Abstract
An estimated 2 million Americans use cocaine, resulting in large personal and societal costs. Discovery of the genetic factors that contribute to cocaine abuse is important for understanding this complex disease. Previously, mutations in the Drosophila LIM-only (dLmo) gene were identified because of their increased behavioral sensitivity to cocaine. Here we show that the mammalian homolog Lmo4, which is highly expressed in brain regions implicated in drug addiction, plays a similar role in cocaine-induced behaviors. Mice with a global reduction in Lmo4 levels show increased sensitivity to the locomotor stimulatory effects of cocaine upon chronic cocaine administration. This effect is reproduced with downregulation of Lmo4 in the nucleus accumbens by RNA interference. Thus, Lmo genes play conserved roles in regulating the behavioral effects of cocaine in invertebrate and mammalian models of drug addiction.
Keywords: Cocaine sensitization, LMO4, nucleus accumbens, RNA interference
Cocaine is a highly addictive and extensively abused drug that exerts its psychostimulant effects primarily by increasing signaling by dopamine and other monoamines in mesocorticolimbic systems, an effect that is achieved by direct inhibition of plasma membrane monoamine transporters (Amara & Kuhar 1993; Uhl et al. 2002). Repeated exposure to psychostimulants, such as cocaine, results in a progressive and enduring increase in locomotor activation, a phenomenon known as behavioral sensitization (Nestler 2001; Robinson & Berridge 1993; White & Kalivas 1998). The long-lasting neuroadaptations underlying sensitization are thought to be related to those responsible for addictive behavior in humans (Kalivas & Volkow 2005; Robinson & Berridge 2001), and to occur as a result of drug-induced changes in the transcriptional program in key brain regions, such as the striatum (Nestler 2001). For example, the transcription factors CREB and ΔFosB have been implicated in the rewarding and psychostimulant properties of cocaine (Carlezon et al. 2005; Kelz et al. 1999; Nestler et al. 2001). In addition, chromatin modifications occur in striatum upon chronic cocaine exposure leading to transcriptional activation of BDNF and Cdk5, two genes whose activity also affects behavioral responses to cocaine (Kumar et al. 2005). Activity of the histone deacetylase HDAC5 decreases with chronic cocaine exposure, leading to increased acetylation and transcription of HDAC5 target genes involved in cocaine sensitization and reward (Renthal et al. 2007).
Previously we performed an unbiased genetic screen in Drosophila and identified the gene dLmo as an important regulator of acute cocaine sensitivity (Tsai et al. 2004). dLmo encodes a LIM domain-only (LMO) nuclear protein consisting of two tandem LIM motifs, which are cysteine-rich zinc co-ordinating domains that mediate protein–protein interactions (Kadrmas & Beckerle 2004). LMO proteins regulate gene transcription by interacting with transcriptional cofactors and regulatory DNA-binding proteins (Bach 2000; Milan & Cohen 1999). Mammalian genomes encode four Lmo genes, Lmo1–4. In the adult mouse brain, Lmo4 is highly expressed in cortex, hippocampus and striatum (Bulchand et al. 2003; Hermanson et al. 1999), brain regions that have been implicated in cocaine-induced behaviors (Nestler 2001). We therefore investigated the role of Lmo4 in cocaine sensitization in the mouse. Mice carrying a gene-trap insertion in Lmo4, resulting in a 50% reduction in Lmo4 expression, showed increased responsiveness to cocaine upon repeated cocaine exposure. Moreover, this behavioral effect was localized to Lmo4 function in the nucleus accumbens (Acb) by virally mediated RNA interference. These data implicate a role for Lmo4 in the long-lasting behavioral plasticity that occurs with repeated cocaine treatment.
Materials and methods
Subjects
Mice used for experiments were male C57BL/6J aged 8–12 weeks. Mice were group-housed until they underwent stereotaxic surgery, after which they were singly housed, throughout all subsequent testing. Food and water were provided at all times, and animals were on a 12-h light/dark cycle. All animal protocols were approved by the Ernest Gallo Clinic and Research Center (EGCRC) institutional animal care and use committee.
