Abstract
Transplantation of pluripotent stem cells has proven beneficial in heart failure, yet the proteomic landscape underlying repair remains largely uncharacterized. In a genetic model of dilated cardiomyopathy elicited by pressure overload in the KCNJ11 (potassium inwardly rectifying channel, subfamily J, member 11) null mutant, proteome-wide profiles were here resolved by means of a systems approach prior to and following disease manifestation in the absence or presence of embryonic stem cell treatment. Comparative two-dimensional gel electrophoresis revealed a unique cardiomyopathic proteome in the absence of therapy, remodeled in response to stem cell treatment. Specifically, linear ion trap quadrupole-Orbitrap mass spectrometry determined the identities of 93 and 109 differentially expressed proteins from treated and untreated cardiomyopathic hearts, respectively. Mapped protein– protein relationships and corresponding neighborhoods incorporated the stem cell-dependent subproteome into a nonstochastic network with divergent composition from the stem cell-independent counterpart. Stem cell intervention produced a distinct proteome signature across a spectrum of biological processes ranging from energetic metabolism, oxidoreductases, and stress-related chaper-ones to processes supporting protein synthesis/degradation, signaling, and transport regulation, cell structure and scaffolding. In the absence of treatment, bioinformatic interrogation of the disease-only proteome network prioritized adverse cardiac outcomes, ablated or ameliorated following stem cell transplantation. Functional and structural measurements validated improved myocardial contractile performance, reduced ventricular size and decreased cardiac damage in the treated cohort. Unbiased systems assessment unmasked “cardiovascular development” as a prioritized biological function in stem cell-reconstructed cardiomyopathic hearts. Thus, embryonic stem cell treatment transformed the cardiomyopathic proteome to demote disease-associated adverse effects and sustain a procardiogenic developmental response, supplying a regenerative substrate for heart failure repair.
Keywords: ATP-sensitive K+ channel, Bioinformatics, KATP channel, Dilated cardiomyopathy, Protein expression, Genetics, Networks, Proteomics, Regenerative medicine
INTRODUCTION
Cardiomyopathy is an intrinsic, progressive disorder of the myocardium resulting in impaired function of the heart [1]. The clinical entity of dilated cardiomyopathy is characterized by ventricular dilation and reduced contractile performance, precipitating heart failure and poor outcome [2, 3]. Discovery of monogenic forms of heritable dilated cardiomyopathy has revealed the intricate nature of corrupted pathways independent of common risk factors such as ischemic disease, underlying endocrine disorders or cardiotoxic insults [4–7]. A case in point is the recent genetic studies linking defects in cardioprotective ATP-sensitive K+ (KATP) channels to an aberrant stress response in the pathogenesis of a malignant form of dilated cardiomyopathy [8–12]. Severity of disease progression in dilated cardiomyopathy often mandates cardiac transplantation as the only remaining treatment option, a constraint compounded by donor organ shortage, warranting exploration of alternative management strategies [12, 13].
Beyond the reach of current therapies, stem cell technology provides a foundation for heart repair without the need for organ replacement [14–18]. Although cardiac rejuvenation has been recognized as a homeostatic self-repair mechanism, the innate regenerative reserve is insufficient to salvage failing myocardium [19–22]. In fact, initiation and evolution of the cardiomyopathic process was linked to depletion of the resident cardiac stem cell pool [23]. Introduction of progenitor cells into the diseased heart may therefore offer a means of promoting the healing process. Indeed, in the context of KATP channel ablation that recapitulates salient traits of the human dilated cardiomyopathy 1O syndrome (CMD 1O), functional repair was recently achieved by an embryonic stem (ES) cell therapy regimen [24]. Moreover, it has been established that embryonic stem cell progeny consistently acquire a cardiogenic phenotype with functional excitation-contraction coupling associated with maturation of the cellular energetic matrix, and when transplanted into damaged heart contribute to repopulation of dysfunctional myocardium improving contractile performance [25–29]. Stem cell lineage commitment and integration within diseased host myocardium has been further documented [30– 33]. Deciphering the molecular substrate underlying the regenerative process would however require mapping the protein landscape of the recipient heart in response to stem cell intervention. So far, however, the proteomic response of cell therapy in heart disease remains unknown.
Enabled by high-throughput technologies for large scale identification of proteins and associated networks, systems platforms offer an integrative, unbiased approach to address the complexity of pathobiologic change in disease and in response to therapy [34–38]. Here, in the stressed KATP channel knockout, a prototype of genetic dilated cardiomyopathy, proteome-wide profiling decoded the global manifestation of disease and the signature of embryonic stem cell therapy. In the stem cell-dependent proteome, network analysis demonstrated a prioritized cardiovascular development function and demotion of disease-associated adverse effects, validated by improved functional and structural outcome with treatment. Stem cell-based proteomic remodeling is thus resolved in a genetic model of dilated cardiomyopathy, mapping molecular underpinnings of regenerative outcome.
MATERIALS AND METHODS
Genetic Model of Dilated Cardiomyopathy and Intervention Protocols
Protocols were carried out in accordance with NIH guidelines, and with approval of the Institutional Animal Care and Use Committee. Eight- to twelve-week-old male KATP channel knockout mice (Kir6.2-KO), generated by disruption of the KCNJ11 (potassium inwardly rectifying channel, subfamily J, member 11) gene encoding the Kir6.2 channel pore [9, 24], served as the prestress Control cohort (experimental group 1). Separate cohorts of syngeneic age- and sex-matched Kir6.2-KO mice underwent transverse aortic constriction (TAC) to induce continuous hemodynamic pressure overload upon the left ventricle [39, 40]. At 2 weeks following aortic constriction, pressure-overloaded Kir6.2-KO hearts were exposed by thoracotomy, and animals were further randomly separated into two disease groups, that is, untreated [ES(−), experimental group 2] and embryonic stem cell treated [ES(+), experimental group 3]. Study subjects uniformity across cohorts ensured controlled head-to-head comparisons of experimental interventions. For the ES(+) cohort, epicardial injection of 200,000 murine lacZ-labeled R1 embryonic stem cells, in 15 µl propagation medium (Glasgow’s Minimum Essential Medium, Lonza, Basel, Switzerland), was performed at five separate sites (40,000 cells in 3 µl per site) in the anterior wall of the left ventricle [24]. The R1 embryonic stem cell line was selected as it is the only cell type demonstrated to ensure repair in this model of genetic dilated cardiomyopathy [24]. End points included survival, cardiac function and structure evaluated by echocardiography and pathological examination [24], as well as proteome-wide and network analysis. Functional parameters were measured at a series of time points, including at baseline and at 1.5, 2, 4, and 8 weeks postaortic constriction, with additional comparative tracking of contractile and structural changes in ES(−) versus ES(+) cohorts at 0, 2, and 6 weeks postrandomization for cell intervention. The time point of 8 weeks was selected for proteomic analysis as indices of disease progression were consistent with chronic end-stage heart failure. Because of high mortality of KATP channel knockout animals [24, 40], a total of 90 animals were required in the study to assure sufficient survivorship up to 8 weeks of sustained stress load.
Prospective Evaluation of Cardiac Function and Structure
To evaluate progression of disease and the impact of stem cell therapy, transthoracic echocardiography (30-MHz MS400 transducer, Vevo2100, Visual Sonics, Toronto, Canada; 15L8 transducer, Sequoia 512, Siemens, Concord, CA) [41] was performed at multiple time points to prospectively assess cardiac function and structure. For all experimental groups, left ventricular fractional shortening (%) was calculated as ([LVDd – LVDs]/ LVDd) × 100, where LVDd is left ventricular end-diastolic dimension (mm) and LVDs, left ventricular end-systolic dimension (mm) [42]. Ejection fraction (%) was calculated as ([LVVd – LVVs]/LVVd) × 100, where LVVd is left ventricular end-diastolic volume (µl) and LVVs, left ventricular end-systolic volume (µl). Left ventricular weight (mg) was derived as [(LVDd + IVST + PWT)3 – LVDd3] × 1.055, where IVST is interventricular septum thickness (mm), and PWT, posterior wall thickness (mm). Left ventricular wall thickness to dimension ratio was calculated as a sum of IVST and PWT divided by LVDd, and was used to monitor the evolution of heart geometry [24, 33, 40]. Total heart weight was measured at the time of autopsy.
Protein Extraction and Quantitation
For proteomic analysis, disease-untreated [ES(−)] and disease-treated [ES(+)] Kir6.2-KO, as well as prestress unconstricted (control) animals, were sacrificed under isoflurane anesthesia, and hearts excised and rinsed in phosphate-buffered saline. Left ventricles including septum were removed, weighed ex vivo, snap-frozen in liquid N2, and stored at −80°C. Cytosolic tissue extracts, comprising the majority of cellular proteins compatible with isoelectric focusing (IEF) solubilization and resolution, were prepared by homogenization at 4°C in four volumes of extraction buffer, consisting of (in mM) 4-(2-hydroxyethyl)-1-ppiperazineethanesulfonic acid (HEPES) 25 (pH 7.4), phenyl-methylsulfonyl fluoride (PMSF) 0.25, and dithiothreitol (DTT) 50, 1.25 µM pepstatin A, Mini-Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), and 1% phosphatase inhibitor cocktails 1 and 2 (Sigma, St. Louis, MO) [43]. Samples were centrifuged (16,000g) at 4°C for 10 minutes, supernatants were transferred to fresh tubes, and protein was quantified in triplicate by Bio-Rad (Hercules, CA) protein assay using the microassay procedure with a bovine γ-globulin standard [44], with equivalent protein amounts from all samples resolved simultaneously by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained to independently assess protein quantitation prior to two-dimensional (2D) gel electrophoresis.
