Abstract
Many developmental control genes contain paused RNA Polymerase II (Pol II) and are thereby “poised” for rapid and synchronous activation in the early Drosophila embryo. Evidence is presented that Polycomb group (PcG) repressors can influence paused Pol II. ChIP-Seq and GRO-Seq assays were used to determine the genome-wide distributions of Pol II, H3K27me3 and H3K4me3 in extra sex combs (esc) mutant embryos. ESC is a key component of the Polycomb Repressive complex 2 (PRC2), which mediates H3K27me3 modification. Enhanced Pol II occupancy is observed for thousands of genes in esc mutant embryos, including genes not directly regulated by PRC2. Thus, it would appear that silent genes lacking promoter-associated paused Pol II in wild-type embryos are converted into “poised” genes with paused Pol II in esc mutants. We suggest that this conversion of silent genes into poised genes might render differentiated cell types susceptible to switches in identity in PcG mutants.
INTRODUCTION
Recent studies have identified many instances of paused Pol II, whereby the Pol II machinery is pre-loaded onto the promoters of genes prior to their expression. Paused Pol II was characterized at the Drosophila heat shock genes (Rougvie and Lis, 1988). It is associated with short, 25-50 nucleotide (nt) nascent transcripts that interact with NELF and DSIF complexes to inhibit Pol II elongation. This paused Pol II is poised for rapid activation upon heat induction (Saunders et al., 2006).
Prior to 2007, promoter-proximal pausing was established as a regulated step for the transcription of just a handful of genes, including a few stress-induced genes such as Drosophila heat shock genes and mammalian myc genes (Kerppola and Kane, 1988; Saunders et al., 2006). However, whole-genome ChIP-chip assays identified Pol II at the promoter regions of many or most developmental control genes in the early Drosophila embryo (Muse et al., 2007; Zeitlinger et al., 2007). Moreover, genome-wide nuclear run-on assays (GRO-Seq) indicate that ~30% of all mammalian genes accumulate paused Pol II in a rate-limiting step early during transcription elongation (Core et al., 2008).
It is possible that Polycomb group (PcG) repressors influence Pol II pausing at inactive genes (Breiling et al., 2001; Brookes and Pombo, 2009; Chopra et al., 2009; Enderle et al., 2011; Stock et al., 2007). PcG repressors have been shown to maintain the silent state of target genes for extended periods during the lifecycle (Ringrose and Paro, 2004; Simon and Kingston, 2009). PcG proteins form four multi-subunit enzymatic complexes: PRC1, PRC2, PHO-RC and PR-DUB (Klymenko et al., 2006; Papp and Muller, 2006; Scheuermann et al., 2010; Simon and Kingston, 2009). The PRC2 complex contains the Extra Sex Combs (ESC) and Enhancer of Zeste [E(Z)] proteins, which mediate trimethylation of lysine-27 of histone H3 (H3K27me3) (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002; Simon and Kingston, 2009). Removal of ESC from the PRC2 complex results in a significant diminishment of methyl transferase activity (Ketel et al., 2005; Nekrasov et al., 2005).
The four PcG complexes function in an interdependent fashion to silence gene expression. The PHO-RC complex binds specialized DNA sequences, Polycomb Response Elements (PREs), and subsequently recruits the PRC2 complex. PRC1 binds to H3K27me3 and limits access of the Pol II machinery to silent genes by compacting chromatin at the core promoter (Francis et al., 2004; Simon and Kingston, 2009). Polycomb proteins are required for the stable repression of key developmental control genes in Drosophila (e.g. Hox genes), and are also important for maintaining stem cell fate (Pietersen and van Lohuizen, 2008; Ringrose and Paro, 2004).
We used ChIP-Seq and GRO-Seq assays to examine chromatin modifications and Pol II binding in esc mutant embryos (Fig. 1-3, Fig. S1, Fig.S2G and Table S1). ChIP-seq identifies chromatin modifications and Pol II occupancy, while GRO-seq (Core et al., 2008) distinguishes promoter-proximal Pol II peaks as transcriptionally engaged and paused. Most paused genes exhibit enhanced Pol II binding in esc mutants as compared with wild-type embryos. Moreover, many non-paused genes display Pol II pausing in mutant embryos. Thus, PRC2 appears to maintain gene silencing, at least in part, by inhibiting the establishment of promoter-proximal paused Pol II.