Generation of Lmo4 gene-trap mice
To confirm the position of the gene-trap insertion in ES cell line RR0142, we isolated genomic DNA by standard methods. Polymerase chain reaction (PCR) amplification was performed using a forward primer designed from exon 4 of Lmo4 and a reverse primer that recognizes the gene-trap vector (Table S1) to generate a product of ~600 bp. Blastocysts derived from C57BL/6J (B6) females were injected with Lmo4Gt ES cells (RR0142, http://baygenomics.ucsf.edu) according to standard protocols (Hogan 1994) and implanted into pseudopregnant CD1 females to generate chimeric offspring. Several chimeras were crossed to wild-type C57BL/6J mice, and transmission of the targeted allele, designated Lmo4Gt(pGt2Lxf)1Heb (abbreviated Lmo4Gt), was confirmed by PCR (see Table S1 for primer sequences). Intercrosses of the resulting F1 mice (Lmo4Gt129/+B6) were established to generate LmoGt/+ heterozygotes and wild-type controls for experiments involving chronic cocaine exposure.
Design and cloning of short-hairpin RNAs
Three 19-nucleotide small interfering RNAs (siRNAs) targeting Lmo4 messenger RNA (mRNA) (Genbank accession NM-010723.2) were designed according to published criteria (Elbashir et al. 2001; Reynolds et al. 2004), using the siRNA selection program at the Whitehead Institute (http://jura.wi.mit.edu/siRNAext/). The 19-nucleotide targeting sequences are listed in Table S1. Target sequences were incorporated into hairpins and cloned into the lentiviral vector pLL3.7 as described previously (Lasek et al. 2007). The control vector for infection produced a scrambled (Scr) short-hairpin RNA (shRNA) (Table S1). The Scr sequence shows no significant alignment to any mouse mRNA.
Testing RNAi in cell culture
Neuro-2a cells (American Type Culture Collection, Rockville, MD, USA) were grown in Dulbecco’s modified Eagle’s minimal essential medium plus 10% fetal bovine serum and 10% CO2. Cells were seeded into six-well dishes and transfected with pLL3.7 plasmids containing Lmo4 shRNA sequences using Lipofectamine™ 2000 and Opti-MEM media (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Forty-eight hours after transfection, media was removed, 0.5 ml of Trizol® reagent (Invitrogen) added to each well, and total RNA isolated according to the Trizol® instructions. Total RNA was treated with RNAse-free DNAse (Promega, Madison, WI, USA) to remove genomic DNA contamination.
Quantitative Real-time-PCR
Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using reverse transcription reagents from Applied Biosystems (Foster City, CA, USA). Following synthesis, cDNA was diluted 1:10 in water. TaqMan qPCR was performed using standard thermal cycling conditions on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Amplification reactions contained 5 μl of cDNA template, 1× Universal PCR Master Mix, 100 nM each of forward and reverse primers and 200 nM of FAM or VIC-labeled probe in a final volume of 10 μl. The sequences of the mouse Lmo4 probe and primers are listed in Table S1. Mouse Gapdh probe and primers (Applied Biosystems) were used as controls for the PCRs.
Lentivirus production
Lentivirus was produced in 293FT cells (Invitrogen) as described previously (Lasek et al. 2007).
Stereotaxic surgeries
Male, 8- to 12-week-old C57BL/6J mice weighing 23–28 g were infused with lentivirus as described previously (Lasek et al. 2007), except co-ordinates for Acb were A/P +1.7 mm, M/L ±0.9 mm, D/V −4.6 mm (from the top of the skull), in reference to bregma (Paxinos & Franklin 2001). Co-ordinates for caudate putamen (Cpu) were A/P +1.5 mm, M/L ±1.5 mm, D/V −3.2 mm. Mice were allowed to recover for at least 9 days prior to commencing behavioral experiments.