2D Electrophoresis and Gel Imaging
Protein extracts (100 µg) were resolved by immobilized pH gradient (IPG) 2-DE following addition to IEF rehydration buffer (7 M urea, 2 M thiourea, 2% w/v CHAPS, 50 mM DTT, 1 × Bio-Rad pH 3–10 ampholytes). Same lot IPG Ready Strips (pH 3–10, 170 mm, Bio-Rad) were actively rehydrated at 50 V for 10 hours, followed by rapid voltage ramping with a series of 15 minutes steps at 100, 500, and 1,000 V, and a final step at 10,000 V for 60 kVh at 20°C [45]. Focused IPG strips were rinsed with distilled, deionized water, and incubated for 15 minutes in equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% v/v glycerol, 2% w/v SDS) containing 10 mg/ml DTT, followed by 15 minutes in equilibration buffer containing 25 mg/ml iodoacetamide. After horizontal positioning on freshly prepared 12.5% SDS-PAGE gels, strips were overlaid with SDS buffer (25 mM Tris, 192 mM glycine, 0.1% w/v SDS) containing 0.5% w/v agarose, and resolved orthogonally by SDS-PAGE in a Protean II XL system (Bio-Rad). Resolved 2D gels were silver stained and digitized at 400 dpi for spot image analysis, including spot detection, matching, normalization, and quantification, conducted with Bio-Rad PDQuest v.7.4.0 following subtraction of background and horizontal/vertical streaking intensities [36, 43]. Individual gel images were normalized by total intensity of valid spots. Fold change ratios were calculated as [mean ES(−)]:[mean Control], [mean ES(+)]:[mean Control], and [mean ES(+)]:[mean ES(−)] for protein spots increasing in the numerator group, or the negative inverse for protein species decreasing in the numerator group. For proteins identified in more than one spot, the sum of values for all spots was used to determine a weighted average treatment ratio, unless otherwise indicated.
Nanoelectrospray Linear Ion Trap Tandem Mass Spectrometry
Significantly altered protein species were isolated from resolved gels, destained, and prepared for liquid chromatography-tandem mass spectrometry (LC-MS/MS) by reduction, alkylation, tryptic digestion, peptide extraction, and drying [32, 38, 43]. Peptides were reconstituted in 0.15% formic acid, 0.05% trifluoroacetic acid (TFA), and trap injected onto a 75 µm × 10 cm ProteoPep C18 PicoFrit nanoflow column (New Objective, Woburn, MA). Chromatography was performed using 0.2% formic acid in solvent A (99% water, 1% acetonitrile) and B (80% acetonitrile, 5% isopropanol, 15% water), with peptides eluted over 30 minutes with a 5%–45% solvent B gradient using an Eksigent nanoHPLC system (MDS Sciex, Toronto, Canada) coupled to a linear ion trap quadrupole (LTQ)-Orbitrap mass spectrometer (Thermo Fisher Scientific, Barrington, IL). Continuous scanning of eluted peptide ions was carried out between 375 and 1600 m/z, automatically switching to MS/MS collision induced dissociation mode on ions exceeding an intensity of 8,000. Raw MS/MS spectra were converted to .dta files using Bioworks 3.2 (Thermo Fisher Scientific), and merged files matching +1, 2, or three peptide charge states were correlated to theoretical tryptic fragments in Swiss-Prot (v.53.0) using Mascot v.2.2 [46]. Searches were conducted on mammalian sequences (53,539 entries), tolerating up to two missed cleavages, a mass tolerance of 60.01 Da for precursor ions (including 13C peak detection) and 60.6 Da for MS/MS product ions allowing for protein N-terminal acetylation, methionine oxidation, and cysteine carbamidomethylation. Protein identities were confirmed by matching multiple peptide spectra at p < .05, with proteins accepted at p < .01. Proteins identified by a single peptide were thus subjected to a higher stringency level (p < .01), and were confirmed by manual spectrum inspection with detected fragment ions from the MS/MS spectrum required to be above baseline noise, have demonstrable continuity in b-or y-ion series, and proline residues yielding intense y-ions [47]. Protein assignments were further validated by congruence of observed versus predicted pI/Mr, using the ExPASy pI/Mr tool (http://us.expasy.org/tools/pi_tool.html), taking into consideration protein processing and post-translational modifications.
Interactome Network Analysis
Differentially expressed proteins, with fold change ratios, were submitted as focus proteins for network analysis using Ingenuity Pathways Knowledge Base (Ingenuity Systems, www.ingenuity.com) to identify associated functional networks. An overview of interactions was obtained by merging functional subnetworks into composite interactomes. The composites were depicted using the molecular interaction network visualization program Cytoscape 2.6.2 [48], with paired network layouts coordinated using the ReOrientPlugin to localize common nodes in the same spatial location within multiple networks, and topological properties characterized using Network Analyzer [49]. Computed properties included node degree (k), the number of links connected to the node, and node degree distribution [P(k)], the probability that a specified node has k links, defined as P(k) = X(k)/n, where X(k) is the number of nodes with degree k and n is the total number of network nodes [50, 51]. P(k) versus k discriminates between random and scale-free topographies, defined by normal and power law distributions, respectively [50]. The Anderson-Darling normality test ruled out a normal distribution, so P(k) versus k was calculated as a power law relationship using a cumulative distribution function [52] to determine γ in the power law distribution [P(k) ~ k−γ] according to Equation (1):
| (1) |
where γ is the power law exponent, n the number of network nodes, xi node degree, and xmin the minimum node degree within the network, with statistical error σ [52] for Equation (1) defined by Equation (2):
| (2) |
To link expression data with systems functions, resolved networks were interrogated with Ingenuity Pathways Analysis (IPA) [36, 43].
Statistical Analysis
Comparison between groups was performed using a standard t-test of variables with 95% confidence intervals, with data expressed as mean ± standard error. Wilcoxon test was used to evaluate cardiac physiological parameters (JMP 8, SAS Institute, Cary, NC). Kaplan-Meier analysis with log-rank testing was applied to evaluate survivorship. Comparison of 2D gel spot intensities between experimental groups was carried out by t-test. A p < .05 was considered significant, unless otherwise indicated.