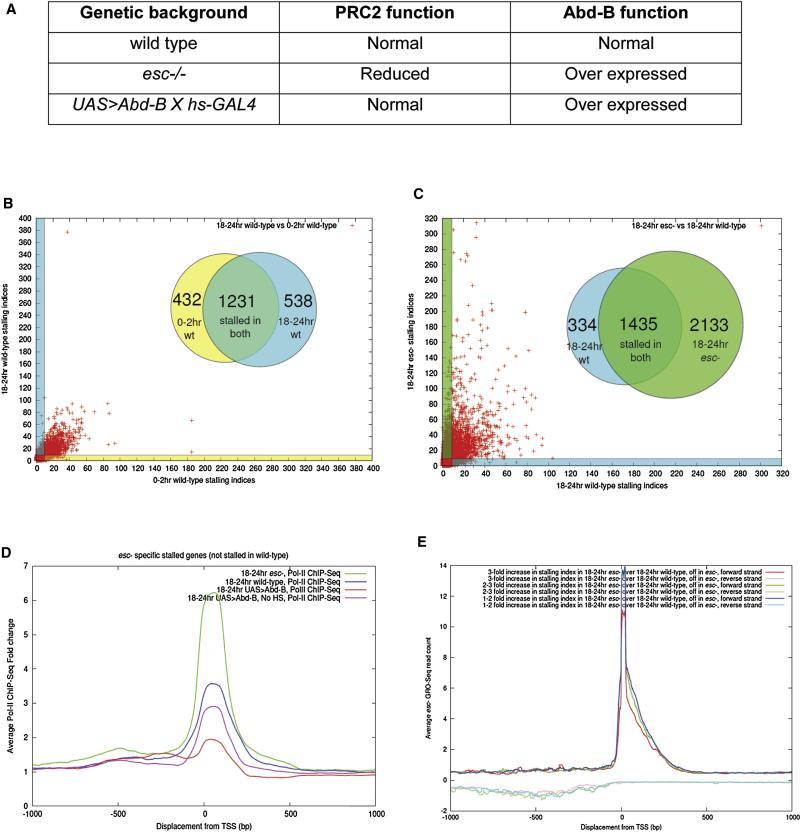
FIGURE 1. Augmented Pol II occupancy in esc mutant embryos.
A. The three different genetic backgrounds used for ChIP-Seq assays. Wild type (wt) embryos have normal function of PRC2 and Abd-B. There is diminished PRC2 function in esc mutant, leading to ectopic expression of Abd-B and the conversion of all embryonic segments to an eighth abdominal segment (A8) morphology. Abd-B was over-expressed in wt embryos via heat shock using hs-GAL4 and UAS-Abd-B transgenes (also see supp. methods section).
B. Pol II ChIP-Seq assays in early (0-2hr) and late (18-24hr) wt embryos. They contain overlapping subsets of stalled genes (432 early, 538 late, and 1231 overlap). These genes contain a stalling index of at least 10 (see Fig. S1). Each red mark represents a single gene. Marks located within the blue (18-24 hr) or yellow (0-2 hr) shaded regions correspond to genes that are stalled/paused in just one of the genotypes. The red marks located within non-shaded regions of the plot are stalled/paused in both genotypes.
C. Pol II stalling/pausing in late (18-24 hr) wt and esc mutant embryos. There are ~3500 stalled genes in esc mutants, including 2133 genes which show “ectopic stalling” only in mutant embryos (class II genes). As in (B), red marks falling within the shaded regions (green, esc; blue, wt) represent genes that are specifically stalled in only one of the genotypes.
D. Relative levels of Pol II ChIP-Seq binding in wt and esc embryos for class II genes (stalled/paused in esc mutants, but not wt embryos). 18-24 hr esc mutant embryos exhibit a 6.5-fold increase over background levels (green). A 3.5-fold (blue) increase was observed for 18-24 hr wt embryos. Just 3-fold (pink) and 2-fold (red) increases were observed for 18-24 hr embryos with or without heat shock Abd-B, respectively.