Laser-capture microdissection
Mice were infected with lentivirus expressing shLmo4.3 or shScr in the Acb and allowed to recover for 12 days. Mice were euthanized with CO2, and brains were quickly removed and frozen in −50°C isopentane; 8 μm sections were cut with a cryostat and mounted on RNAse-free silane-prep slides (Sigma, St. Louis, MO, USA) and dehydrated with graded alcohol and xylene. Infected cells were visualized by green fluorescence protein (GFP). Approximately 200 pulses of 20 μm each were used to collect infected tissue using a PixCell IIe apparatus (Molecular Devices, Sunnyvale, CA, USA). RNA was isolated using the PicoPure RNA isolation kit (Molecular Devices) and subjected to two rounds of linear amplification using the RiboAmp HS kit (Molecular Devices). Complementary DNA synthesis and quantitative PCR were performed as described above.
β-Galactosidase detection
Mice were either killed with CO2 or Euthasol, injected intracardially with heparin (250 U) to prevent blood coagulation and perfused using phosphate-buffered saline (PBS) containing 0.2% glutaraldehyde. Brains were removed and left in 30% sucrose/0.2% glutaraldehyde overnight, and then cut frozen on a sliding microtome (Leica Microsystems, Bannockburn, IL, USA) into 80-μm-thick sagittal or coronal sections. Sections were processed for LacZ expression using standard X-Gal histochemistry at 37°C overnight in solution containing 1 mg/ml X-Gal, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide and 2 mM MgCl2. Sections were then postfixed in 4% paraformaldehyde (PFA) for 1 h, mounted on gelatinated slides, lightly stained with neutral red, dehydrated in graded alcohols, cleared with xylene and coverslipped with DPX mounting media (Sigma).
Immunohistochemistry
Mice were quickly euthanized as described above and were perfused intracardially with 0.9% NaCl for 5 min, followed by 4% PFA in PBS for 10 min. The brain was removed, fixed in 4% PFA for 2 h and transferred to PBS. Sections (40 μm) were cut on a vibratome. Free-floating sections were pretreated with 3% H2O2 for 10 min followed by 50% ethanol for two times 10 min. Sections were blocked with 10% normal donkey serum for 30 min. Sections were incubated with mouse anti-GFP monoclonal antibody 3E6 (Invitrogen) diluted 1:1500 in PBS/0.1% Triton-X-100 for 48–60 h. Sections were washed three times for 5 min with PBS and then incubated with 10% normal donkey serum for 10 min. Biotin-conjugated donkey anti-mouse secondary antibodies (diluted 1:250; Jackson ImmunoResearch, West Grove, PA, USA) were incubated with sections for 2–3 h, followed by ExtrAvidin (1:2500; Sigma) for 1–2 h. Diaminobenzidine was used for brown color detection of the GFP immunostaining. Sections were washed with PBS, mounted on gelatin-coated slides and dried. Slides were stained with cresyl violet according to standard protocols.
For immunofluorescent detection of LMO4 and GFP, sections or cells were treated as above, except fluorescently labeled secondary antibodies were used. Goat anti-LMO4 C-15 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was diluted 1:200, and mouse anti-GFP was diluted 1:1000. Secondary antibodies (Jackson ImmunoResearch) were Cy3-labeled donkey anti-goat and FITC-labeled donkey anti-mouse, both diluted 1:250. After staining, sections were mounted on slides with Vectashield fluorescent mounting medium containing DAPI (Vector Laboratories, Burlingame, CA, USA). Images were acquired using a Zeiss confocal microscope and visualized using Zeiss LSM software.
Cocaine treatments
Locomotor activity measurements were performed in Plexiglas locomotor activity chambers (Med Associates, St. Albans, VT, USA). To test locomotor sensitization to cocaine, on the first 2 days mice were injected i.p. with saline (10 ml/kg) and placed into the activity chambers for 15 min to habituate them to the testing procedures. On days 3–7, drug-induced locomotor activity was measured by dividing animals into two groups and treating with either saline or cocaine HCl (15 mg/kg; Sigma) in saline. Mice were then immediately placed into the activity chambers, and horizontal distance traveled (in cm) was recorded for 15 min. Cocaine sensitization was also measured 10 days later (day 17), by injecting both saline- and cocaine-treated groups with 15 mg/kg cocaine. In a separate group of mice, locomotor activity in response to a saline injection was measured (Fig. 2b). Mice were placed in activity chambers for 1 h to measure baseline locomotor activity. They were then injected with saline (10 ml/kg) and placed back into the chambers for an additional 30 min. Horizontal distance traveled (in cm) was graphed in 5 min bins over the 80-min period.