RESULTS
Global Protein Profiling in Genetic Cardiomyopathy and Stem Cell Intervention
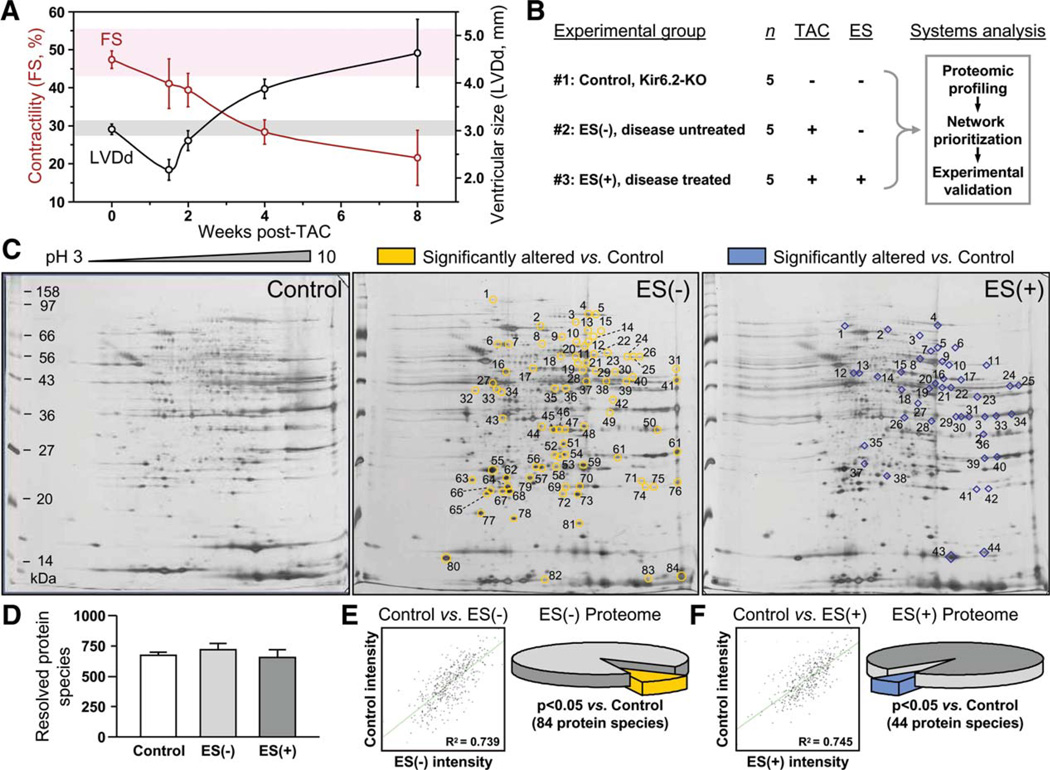
TAC imposes sustained pressure overload on the left ventricle resulting over 8 weeks in progressive contractile dysfunction and cardiomegaly, characteristic of congestive heart failure in the KCNJ11 knockout, Kir6.2-KO model (Fig. 1A), a surrogate of human genetic KATP channel-deficient cardiomyopathy [24, 40]. To assess, in the setting of KATP channel deficiency, molecular consequences of TAC-imposed stress in the absence and presence of stem cell intervention, prestressed KCNJ11 knockouts (control, experimental group 1) were compared head-to-head to pressure-overloaded Kir6.2-KO randomized at 2 weeks post-TAC into untreated [ES(−), experimental group 2] and embryonic stem cell-treated [ES(+), experimental group 3] cohorts (Fig. 1B). To this end, left ventricle cytosolic proteomes were extracted from control (n = 5), and 8 weeks after TAC from ES(−) (n = 5) and ES(+) (n = 5) cohorts, and profiled by differential proteomics. In broad pH range silver-stained 2D gels (Fig. 1C), over 700 protein species were consistently resolved (Fig. 1D). Reproducibility across cohorts was documented by positive correlation of average normalized intensities of matching protein spots in control versus ES(−) and control versus ES(+) gels (Fig. 1E and 1F, scatter plots). Densitometric quantification revealed that a subset of 84 unique protein spots (12% of resolved proteome) was significantly altered in response to TAC-induced pressure overload in the cardiomyopathic KATP channel knockout hearts [control vs. ES(−); Fig. 1E, pie chart]. Following 6 week-long cell therapy, 44 protein spots (7% of resolved proteome) differed between prestressed controls and stem cell-treated TAC Kir6.2-KO [control vs. ES(+); Fig. 1F, pie chart]. Thus, differential expression profiling of Kir6.2 KATP channel knockout hearts demonstrated the global proteomic impact of imposed stress, with cell therapy eliciting distinct remodeling of the cardiomyopathic subproteome in stressed failing hearts.
Figure 1.
Stem cell intervention remodels heart proteome in genetic dilated cardiomyopathy. (A): Malignant dilated cardiomyopathy in KATP channel-deficient (Kir6.2-KO) hearts under stress imposed by TAC characterized by decreased contractility (FS) and increased LVDd following initial compensatory hypertrophy observed at 1.5 weeks post-TAC. Shaded background indicates 95% confidence interval for FS (pink) and LVDd (gray) in age- and sex-matched Kir6.2-KO (Control) hearts. (B): The experimental protocol involved systems analysis combining proteomic comparison of left ventricular (LV) extracts obtained 8 weeks post-TAC from three separate experimental groups, unstressed Kir6.2-KO (#1 - Control), disease untreated TAC Kir6.2-KO [#2 – ES(−)] and disease treated TAC Kir6.2-KO [#3 – ES(+)] treated by 6 weeks of R1 embryonic stem cell therapy, followed by network analysis and in silico prioritization of proteomic findings in conjunction with in vivo and ex vivo functional validation. (C): Representative silver stained two-dimensional gels (pH 3–10 IEF, 12.5% SDS-PAGE) of LV tissue extracts (100 µg protein) from unstressed Kir6.2-KO (control), and from aortic-constricted untreated [ES(−)] or stem cell-treated [ES(+)] counterparts. Spots identified as differentially expressed relative to those in unstressed hearts are circled and numbered on ES(−) and ES(+) gels. (D): Gel reproducibility demonstrated by no significant difference in number of resolved protein species. (E, F): Densitometric spot quantitation, with scatter plots of average normalized intensities of matching protein spots showing correlation for control versus ES(−) and control versus ES(+) gels, and pie charts indicating significant differences in 84 and 44 protein species for control versus ES(−) and control versus ES(+), respectively (p < .05). Abbreviations: ES, embryonic stem cells; FS, fractional shortening; LVDd, left ventricular end-diastolic dimension; TAC, transverse aortic constriction.
Distinct Protein Signatures in Cardiomyopathic Hearts in Absence and Presence of Stem Cell Therapy
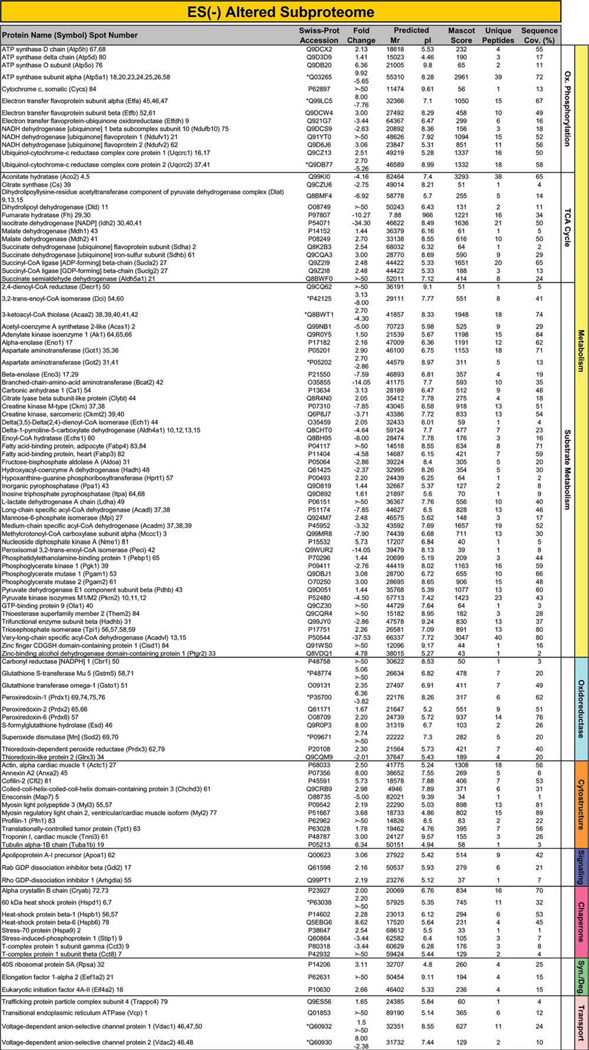
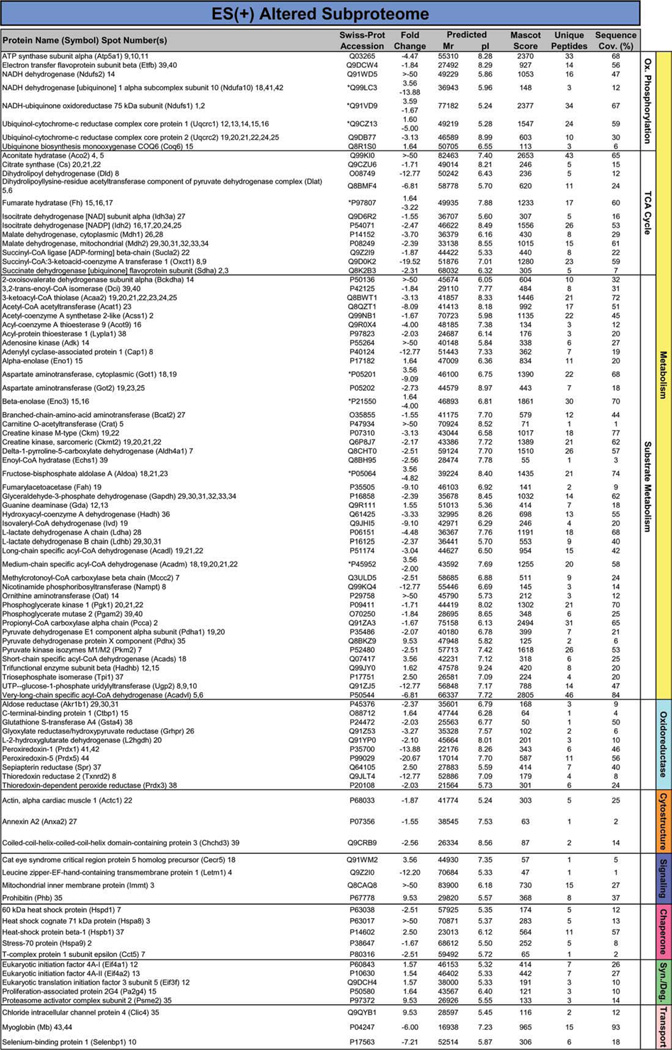
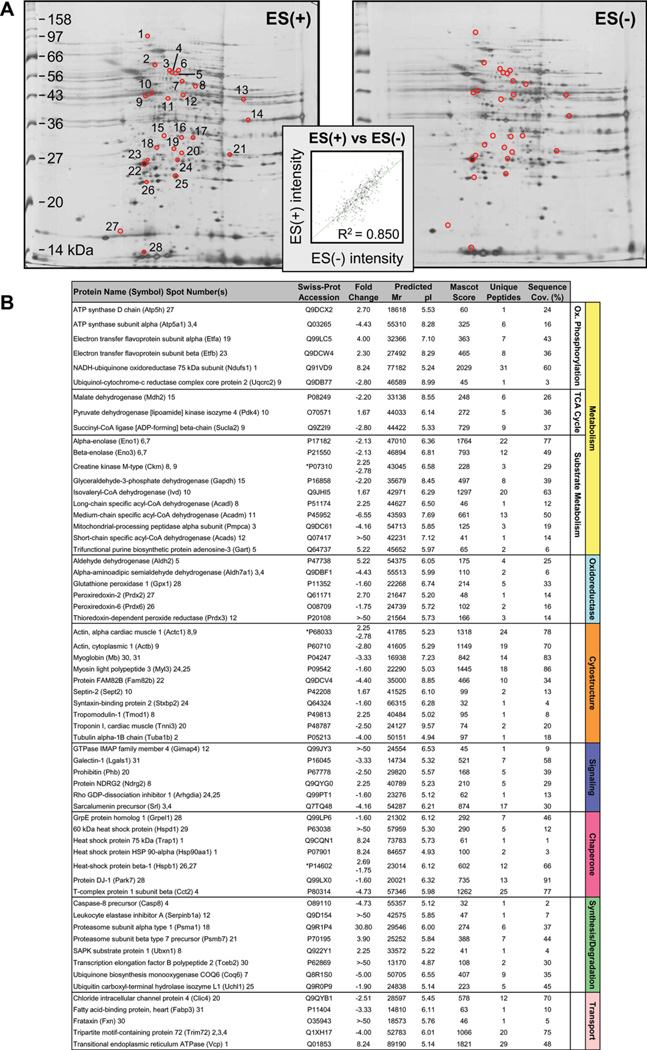