E. The stalled and inactive (“off”) genes seen in 18-24 hr esc embryos were divided into three subsets, those showing 1-2 fold (blue), 2-3 fold (green) and greater than a 3-fold (red) increase in Pol II stalling as compared with wt embryos (see Fig.3A). Genes in the 3-fold category [1485/2133 (70%) of class II genes] are enriched for putative targets of PRC2, including Hox genes and head patterning genes (e.g., orthodenticle and aristaless see Fig. S2A). All three subsets of stalled genes also exhibit strong GRO-Seq signals, confirming that they are “paused” and capable of productive elongation.
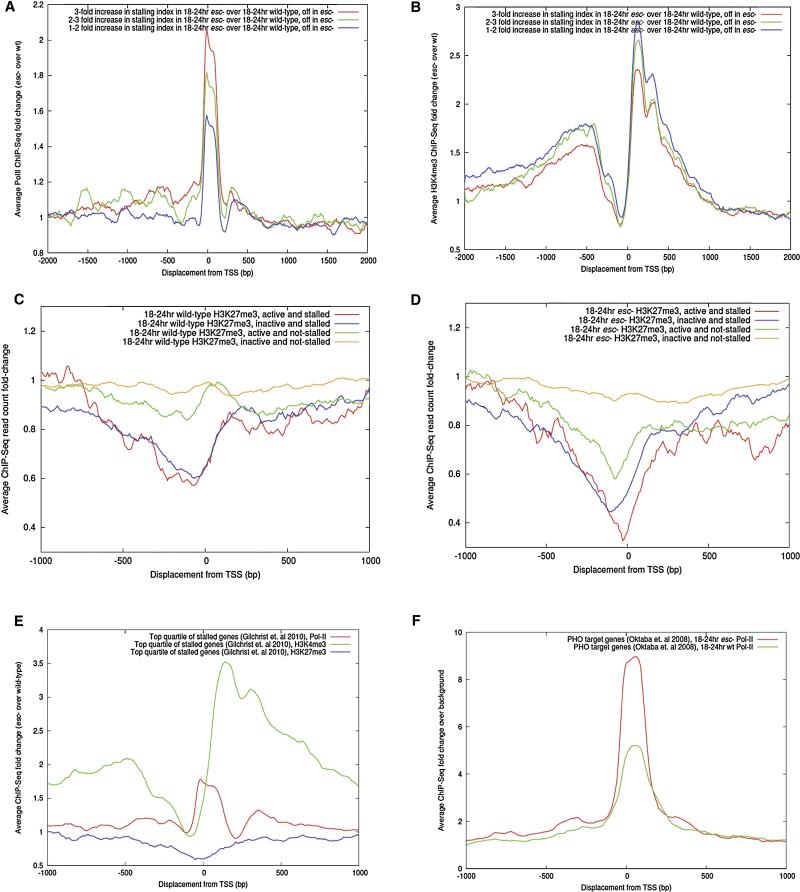
Figure 3. Transcriptional profiling of wt and esc mutant embryos.
(A) The stalled genes that are inactive in esc mutants were divided into three subsets (similar to Fig.1D): 1-2 fold (blue), 2-3 fold (green) and more than a 3-fold (red) increase in Pol II stalling in esc as compared with wt embryos.
(B) The same three subsets of genes exhibit increased H3K4me3 binding (a trxG mark) when normalized to wt embryos. This modification is most enriched ~200-300 bp downstream of the +1 transcription start site.
(C) H3K27me3 ChIP-Seq read counts at stalled and non-stalled promoters in 18-24hr wt embryos. There is a slight diminishment of H3K27me3 read counts at the promoter regions of stalled genes (red and blue lines) as compared with non-stalled genes (yellow and green lines).
(D) H3K27me3 ChIP-Seq read counts at stalled and non-stalled promoters in esc mutants. There is a significant depletion of H3K27me3 read counts as compared with wt embryos (Fig. 3C). There are also diminished read counts in the promoter regions of active non-stalled genes (green line) as compared with inactive non-stalled genes (yellow line).
(E) Analysis of the ~1500 “top quartile” genes (Gilchrist et al., 2010). Relative changes in Pol II (red), H3K4me3 (green) and H3K27me3 levels in esc vs. wt embryos. There is a ~1.8 fold increase in Pol II, 3.5 fold increase in H3K4me3, and reduced levels of H3K27me3 in esc embryos as compared with wt embryos.
