj.ejhg.5201617 [DOI] [PubMed] [Google Scholar]
- 43.Kriek M, Knijnenburg J, White SJ, Rosenberg C, den Dunnen JT, van Ommen GJ, et al. Diagnosis of genetic abnormalities in developmentally delayed patients: a new strategy combining MLPA and array-CGH. Am J Med Genet. 2007;143:610–14. doi: 10.1002/ajmg.a.31593. 10.1002/ajmg.a.31593 [DOI] [PubMed] [Google Scholar]
- 44.van Hagen JM, Eussen HJ, van Schooten R, van Der Geest JN, Lagers-van Haselen GC, Wouters CH, et al. Comparing two diagnostic laboratory tests for Williams syndrome: fluorescent in situ hybridization versus multiplex ligation-dependent probe amplification. Genet Test. 2007;11:321–27. doi: 10.1089/gte.2007.0007. 10.1089/gte.2007.0007 [DOI] [PubMed] [Google Scholar]
- 45.Cho EH, Park BY, Cho JH, Kang YS. Comparing two diagnostic laboratory tests for several microdeletions causing mental retardation syndromes: multiplex ligation-dependent amplification vs fluorescent in situ hybridization. Korean J Lab Med. 2009;29:71–76. doi: 10.3343/kjlm.2009.29.1.71. 10.3343/kjlm.2009.29.1.71 [DOI] [PubMed] [Google Scholar]
- 46.Baris HN, Tan WH, Kimonis VE, Irons MB. Diagnostic utility of array-based comparative genomic hybridization in a clinical setting. Am J Med Genet. 2007;143:2523–33. doi: 10.1002/ajmg.a.31988. 10.1002/ajmg.a.31988 [DOI] [PubMed] [Google Scholar]
- 47.Pickering DL, Eudy JD, Olney AH, Dave BJ, Golden D, Stevens J, et al. Array-based comparative genomic hybridization analysis of 1176 consecutive clinical genetics investigations. Genet Med. 2008;10:262–66. doi: 10.1097/GIM.0b013e31816b64ad. 10.1097/GIM.0b013e31816b64ad [DOI] [PubMed] [Google Scholar]