Proteins specifically altered in failing KATP channel knockout hearts, in the absence and presence of stem cell therapy, were determined by LTQ-Orbitrap MS/MS analysis of in-gel tryptic digests. In ES(−) hearts, 109 altered proteins were identified by this high-throughput approach. Individual protein identities, along with extent of fold-change and supporting gel and mass spectrometry metrics, encompass a spectrum across metabolism and signaling infrastructure reflecting the complex pathologic substrate of the cardiomyopathic state (Fig. 2; Supporting Information Table S1 and Table S2). Cellular metabolism related identities were responsible for 64% of disease-induced proteome change, and included 13 oxidative phosphorylation, 13 tricarboxylic acid (TCA) cycle, and 44 substrate metabolism proteins (Fig. 2). Ontological annotation revealed that the additional 39 identified proteins formed a metabolism-related signaling infrastructure encompassing oxidoreductases (10 proteins), cellular structure and scaffolding (11), signaling regulation (3), stress-related chaperones (8), protein synthesis and degradation (3), as well as transport regulation (4) (Fig. 2). In ES(+) hearts, a distinct set of 93 proteins were identified in response to stem cell treatment (Fig. 3; Supporting Information Table S1 and Table S3). These were primarily associated with metabolism (63 proteins, 68%), including oxidative phosphorylation (8 proteins), TCA cycle (12) and substrate metabolism (43), and a metabolism-related infrastructure of oxidoreductases (10), cellular structure and scaffolding (3), signaling regulation (4), stress-related chaperones (5), protein synthesis and degradation (5), as well as transport regulation (3) (Fig. 3). When compared with untreated hearts (Fig. 2), cell therapy (Fig. 3) nullified disease-induced changes in 68% of proteins and reduced the extent of fold-change in an additional 16%, reorganizing the proteome landscape of failing hearts. Thus, resolving individual identities of protein changes establishes a signature of KATP channel-deficient cardiomyopathy, extensively restructured by stem cell intervention.
Figure 2.
Identity of cardiomyopathy-induced subproteome. The 109 significantly altered proteins induced by progressive cardiomyopathy [ES(−) altered subproteome] and identified by LTQ-Orbitrap MS/MS analysis, were functionally categorized and color-coded by Swiss-Prot ontological annotation. Protein names are listed with their symbol (Swiss-Prot gene abbreviation) and spot numbers to locate corresponding 2D gel position(s) in Figure 1. Mascot score, number of unique identified peptides, % sequence cov. (coverage), predicted Mr and pI for each protein (following expected post-translational processing, for example, removal of a mitochondrial signal peptide), and fold change of ES(−) over control are indicated. For proteins detected in more than one spot, maximum score and corresponding number of unique peptides are reported. Fold change was calculated as described in experimental procedures, and for proteins detected in both increasing and decreasing spots (*), both values are indicated. Abbreviations: 2D, two-dimensional; CDGSH, Unique 39 amino acid CDGSH domain [C-x-C-x2-(S/T)-x3-P-x-C-D-G-(S/A/T)-H]; ES, Embryonic stem cells; LTQ, Linear ion trap quadrupole; MS/MS, Tandem mass spectrometry; Ox., oxidative; Syn./Deg., protein synthesis/degradation; TCA cycle, tricarboxylic acid cycle.
Figure 3.
Identity of cardiomyopathic subproteome following stem cell therapy. The 93 significantly altered proteins induced by stem cell therapy in the setting of dilated cardiomyopathy [ES(+) altered subproteome], and identified by LTQ-Orbitrap MS/MS analysis, were functionally categorized and color-coded by Swiss-Prot ontological annotation. Protein names are listed with their symbol (Swiss-Prot gene abbreviation) and spot numbers to locate corresponding 2D gel position(s) in Figure 1. Mascot score, number of unique identified peptides, % sequence cov. (coverage), predicted Mr and pI for each protein (following expected post-translational processing, for example, removal of a mitochondrial signal peptide), and fold change of ES(+) over control are indicated. For proteins detected in more than one spot, maximum score and corresponding number of unique peptides are reported. Fold change was calculated as described in experimental procedures, and for proteins detected in both increasing and decreasing spots (*), both values are indicated. Abbreviations: 2D, two-dimensional; Ox., oxidative; Syn./Deg., protein synthesis/ degradation; TCA cycle, tricarboxylic acid cycle.