(F) PHO regulated genes (~500 genes) (Oktaba et al., 2008). Relative changes in Pol II binding in esc mutants and wt embryos, as compared with background controls. There is a 9 fold (red) increase in Pol II binding in esc mutants and a 5 fold (green) increase in wt embryos (also see Table S4 and Fig.S3F).
RESULTS
Dynamics of Pol II pausing during embryogenesis
We examined the distribution of Pol II in early (0-2hr) and late (18-24hr) wild-type (wt) embryos via Pol II ChIP-Seq assays (Fig. 1B). There are over 2000 genes that exhibit disproportionate association of Pol II at the core promoter as compared with associated transcription units (“stalling” index greater than 10; see Fig. S1 for explanation). We observe promoter–proximal Pol II occupancy at ~400 genes in 0-2 hr embryos, ~500 genes in 18-24 hr embryos, and ~1200 genes are common to both datasets (Fig. 1B, Fig. S1). These observations suggest that there are subsets of genes that do not contain promoter-proximal Pol II during early stages of development but become stalled at later stages and vice-versa. Perhaps Pol II stalling “anticipates” gene activation at subsequent stages of development.
Augmented Pol II binding in esc embryos
To determine whether PcG influences Pol II activity, whole-genome Pol II ChIP-Seq assays were performed with esc mutant embryos (Fig. 1A and C-D). These mutants exhibit the classic Polycomb phenotype; the transformation of all body segments into the posterior-most eighth abdominal (A8) segment (Struhl, 1981; Struhl and Brower, 1982) (see supp. methods section). Approximately 3500 genes (25% of all genes in the Drosophila genome) exhibit a significant increase in the levels of promoter-proximal Pol II in esc mutants as compared with wt embryos. These genes fall into two broad categories. Over half of the genes (2100 of 3500) exhibit stalled Pol II only in esc mutants and not in comparably staged wt embryos (Fig. 1C). We refer to these genes as “ectopically stalled or paused” (class II, Fig. 4). The remaining genes (1400 of 3500) contain stalled Pol II in both wt and esc embryos (Fig. 1C, class I in Fig.4), but there is at least a 2-fold increase in the levels of promoter-proximal Pol II in mutant embryos (summarized in Fig. 1D, Fig.S3A and S3E). Many of the Pol II peaks exhibit a 5’ shift of ~9 bp in esc mutants as compared with wt embryos (Fig. S3G). This might indicate regulation at an early stage of elongation, immediately following initiation.
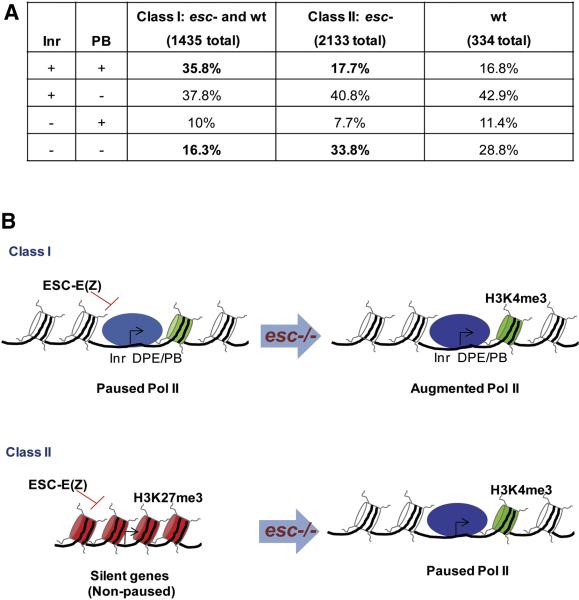
Figure 4. Binding threshold model for paused Pol II.
A.Core promoter pausing elements in different classes of stalled genes. Nearly 36% of class I genes (Pol II stalling in both wt and esc embryos) contain both an initiator element at the transcription start site (Inr) and a pause button (PB) motif located within 50 bp downstream of the start site, most often at ~+30 bp. Only ~16% of class I genes lack both promoter elements. A flip-flop correlation was observed for class II genes (Pol II stalling only in esc mutants). Only ~18% contain both Inr and PB elements, while ~34% of all class II genes lack both elements. Thus, there is a 2-fold enrichment in the occurrence of Inr and PB elements in class I vs. class II genes.