Stem Cell Treatment Restructures Cardiomyopathic Proteome Network
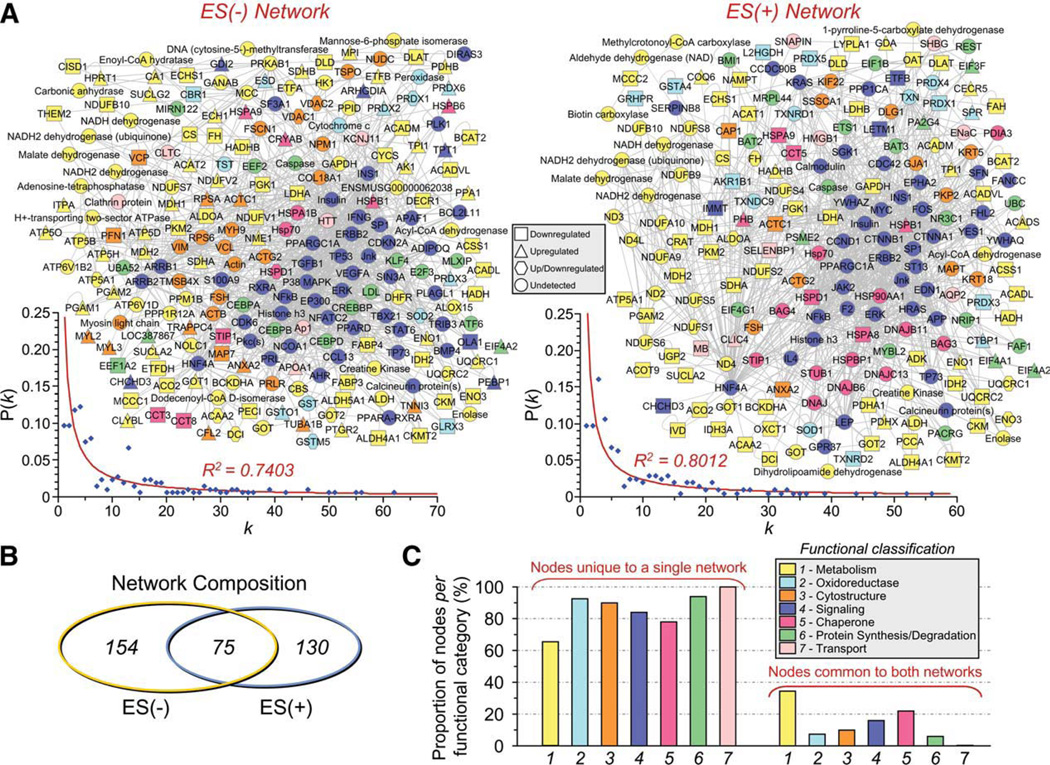
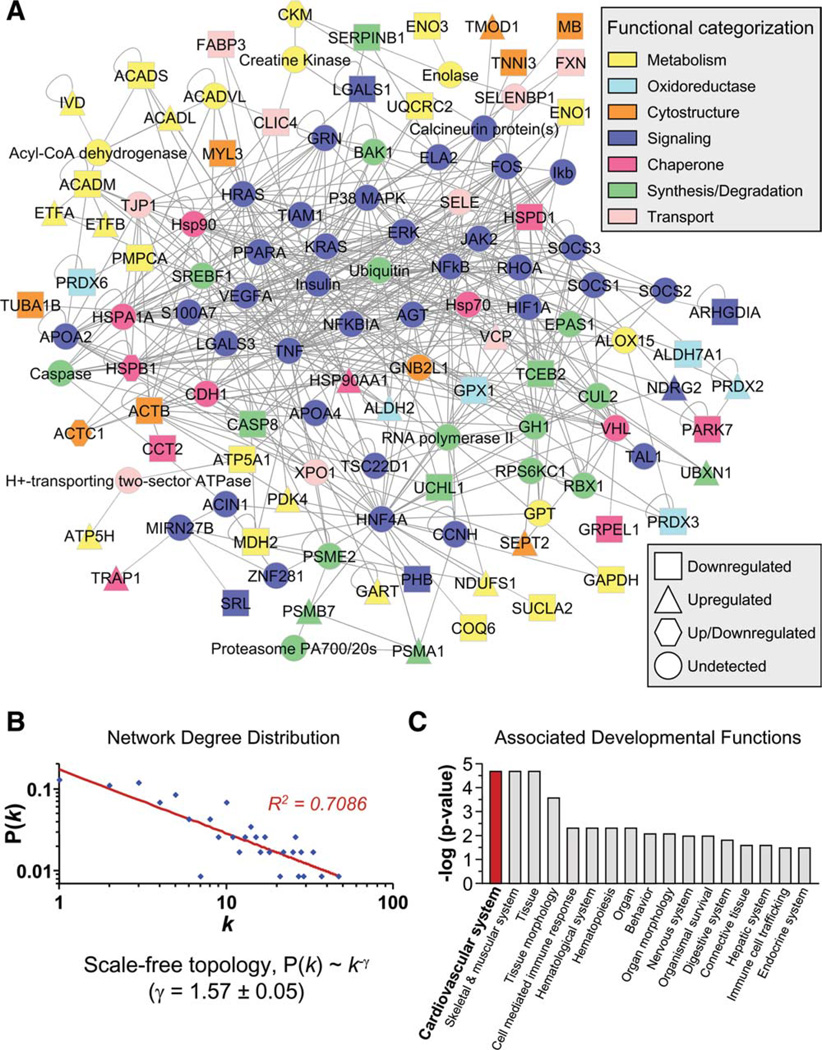
To obtain a collective understanding of processes associated with cell therapy-induced proteome remodeling in the setting of KATP channel deficient cardiomyopathy, network analysis was implemented to compare untreated ES(−) versus treated ES(+) subproteomes. Protein–protein interaction mapping clustered the 109 altered proteins from untreated cardiomyopathic hearts into an organized network comprised of 229 nodes linked by 1,207 interactions [or edges; Fig. 4A, left; ES(−) network]. The 93 proteins specific to the stem cell-treated diseased hearts assembled into a network of 205 nodes connected by 975 edges [Fig. 4A right, ES(+) network]. Network topologies for both ES(−) and ES(+) networks displayed a nonstochastic pattern, excluding a random association among changing proteins. Indeed, the inter-relationship between node degree (k) and degree distribution P(k) followed power law distributions indicative of scale-free nonrandom architecture (Fig. 4A, insets left and right). Although similar in size, the two networks shared only 75 common proteins (Fig. 4B), underscoring the distinct molecular composition of the ES(−) and ES(+) proteomes expanded to a broader network neighborhood context. The majority of proteins in every functional category were unique to each of the respective networks, as only 35% of metabolic proteins and fewer than 22% of nodes for any metabolism-related infrastructure category were shared between ES(−) and ES(+) networks (Fig. 4C). Thus, mapped ES(−) and ES(+) heart subproteome-dependent networks exhibit limited overlap, suggesting different functional consequences in the absence and presence of stem cell therapy.
Figure 4.
Stem cell therapy transforms the cardiomyopathy-associated protein interaction network. (A): The 109 ES(−) and 93 ES(+) differentially expressed proteins were submitted to IPA as focus nodes, generating broader interaction networks of 229 and 205 proteins, respectively. Proteins are designated by symbols corresponding to Swiss-Prot gene abbreviations or by family name for nodes representing protein families, and are colored by the functional ontology detailed in Figures 2 and 3, with node shape indicating directionality of focus protein expression change (legend) and nodes common to both networks maintained in the same spatial location. Plots of degree distribution [P(k)] versus degree (k) followed power law distributions, where P(k) ~k−γ, with γ= 1.55 ± 0.04 for ES(−) and 1.57 ± 0.04 for ES(+) networks, respectively, indicating scale-free, nonstochastic network architecture. (B): Limited overlap was found between ES(−) and ES(+) networks (total unique and common nodes indicated), with (C) the majority of proteins from each functional category residing within only one of the two networks. Abbreviations: ES, embryonic stem cells; P(k), degree distribution; k, node degree.
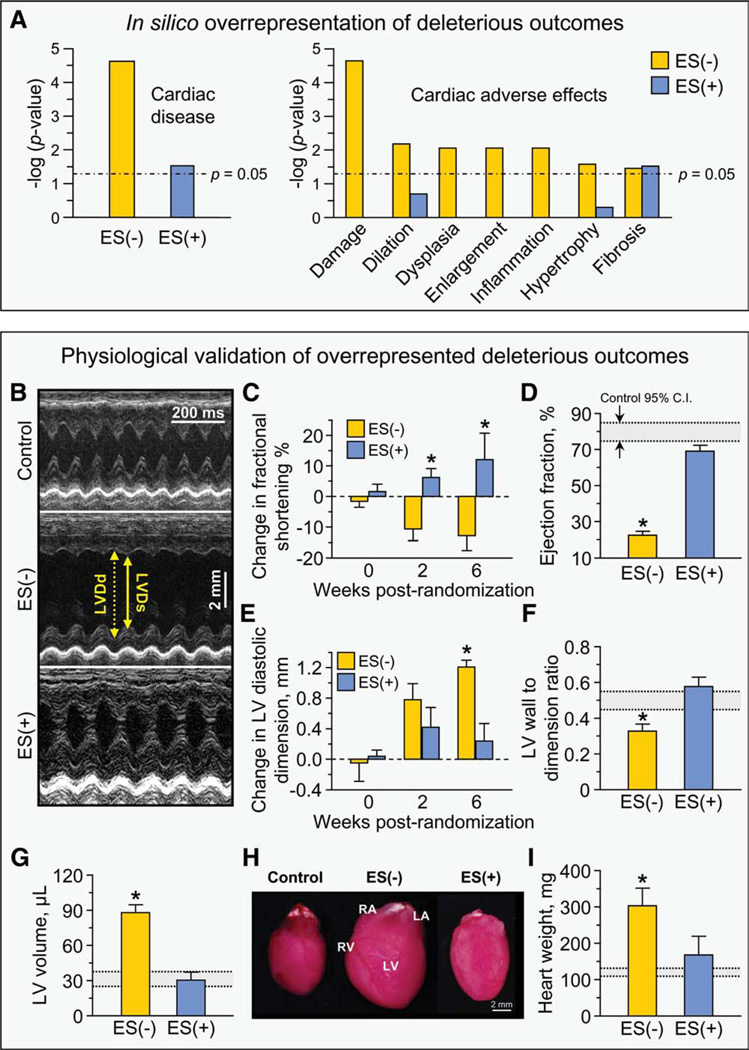
In Silico Phenotypic Patterns Induced by Stem Cell Therapy Validated In Vivo
Network analysis provides a systems framework to delineate, in an impartial manner, putative patterns arising from a remodeled proteome based on extracted core signatures [35, 38]. Collective bioinformatic interrogation demonstrated a dramatically overrepresented “Cardiac Disease” category associated with the ES(−) network (p = 2.35 × 10−5), consistent with heart disease susceptibility, which was reduced by three orders of magnitude (p = 3.11 × 10−2) within the ES(+) network (Fig. 5A, left). Further interrogation of the ES(−) network against a broad spectrum of 134 curated pathological conditions and toxicological pathways within the Ingenuity Pathways Knowledge Base extracted exclusively cardiac structural and functional adverse effects, namely “cardiac damage,” “cardiac dilation,” “cardiac dysplasia,” “cardiac enlargement,” “cardiac inflammation,” “cardiac hypertrophy,” and “cardiac fibrosis” (Fig. 5A, right). Bioinformatic survey predicted that the six of these seven detrimental outcomes in the ES(−) network are eliminated from the ES(+) network, suggesting functional and structural benefit of stem cell intervention in the setting of cardiomyopathy (Fig. 5A, right). To validate this core prediction, cardiac function and structure were assessed in vivo in untreated and stem cell-treated cohorts by prospective echocardiography (Fig. 5B–5G) and pathoanatomical analysis (Fig. 5H–5I). Untreated Kir6.2-KO-stressed mice [ES(−)] demonstrated significant and progressive cardiac dysfunction (Fig. 5B–5G) and associated heart chambers enlargement (Fig. 5H–5I), precipitating without therapy high mortality rates with 53% survivorship between 2 and 8 weeks poststress and poor overall survival (17%). In contrast, throughout the follow-up period, stem cell intervention [ES(+)] significantly improved cardiac contractility and prevented cardiac dilation and enlargement, nullifying premature mortality. Specifically, serial monitoring of fractional shortening from the point of randomization 2 weeks post-TAC indicated continuous deterioration without cell therapy, but improved fractional shortening with cell therapy at multiple time points (Fig. 5C). Ejection fraction, an index of cardiac contractility that prognosticates outcome in heart failure, was reversed towards prestress levels by stem cell treatment (i.e., 22.4 ± 2.4%, n = 5 ES(−) vs. 69.1 ± 3.3%, n = 5 ES(+), p < .01, with control 95% confidence interval indicated for benchmarking; Fig. 5D). The post-TAC increase in cardiac dilation in untreated hearts was marked by 2 weeks and continuously increased by 6 weeks, whereas cell therapy was effective in maintaining diastolic dimension throughout the observation period (Fig. 5E). Significant wall thinning relative to left ventricular diameter without treatment was restored to control levels 6 weeks following cell delivery (i.e., 0.33 ± 0.04, n = 5 ES(−) vs. 0.58 ± 0.05, n = 5 ES(+), p < .01; Fig. 5F). By 6 weeks of therapy, the increased left ventricular volume was normalized (i.e., 88.0 ± 6.8 pl, n = 5 ES(−) vs. 30.4 ± 6.8 µl, n = 5 ES(+), p < .01; Fig. 5G). Functional improvement detected by echocardiography was supported by favorable structural remodeling, demonstrated by marked reduction in cardiac hypertrophy (Fig. 5H) in response to stem cell therapy (i.e., heart weight: 301.8 ± 21.9 mg, n = 5 ES(−) vs. 167.8 ± 7.0 mg, n = 5 ES(+), p < .01; Fig. 5I). Thus, bioinformatic interrogation for prediction of specific cardiomyopathic traits of cardiac function and structure in response to cell therapy were validated by in vivo echocardiography and ex vivo pathology.