B. Threshold model for paused Pol II. In wild-type embryos Pol II (light blue) overcomes repression by PRC2 due to the occurrence of core promoter pausing elements, such as the Inr and DPE/PB (the PB motif is a GC-rich variant of the DPE, which is located +28-+32 bp downstream of the +1 transcription start site). H3K4me3 (light green nucleosome) is detected just downstream of stalled Pol II. In contrast, class II genes contain H3K27me3 at the core promoter and contain reduced levels of H3K4me3 in the downstream nucleosome. In esc mutants, diminished PRC2 activity leads to enhanced Pol II binding at class I promoters (dark blue), and reduced H3K27me3 at the promoter regions of class II genes. Pol II is now able to bind to class II promoters due to diminished levels of repressive nucleosomes. According to this model, the switch in paused Pol II is influenced by the presence or absence of promoter pausing elements and the action of PcG repressive complexes. The reduction in PRC2 activity in esc mutants lowers the threshold of Pol II binding, even for genes lacking core promoter pausing elements.
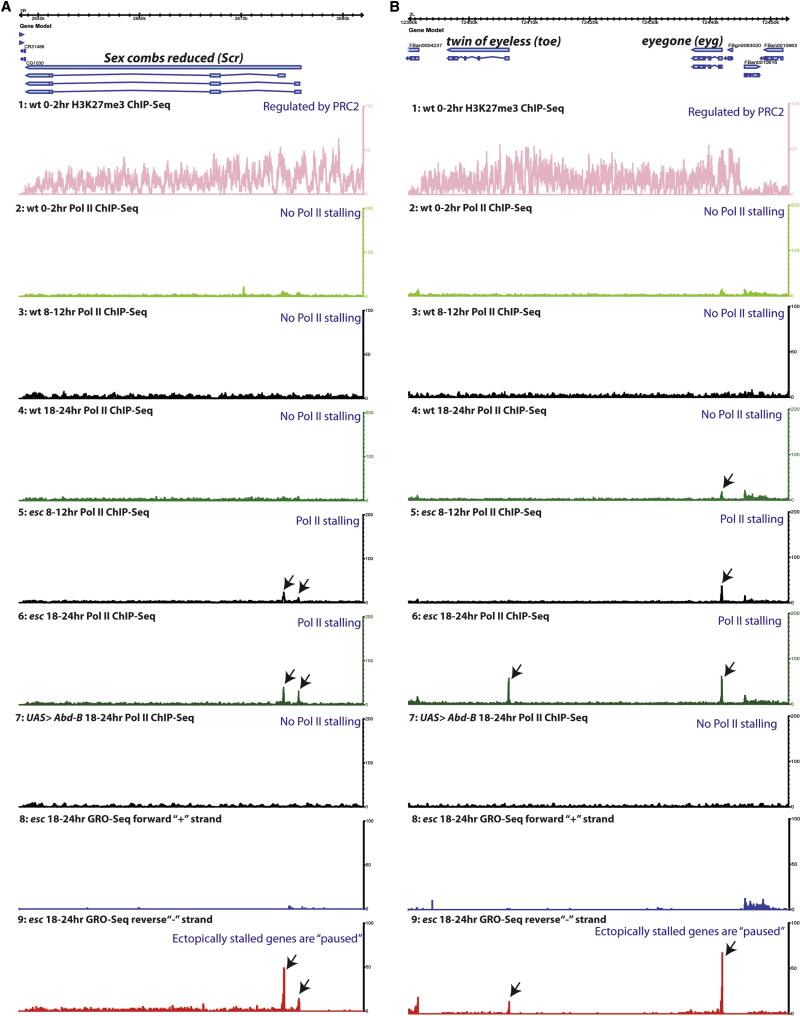
The A8 transformations seen in esc mutant embryos are due, at least in part, to the derepression of the Hox gene, Abdominal-B (Abd-B) (Fig.S2F) (Duboule and Morata, 1994; Wedeen et al., 1986). Surprisingly, Hox genes that are repressed by ABD-B exhibit enhanced, not reduced, levels of Pol II promoter occupancy in esc mutants (Fig. 2A, Fig.S2C and S2E). For example, there is a several-fold increase in the levels of Pol II binding at the proximal Antennapedia (Antp) promoter, even though the expression of the gene is significantly diminished as compared with wt (Fig. S2B and S2D). Augmented promoter occupancy is also observed for other Hox genes in the Antennapedia complex, including Sex combs reduced (Scr) (Fig. 2A) and Deformed (Dfd) (Fig. S2B). Scr lacks promoter-proximal Pol II in wt embryos (0-2 hr, 8-12 hr and 18-24 hr) (Fig. 2A, track 2, 3 and 4), but exhibits increasing levels in esc mutants (Fig. 2A, tracks 5,6). GRO-Seq assays suggest that these peaks coincide with paused Pol II (Fig. 2A, tracks 8,9 and Fig.1E).
Figure 2. Dynamics of Pol II stalling/pausing in wt and esc embryos.
A. The Sex combs reduced (Scr) gene is not paused in early (0-2hr) (track2), mid (8-12hr) (track 3) and late (18-24hr) (track 4) wt embryos. Scr is a likely target of PRC2 regulation, and H3K27me3 signals are observed in early (0-2hr) wt embryos (track 1). Enhanced Pol II occupancy is observed at both Scr promoters in mid-stage (8-12hr) (track 5) and late (18-24hr) (track 6) esc mutant embryos (black arrows). In contrast, no increase in Pol II stalling is observed in wt embryos containing ectopic expression of Abd-B via heat shock (track 7). GRO-Seq assays indicate that the Scr promoters contain elongation competent (paused) Pol II (tracks 8,9).
B. The toe-eyg complex was analyzed for Pol II occupancy and H3K27me3 modification. The toe and eyg genes are not stalled in early (0-2hr) (track 2) and mid (8-12hr) (track 3) stage embryos but exhibit some Pol II pausing (for eyg promoter) in late (18-24hr) (track 4) wt embryos. These genes are likely targets of PRC2 regulation since H3K27me3 signals are distributed throughout the complex in early (0-2hr) wt embryos (track 1). In mid-stage (8-12hr) (track 5) esc mutant embryos we see augmented Pol II stalling at the eyg promoter (black arrow), and both toe and eyg exhibit stalling in older (18-24hr) (track 6) esc mutant embryos. GRO-Seq assays suggest that Pol II is paused at both promoters (arrows, tracks 8,9). There is no significant Pol II stalling of toe and eyg promoters in wt embryos (track 4), nor in embryos containing ectopic Abd-B expression via heat shock (track 7). Note that 0-2 hr embryos are likely to be biased towards 2 hr (or even older) due to the geometric increase in the number of nuclei during precellular development.
To determine whether the observed increase in Pol II occupancy is due to diminished PRC2 activity, or is an indirect consequence of ectopic Abd-B expression, Pol II ChIP-Seq assays were performed with wild-type embryos containing ubiquitous Abd-B expression (Fig. 1D and Fig. S1D). This misexpression was achieved by heat shocking embryos carrying UAS-Abd-B and hsp70-GAL4 (Fig. 2A, track 7 and supp. methods section) (Castelli-Gair et al., 1994). The resulting embryos display homeotic transformations of anterior and posterior segments, but do not exhibit augmented levels of Pol II occupancy at class II genes such as Scr (Fig. 1D). These results suggest that the increase in Pol II occupancy observed in esc mutants is due to diminished PRC2 activity and not repression by Abd-B (Fig. 2A, track 7).
The correlation between PRC2 regulation and paused Pol II is also observed for the eyegone (eyg)/twin of eyegone (toe) gene complex (Fig. 2B). H3K27me3 is detected throughout the complex, but forms sharp boundaries at the neighboring genes (Fig. 2B, track 1). There is no stalled Pol II observed for toe in 0-2 hr (Fig. 2B, track 2), 8-12 hr (Fig. 2B, track 3), or 18-24 hr (Fig. 2B, track 4) wt embryos, and only modest levels of Pol II at the eyg promoter. There is a substantial increase in Pol II binding at both genes in esc mutants (Fig. 2B, tracks 5 and 6), but not when Abd-B is misexpressed via heat shock (Fig. 2B, track 7). Finally, GRO-Seq assays indicate the occurrence of paused Pol II in the promoter regions of both genes in esc mutants (Fig. 2B, tracks 8-9 and Fig. 1E) (more examples in Fig. S2 A-B).