Figure 5.
Stem cell-dependent demotion of disease-associated adverse effects. (A): Bioinformatic interrogation of the ES(−) and ES(+) networks within Ingenuity Pathways Knowledge Base for deleterious outcomes indicated that significant overrepresentation of “Cardiac disease” within the ES(−) network was reduced by three orders of magnitude in the ES(+) network (left panel). Screening against Ingenuity toxicological pathways further indicated seven adverse effects in the ES(−) subproteome, all of which were cardiac specific, that is, damage, dilation, dysplasia, enlargement, inflammation, hypertrophy and fibrosis, and nearly all were abolished in the ES(+) subproteome (right panel). (B): Echocardio-graphic measurements indicated that (C) LV fractional shortening, (D) LV ejection fraction, (E) LV diastolic dimension, (F) LV wall to dimension ratio, and (G) LV end-diastolic volume were all significantly worsened by pressure overload but returned to prestress levels following 6 weeks of cell therapy. (H–I): Measurement of hearts on autopsy indicated that the significant increase in heart mass imposed by pressure overload was reversed by cell therapy. Collectively, these parameters demonstrated cell therapy-induced improvement in myocardial contractile performance, reduction in LV size and decreased cardiac damage (*, p < .01 ES(−) versus ES(+) and ES(−) versus control, with control indicated by 95% C.I. in gray as noted in panel (D). Abbreviations: C.I., confidence intervals; ES, embryonic stem cells; LA, left atrium; LV, left ventricle; LVDd, left ventricular end-diastolic dimension; LVDs, left ventricular end-systolic dimension; RA, right atrium; RV, right ventricle.
Stem Cell Therapy Prioritizes a Cardio-Regenerative Proteome Module
Iterative systems interrogation enables unbiased identification and stratification of functional categories emerging from a perturbed proteome [36]. 2D gel analysis revealed that stem cell therapy induced 28 significantly altered protein species (Fig. 6A). Gel-to-gel reproducibility indicated high correlation of average normalized intensities of matching protein species (Fig. 6A, inset). Mass spectometric analysis of protein species altered by cell therapy resolved 61 unique protein identities (Fig. 6B; Supporting Information Table S1 and Table S4). Proteins integrated into an organized network of 120 nodes linked by 560 edges (Fig. 7A). A nonrandom, scale-free topology was deduced based on network degree distribution properties (Fig. 7B). Ontological assessment of the stem cell-induced interactome against curated biological processes extracted categorically overrepresented functions, with “cardiovascular system development” prioritized among the ranking of all developmental functions (Fig. 7C). In contrast, “cardiovascular system development” was not prioritized in the untreated cardiomyopathic network, and was reduced in significance by more than one order of magnitude, that is, p = 1.98 × 10−5 and 4.14 × 10−4 relative to cell therapy, indicating a cardio-rejuvenative substrate induced by stem cell intervention. Thus, iterative proteome-wide network resolution unmasked a regenerative signature induced by embryonic stem cell treatment of failing heart in the context of KATP channel-deficient cardiomyopathy.
Figure 6.
Stem cell therapy-specific subproteome. (A): Comparison of stem cell-treated [ES(+)] versus untreated [ES(−)] left ventricular tissue extracts by two-dimensional (2D) electrophoresis. Differentially expressed spots isolated for identification by LTQ-Orbitrap mass spectrometric analysis are circled, and numbered on the ES(+) gel. Inset: Gel-to-gel reproducibility indicated by correlation of scatter plot for average normalized densitometric intensities of matching protein spots from ES(+) versus ES(−) gels. (B): Identities of the 61 proteins significantly altered by cell therapy are listed with their symbol (Swiss-Prot gene abbreviation) and spot numbers to locate corresponding 2D gel position(s) in panel (A). Mascot score, number of unique identified peptides, % sequence cov. (coverage), predicted Mr and pI for each protein (following expected post-translational processing, for example, removal of a mitochondrial signal peptide), and fold change in ES(+) versus ES(−) are indicated. For proteins detected in more than one spot, maximum score and corresponding number of unique peptides are reported. Fold change was calculated as described in experimental procedures, and for proteins detected in both increasing and decreasing spots (*), both values are indicated. Abbreviations: ES, embryonic stem cells; Ox., oxidative; TCA cycle, tricarboxylic acid cycle; SAPK, stress activated protein kinase.
Figure 7.
Cardiovascular system development prioritized in stem cell therapy-specific network. (A): Submission to Ingenuity Pathways Analysis (IPA) of differentially expressed proteins in the ES(+)-treated hearts generated a 120-protein interaction network, functionally categorized and color-coded by Swiss-Prot ontological annotation (upper legend), with proteins designated by symbols corresponding to Swiss-Prot gene abbreviations or by family name for nodes representing protein families, and shape indicating directionality of focus protein expression change (lower legend). (B): A log-log plot of ES(+)-specific degree distribution [P(k)] versus degree (k) followed a power law distribution, indicating scale-free, nonstochastic network architecture. (C): Bioinformatic interrogation of IPA for associated developmental functions revealed prioritization of cardiovascular system development in the ES(+)-de-pendent proteome in response to cell therapy. Abbreviations: P(k), degree distribution; k, node degree.
DISCUSSION
Transplantation of stem cells shows promise in the treatment of cardiovascular disease, yet the molecular foundation that underlies repair remains largely unknown [16, 17]. The present study deciphers the proteomic landscape induced by embryonic stem cell intervention in a surrogate of type 1O human genetic dilated cardiomyopathy produced by stress in the setting of KATP channel deficiency. In this nonischemic heart failure model, using proteomics and network analysis, we demonstrate that pluripotent stem cell therapy downgrades adverse consequences of disease, as a result of global remodeling of the pathologic heart, inducing a distinct cardio-regenerative proteome featuring a prioritized cardiovascular development signature. The resolved, stem cell-dependent subproteome identifies a previously undefined regenerative substrate of heart failure repair.
Systems Pathology of Genetic Cardiomyopathy
Traditional means of risk assessment have focused on single candidate pathways [53]. The emergence of systems approaches now enables unbiased, integrative surveys of complex disease states [54–56]. In this regard, proteome-wide profiling provides a high-throughput/high-specificity platform to extract disease processes at a global scale [57, 58]. As applied in this study, proteomics in conjunction with network-based systems analysis incorporates all measured changes for in silico bioinformatic interrogation of disease and therapy phenotypes, avoiding potential bias that may be introduced by selectively focusing only on a particular class of altered protein, for example, those of greatest differential expression, or selection of suspected candidate proteins regardless of observed change in expression [59]. Multiple etiologies cause cardiomyopathies, with complex disease phenotypes arising through interactions between individual genetic make-up and environmental stress challenges [1–8, 60]. Moreover, cardiomyopathic triggers may corrupt the resident cardiac stem cell compartment jeopardizing the innate repair potential of the myocardium [23]. Here, proteomic analysis was implemented within an established paradigm of genetic cardiomyopathy, that is, the stressed KATP channel knockout model that recapitulates salient traits of human disease [24, 40]. The KCNJ11 null mutant under imposed hemo-dynamic load demonstrates compromised contractility and ventricular dilation predisposing to premature death [40], features of the life-threatening cardiomyopathic CMD 1O syndrome associated with mutations in genes encoding for subunits of the KATP channel [8, 11]. Compared to nonfailing unstressed counterparts, failing KATP channel knockout hearts, exposed to the stress of chronic pressure overload, displayed a pronounced rearrangement of the cardiac proteome. The primary impact of the 109 significantly altered protein changes centered on energy metabolism with 70 unique proteins stratified into the “cellular metabolism” category, consistent with impaired metabolic homeostasis a recognized hallmark underlying initiation and maintenance of heart failure [9, 61–64]. Accumulation of defects along energetic pathways precipitates failure of the whole cardiac bioenergetic system leading to myocardial dysfunction [61]. The present systems pathology approach thus pinpoints identities of affected proteins, revealing the broad landscape underlying the KATP channel-deficient CMD1O cardiomyopathic proteome.
Stem Cell Therapy Restructures Cardiomyopathic Proteome
Current pharmacotherapy for dilated cardiomyopathy is largely designed to address symptom reduction or limit disease progression [3, 7, 65]. Targeting heart repair mandates a regimen with the capacity to replace dysfunctional heart muscle [17, 66], as recently demonstrated with tissue reconstruction by stem cell-based therapy in the setting of cardiomyopathy [23, 24]. Beyond dissecting the pathobiological complexity of disease, proteomic approaches could decode responses triggered by applied therapies and ensure a holistic perspective by network integration [35, 67]. It should be noted, however, that global relationships between protein targets and treatment algorithms remain largely uncharacterized [68]. In this first proteomic examination of the large-scale impact of stem cell therapy in disease, a comprehensive protein registry was collected at 6 weeks post-transplantation of embryonic stem cells into failing KATP channel-deficient hearts. Cell intervention was found to produce a significant imprint upon the host myocardium, particularly affecting components of energetic metabolism and metabolism-related infrastructure. Operating as part of highly interconnected cellular networks, the stem cell-dependent interactome was encompassed within a nonstochastic network of protein–protein relationships that diverged from that of the stem cell-independent counterpart. These data indicate that embryonic stem cell treatment orchestrated a wide spectrum of change that collectively restructured the diseased proteome architecture underlying the active process of repair.
Cardio-Regenerative Network Signature Underlies Stem Cell-Based Repair
The processes of protein identification, ontological stratification, and iterative interrogation create a systems tool to project phenotypic outcome [38, 69]. Cytotherapy here canceled six of seven prominent cardiac-specific adverse outcomes prioritized by disease, providing a new proteome identity to the healing myocardium. Verifying bioinformatic output, independent functional and structural measurements documented increased ejection fraction and fractional shortening, reduced ventricular dilation and reversal of cardiomegaly affirming benefit in vivo. Moreover, the in silico prediction of overrepresentation of fibrosis was experimentally verified in tissue samples, and was significantly reduced in treated versus untreated hearts [24]. This unprecedented proteomic perspective offers thereby a reliable resource to diagnose a reparative response at a systems level. Further deconvolution of the stem cell-dependent subproteome exposed a cardiovascular development-enriched module absent from untreated cohorts. In line with the documented formation of new cardiac tissue and cell cycle activation in ~15% of cells within embryonic stem cell-treated cardiac tissue, normalizing sarcomeric and gap junction organization [24], the present findings map a dynamic host–graft interaction that transitions the proteome from a diseased towards a regenerative state. The multifaceted nature of regenerative repair mechanisms includes contributions from both endogenous and exogenous sources [70]. Notwithstanding, here the source of resolved proteins could not be distinguished as transplanted cells came from the same species as the endogenous tissue to avoid rejection and maximize engraftment.
CONCLUSION
Current experience with stem cell-based therapy has been primarily focused on ischemic forms of heart disease, whereas less is known of the potential benefit in the setting of nonischemic cardiomyopathy. Here, global protein expression profiling and comparative network analysis extracted from dilated, nonischemic cardiomyopathic ventricles distinct interactomes in the absence versus presence of embryonic stem cell treatment, establishing a proteomic fingerprint characterizing cell-based therapy in the context of KATP channel dysfunction and heart failure. The decoded proteomic profile provides a systems framework to diagnose regenerative patterns imposed by stem cell-based intervention.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonathan Nesbitt (Mayo Clinic) for surgical expertise, and the Mayo Proteomics Research Center staff for guidance in mass spectrometry analysis. We are particularly grateful to Dr. Takashi Miki (Chiba University, Chiba, Japan) and Dr. Susumu Seino (Kobe University, Kobe, Japan) for the initial derivation of the Kir6.2 knockout mice. This work was supported by the National Institutes of Health (R01HL064822, R01HL083439, T32HL007111), Marriott Heart Disease Research Program, and Mayo Clinic. J.Z.L. holds a Mayo Graduate School fellowship. D.K.A. is the recipient of a Marriott Individualized Medicine Career Development Award. S.Y. is the recipient of an American Society for Clinical Pharmacology and Therapeutics Young Investigator Award and a Marriott Mitochondrial Medicine Award. T.J.N. is a recipient of a PhRMA Foundation Faculty Development Award. A.T. holds the Marriott Family Professorship in Cardiovascular Research at Mayo Clinic.
Footnotes
Author contributions: J.Z.L.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; D.K.A.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; S.Y.: conception and design, provision of study material, collection and/ or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; T.J.N.: conception and design, provision of study material, data analysis and interpretation, manuscript writing, final approval of manuscript; A.T.: conception and design, financial support, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest.
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 2.Hershberger RE, Lindenfeld J, Mestroni L, et al. Genetic evaluation of cardiomyopathy: A Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Luk A, Ahn E, Soor GS, et al. Dilated cardiomyopathy: A review. J Clin Pathol. 2009;62:219–225. doi: 10.1136/jcp.2008.060731. [DOI] [PubMed] [Google Scholar]
- 4.Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature. 2000;407:227–232. doi: 10.1038/35025196. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 6.Olson TM. Monogenic dilated cardiomyopathy. In: Walsh RA, editor. Molecular Mechanisms of Cardiac Hypertrophy and Failure. 1st ed. Boca Raton, Florida: Taylor & Francis; 2005. pp. 525–540. [Google Scholar]
- 7.Hershberger RE, Cowan J, Morales A, et al. Progress with genetic cardiomyopathies: Screening, counseling, and testing in dilated, hy-pertrophic, and arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Heart Fail. 2009;2:253–261. doi: 10.1161/CIRCHEARTFAILURE.108.817346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienengraeber M, Olson TM, Selivanov VA, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kane GC, Liu XK, Yamada S, et al. Cardiac KATP channels in health and disease. J Mol Cell Cardiol. 2005;38:937–943. doi: 10.1016/j.yjmcc.2005.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes S, Park S, Johnson BD, et al. KATP channel Kir6.2 E23K variant overrepresented in human heart failure is associated with impaired exercise stress response. Hum Genet. 2009;126:779–789. doi: 10.1007/s00439-009-0731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson TM, Terzic A. Human KATP channelopathies: Diseases of metabolic homeostasis. Pflugers Arch Eur J Physiol. 2010;460:295–306. doi: 10.1007/s00424-009-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashrafian H, Watkins H. Cardiomyopathies: Therapeutics based on molecular phenotype. J Am Coll Cardiol. 2007;49:1251–1264. doi: 10.1016/j.jacc.2006.10.073. [DOI] [PubMed] [Google Scholar]