Paused Pol II is associated with changes in histone modifications
There is an overall 2-fold increase in the levels of stalled Pol II in 18-24 hr esc mutant embryos as compared with 18-24 hr wt embryos (Fig. 3A, Fig.S3A and S3E). To determine whether this increase is associated with changes in histone modifications, whole-genome ChIP-Seq assays were done with antibodies that recognize H3K27me3 and H3K4me3. These modifications are mediated by the PRC2 PcG complex and the TAC1 trxG complex, respectively (Simon and Kingston, 2009; Smith et al., 2004) . There is a significant increase in the levels of H3K4me3 in esc mutants (Fig. 3B, Fig.S3B and S3E), and a significant diminishment in the levels of H3K27me3 at the promoter proximal regions of stalled genes in esc mutant embryos (Fig. 3C-D and Fig.S3C-S3E).
Gilchrist et al. (2010) recently identified a set of nearly 1500 genes (“top quartile”) that are highly regulated in development and contain either a positioned nucleosome or paused Pol II at the core promoter. The top quartile genes are regarded as those most tightly regulated at the level of release of Pol II from promoter-proximal pause sites. The promoter regions of these genes exhibit a ~2 fold increase in stalled Pol II, a ~3 fold increase in H3K4me3, and diminished H3K27me3 in esc mutants as compared with wt embryos (Fig. 3E).
A significant fraction of Pleiohomeotic (PHO) target genes (Oktaba et al., 2008) exhibit ectopic Pol II stalling in esc mutants (126/536) (Fig. 3F, also see Table S4 and Fig.S3F). Moreover, the analysis of a subset of individual genes by Pol II ChIP-qPCR, RT-qPCR, GRO-Seq assays and in situ hybridization assays is consistent with the general trend that increases in promoter-proximal Pol II occupancy in esc mutants does not correlate with changes in gene expression (Fig. S2C-S2G; see below). Indeed, the majority of genes that exhibit increased Pol II stalling exhibit either reduced levels of gene expression or no change in expression in esc mutants.
DISCUSSION
We have examined the genome-wide distribution of Pol II, H3K4me3, and H3K27me3 using ChIP-Seq and GRO-Seq assays in staged wild-type embryos and esc mutants (Figure S1, table S1). Most such studies have relied on the use of cultured cells (Nechaev et al., 2009; Schwartz et al., 2006), and to a lesser extent, imaginal discs (Chopra et al., 2009; Papp and Muller, 2006). Our choice of esc is constrained by the need for large quantities of staged mutant embryos. Of the different subunits comprising the four PcG repression complexes, only esc mutants have a sufficiently strong maternal contribution to produce homozygous mutant adults that are fully viable and fertile, permitting the isolation of large numbers of mutant embryos (Struhl, 1981). Unfortunately, it is not possible to use this approach to examine mutant embryos deficient in trxG components.
There are two broad classes of genes showing augmented Pol II occupancy in esc mutants: those normally stalled or paused in wt embryos (class I), and those stalled only in esc mutants (class II) (summarized in Fig. 4). Approximately 35% of all class I genes contain an initiator element (Inr) at the transcription start site and a pause button motif (PB) located within 50 bp downstream of the start site (Fig. 4A and table S3) (Hendrix et al., 2008; Juven-Gershon et al., 2008; Nechaev and Adelman, 2010). These elements might permit Pol II binding even in the presence of PRC2 repression. In contrast, less than 18% of all class II genes contain Inr and PB elements (Fig. 4A and table S3). The diminished occurrence of “pausing elements” in the promoter regions of these genes might help explain why they exhibit paused Pol II only in esc mutants and not wt embryos. However, we note that ~40% of the 3500 genes exhibiting enhanced Pol II binding lack H3K27me3 and are therefore likely to be indirect targets of PRC2 (Table S4). The basis for increased promoter-proximal Pol II occupancy is uncertain for such genes, but might arise from a change in the “global balance” of chromatin states.
Gilchrist et al. (2010) have recently proposed a simple model for the regulation of gene expression during development. Promoter regions are occupied by either paused Pol II or positioned nucleosomes. Paused Pol II precludes the binding of repressive nucleosomes and thereby “marks” the promoter for activation upon receipt of appropriate inducing signals. This Pol II/nucleosome switch may be influenced by the presence or absence of core promoter pausing elements, such as the Inr and PB [see (Gilchrist et al., 2010)]. The present study and a recent report (Min et al., 2011) provide evidence that this switch is also influenced by PcG repressors.
It is possible that PcG repressors raise the “threshold” of Pol II binding by facilitating nucleosome compaction at paused promoters (Fig. 4B). This threshold is reduced in esc mutants due to diminished PRC2 activity (decrease of H3K27me3) and hence, non-paused genes come to acquire paused Pol II. This conversion does not usually result in changes in gene expression. The target genes remain inactive (or minimally expressed) in inappropriate tissues, but they are presumably more easily activated when appropriate inducing signals become available. This might explain why PcG mutants exhibit enhanced developmental plasticity in fly imaginal discs and C. elegans embryos (Lee et al., 2005; Yuzyuk et al., 2009). We propose that esc mutant tissues are more susceptible to changes in cell fate due to augmented levels of paused Pol II.
EXPERIMENTAL PROCEDURES
ChIP-Seq and GRO-seq assays
Staged wild-type and mutant Drosophila embryos were cross-linked with formaldehyde followed by isolation of chromatin using standard protocols (Chopra et al., 2009; Zeitlinger et al., 2007). The chromatin was sonicated using a Brandson sonicator and was subjected to ChIP using a cocktail of Pol II antibodies [anti-HWG16 and anti-H14 (COVANCE)] (Zeitlinger et al., 2007) or H3K27me3 antibody (Millipore) and H3K4me3 antibody (Abcam). A fraction of the chromatin was kept aside as input control and a Mock IP was performed without antibodies. Libraries were prepared for ChIP and input samples using standard protocols for the Illumina platform, quantified using a Bioanalyzer and subjected to single-end sequencing (36 cycles) (see supplemental methods section). The sequence reads were aligned back to the release 4.3 version of the Drosophila genome. The GRO-Seq assay was performed as described earlier (Core et al., 2008).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elimbiopharm Inc. (Hayward, CA) and L. Tonkin (UC Berkeley) for high-throughput DNA sequencing. J. Choi and K. Hermens (UC Berkeley) provided valuable help with library quantification. We are particularly grateful to G. Struhl and J. Simon for providing esc mutant stocks, I. Hariharan for the heat shock GAL4 stock, the Bloomington Stock center for providing UAS-Abd-B flies and M. O’Connor for the wit plasmid. Thanks to P. Paliwal for support and help with the Western blot assays. Thanks to J. Müller, D.S. Rokhsar and Levine lab members for helpful discussions and feedback. We also thank K. Adelman for sharing the list of top quartile paused genes. This study was funded by a grant from the NIH (GM34431) to M.L. The experiments were designed by M.L. and V.S.C., the manuscript was prepared by M.L., V.S.C. and D.A.H. with critical feedback from J.T.L and L.J.C. The experiments were performed by V.S.C. and C.T. helped with RNA in situ hybridization assays. The bioinformatics analysis was performed by D.A.H. GRO-Seq assays were performed in collaboration with L.J.C. and J.T.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The ChIP-Seq and GRO-Seq data has been deposited at the Gene Expression Omnibus database (GEO accession number GSE28833).
REFERENCES
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009;10:1213–1219. doi: 10.1038/embor.2009.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Castelli-Gair J, Greig S, Micklem G, Akam M. Dissecting the temporal requirements for homeotic gene function. Development. 1994;120:1983–1995. doi: 10.1242/dev.120.7.1983. [DOI] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 2011;21:216–226. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK, Kane CM. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988;8:4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2009;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Muller J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005;6:348–353. doi: 10.1038/sj.embor.7400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- Struhl G, Brower D. Early role of the esc+ gene product in the determination of segments in Drosophila. Cell. 1982;31:285–292. doi: 10.1016/0092-8674(82)90428-7. [DOI] [PubMed] [Google Scholar]
- Wedeen C, Harding K, Levine M. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell. 1986;44:739–748. doi: 10.1016/0092-8674(86)90840-8. [DOI] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.