- 13.Hunt SA, Abraham WT, Chin MH, et al. 2009 focused update incorporated into the ACC/AHA 2005 Guidelines for the Diagnosis and Management of Heart Failure in Adults: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: Developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119:e391–e479. doi: 10.1161/CIRCULATIONAHA.109.192065. [DOI] [PubMed] [Google Scholar]
- 14.Dimmeler S, Zeiher AM, Schneider MD. Unchain my heart: The scientific foundations of cardiac repair. J Clin Invest. 2005;115:572–583. doi: 10.1172/JCI24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passier R, van Laake LW, Mummery CL. Stem cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 16.Segers VF, Lee RT. Stem cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 17.Hansson EM, Lindsay ME, Chien KR. Regeneration next: Toward heart stem cell therapeutics. Cell Stem Cell. 2009;5:364–377. doi: 10.1016/j.stem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Nelson TJ, Behfar A, Yamada S, et al. Stem cell platforms for regenerative medicine. Clin Transl Sci. 2009;2:222–227. doi: 10.1111/j.1752-8062.2009.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anversa P, Kajstura J, Leri A, et al. Life and death of cardiac stem cells: A paradigm shift in cardiac biology. Circulation. 2006;113:1451–1463. doi: 10.1161/CIRCULATIONAHA.105.595181. [DOI] [PubMed] [Google Scholar]
- 20.Torella D, Ellison GM, Méndez-Ferrer S, et al. Resident human cardiac stem cells: Role in cardiac cellular homeostasis and potential for myocardial regeneration. Nat Clin Pract Cardiovasc Med. 2006;3 suppl 1:8–13. doi: 10.1038/ncpcardio0409. [DOI] [PubMed] [Google Scholar]
- 21.Kajstura J, Hosoda T, Bearzi C, et al. The human heart: A self-renewing organ. Clin Transl Sci. 2008;1:80–86. doi: 10.1111/j.1752-8062.2008.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergmann O, Bhardwaj RD, Bernard S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Angelis A, Piegari E, Cappetta D, et al. Anthracycline cardiomyopathy is mediated by depletion of the cardiac stem cell pool and is rescued by restoration of progenitor cell function. Circulation. 2010;121:276–292. doi: 10.1161/CIRCULATIONAHA.109.895771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada S, Nelson TJ, Crespo-Diaz RJ, et al. Embryonic stem cell therapy of heart failure in genetic cardiomyopathy. Stem Cells. 2008;26:2644–2653. doi: 10.1634/stemcells.2008-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodgson DM, Behfar A, Zingman LV, et al. Stable benefit of embryonic stem cell therapy in myocardial infarction. Am J Physiol Heart Circ Physiol. 2004;287:H471–H479. doi: 10.1152/ajpheart.01247.2003. [DOI] [PubMed] [Google Scholar]
- 26.Me´nard C, Hage`ge AA, Agbulut O, et al. Transplantation of cardiac-committed mouse embryonic stem cells to infarcted sheep myocardium: A preclinical study. Lancet. 2005;366:1005–1012. doi: 10.1016/S0140-6736(05)67380-1. [DOI] [PubMed] [Google Scholar]
- 27.Kolossov E, Bostani T, Roell W, et al. Engraftment of engineered ES cell-derived cardiomyocytes but not BM cells restores contractile function to the infarcted myocardium. J Exp Med. 2006;203:2315–2327. doi: 10.1084/jem.20061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung S, Dzeja PP, Faustino RS, et al. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med. 2007;4 suppl 1:60–67. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laflamme MA, Chen KY, Naumova AV, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 30.Kehat I, Khimovich L, Caspi O, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Bio-technol. 2004;22:1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 31.Baba S, Heike T, Yoshimoto M, et al. Flk1+ cardiac stem/progenitor cells derived from embryonic stem cells improve cardiac function in a dilated cardiomyopathy mouse model. Cardiovasc Res. 2007;76:119–131. doi: 10.1016/j.cardiores.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Behfar A, Perez-Terzic C, Faustino RS, et al. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–420. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson TJ, Martinez-Fernandez A, Yamada S, et al. Repair of acute myocardial infarction with human stemness factors induced pluirpotent stem cells. Circulation. 2009;120:408–416. doi: 10.1161/CIRCULATIONAHA.109.865154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baraba´si AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 35.Weston AD, Hood L. Systems biology, proteomics, and the future of health care: Toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 36.Arrell DK, Niederländer NJ, Faustino RS, et al. Cardioinductive network guiding stem cell differentiation revealed by proteomic cartography of tumor necrosis factor α-primed endodermal secretome. Stem Cells. 2008;26:387–400. doi: 10.1634/stemcells.2007-0599. [DOI] [PubMed] [Google Scholar]
- 37.Van Hoof D, Heck AJ, Krijgsveld J, et al. Proteomics and human embryonic stem cells. Stem Cell Res. 2008;1:169–182. doi: 10.1016/j.scr.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Arrell DK, Zlatkovic J, Kane GC, et al. ATP-sensitive K+ channel knockout induces cardiac proteome remodeling predictive of heart disease susceptibility. J Proteome Res. 2009;8:4823–4834. doi: 10.1021/pr900561g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kane GC, Behfar A, Dyer RB, et al. KCNJ11 Gene knockout of the Kir62 KATP channel causes maladaptive remodeling and heart failure in hypertension. Hum Mol Genet. 2006;15:2285–2297. doi: 10.1093/hmg/ddl154. [DOI] [PubMed] [Google Scholar]
- 40.Yamada S, Kane GC, Behfar A, et al. Protection conferred by myocardial ATP-sensitive K+ channels in pressure overload-induced congestive heart failure revealed in KCNJ11 Kir6.2-null mutant. J Physiol. 2006;577:1053–1065. doi: 10.1113/jphysiol.2006.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Fernandez A, Nelson TJ, Yamada S, et al. iPS programmed without c-MYC yield proficient cardiogenesis for functional heart chi-merism. Circ Res. 2009;105:648–656. doi: 10.1161/CIRCRESAHA.109.203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada S, Nelson TJ, Behfar A, et al. Stem cell transplant into preim-plantation embryo yields myocardial infarction-resistant adult phenotype. Stem Cells. 2009;27:1697–1705. doi: 10.1002/stem.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlatkovic J, Arrell DK, Kane GC, et al. Proteomic profiling of KATP channel-deficient hypertensive heart maps risk for maladaptive cardiomyopathic outcome. Proteomics. 2009;9:1314–1325. doi: 10.1002/pmic.200800718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arrell DK, Niederländer NJ, Perez-Terzic C, et al. Embryonic stem cell cardiac differentiation: A proteomic perspective. Adv Mol Med. 2006;2:149–156. [Google Scholar]
- 45.Arrell DK, Niederländer NJ, Perez-Terzic C, et al. Pharmacoproteomics: Advancing the efficacy and safety of regenerative therapeutics. Clin Pharmacol Ther. 2007;82:316–319. doi: 10.1038/sj.clpt.6100310. [DOI] [PubMed] [Google Scholar]
- 46.Perkins DN, Pappin DJC, Creasy DM, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 47.Link AJ, Eng J, Schieltz DM, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 48.Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assenov Y, Ramírez F, Schelhorn S-E, et al. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 50.Barabasí A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 51.Jeong H, Tombor B, Albert R, et al. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 52.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 53.Braunwald E. Biomarkers in heart failure. N Engl J Med. 2008;358:2148–2159. doi: 10.1056/NEJMra0800239. [DOI] [PubMed] [Google Scholar]
- 54.Bell J. Predicting disease using genomics. Nature. 2004;429:453–456. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 55.Arab S, Gramolini AO, Ping P, et al. Cardiovascular proteomics: Tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–1741. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 56.Asakura M, Kitakaze M. Global gene expression profiling in the failing myocardium. Circ J. 2009;73:1568–1576. doi: 10.1253/circj.cj-09-0465. [DOI] [PubMed] [Google Scholar]
- 57.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteo-mics to genetics, genomics and network biology. Nat Rev Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 58.Merico D, Gfeller D, Bader GD. How to visually interpret biological data using networks. Nat Biotechnol. 2009;27:921–924. doi: 10.1038/nbt.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arrell DK, Terzic A. Network systems biology for drug discovery. Clin Pharmacol Ther. 2010;88:120–125. doi: 10.1038/clpt.2010.91. [DOI] [PubMed] [Google Scholar]
- 60.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 61.Dzeja PP, Redfield MM, Burnett JC, et al. Failing energetics in failing hearts. Curr Cardiol Rep. 2000;2:212–217. doi: 10.1007/s11886-000-0071-9. [DOI] [PubMed] [Google Scholar]
- 62.Neubauer S. The failing heart: An engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 63.Chen CH, Budas GR, Churchill EN, et al. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ping P. Getting to the heart of proteomics. N Engl J Med. 2009;360:532–534. doi: 10.1056/NEJMcibr0808487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waldman SA, Terzic A. Therapeutic targeting: A crucible for individualized medicine. Clin Pharmacol Ther. 2008;83:651–654. doi: 10.1038/clpt.2008.65. [DOI] [PubMed] [Google Scholar]
- 66.Nelson TJ, Behfar A, Terzic A. Strategies for therapeutic repair: The “R3” regenerative medicine paradigm. Clin Transl Sci. 2008;1:168–171. doi: 10.1111/j.1752-8062.2008.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lusis AJ, Weiss JN. Cardiovascular networks. Systems-based approaches to cardiovascular disease. Circulation. 2010;121:157–170. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yildirim MA, Goh KI, Cusick ME, et al. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 69.Hood L, Heath JR, Phelps ME, et al. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 70.Gersh BJ, Simari RD, Behfar A, et al. Cardiac cell repair therapy: A clinical perspective. Mayo Clin Proc. 2009;84:876–892. doi: 10.4065/84.10.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.