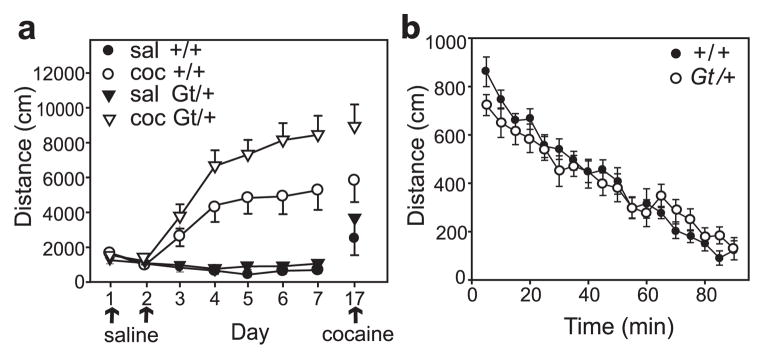
Figure 2. Enhanced cocaine sensitization in Lmo4Gt mice.
(a) Cocaine sensitization in Lmo4Gt/+ mice (Gt/+, triangles) and wild-type littermates (+/+, circles). All mice were injected with saline on days 1 and 2 for habituation to the locomotor chambers. On days 3–7, mice received either saline (filled symbols) or 15 mg/kg cocaine (open symbols), as indicated. On day 17, all mice received a cocaine challenge (15 mg/kg) to test for differences between acute and sensitized responses. Shown is the distance traveled in centimeters in 15 min. (b) Baseline locomotor behavior in Lmo4Gt/+ mice (open circles) and their wild-type littermates (closed circles). Mice were placed in locomotor chambers and tracked for 1 h, followed by a saline injection at 60 min and tracked for an additional 30 min. Each data point represents distance traveled in centimeters in 5 min, n = 13 – 16. All data are presented as mean ± SEM.
Statistical analysis
For qPCR experiments, we compared Gapdh-normalized mRNA levels between shScr- and shLmo4.3-infected Acb (LCM experiments) using a Student’s t-test. To assess cocaine sensitization data, three-way analysis of variance (ANOVA) (genotype × drug × day) and two-way repeated measures ANOVA (genotype × day within each drug treatment) statistical tests were used.
Results
Characterization of Lmo4Gt mice
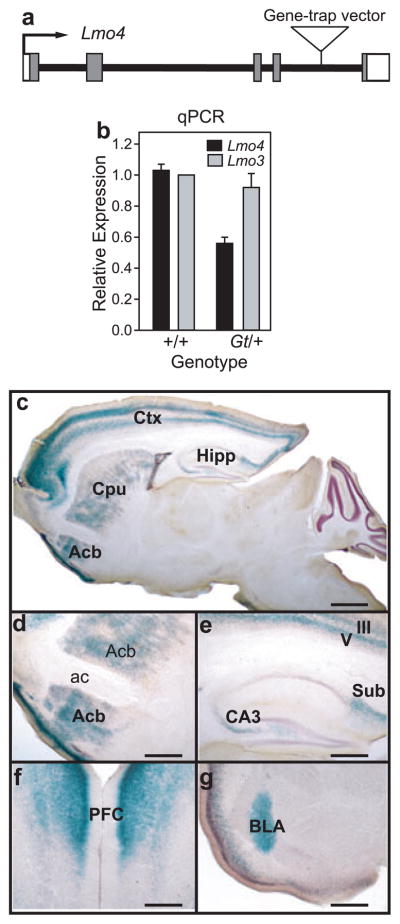
Previous studies from our laboratory indicated that levels of Drosophila dLmo modulate acute behavioral responses to cocaine (Tsai et al. 2004). We therefore examined whether mammalian LMOs affect cocaine sensitivity. Lmo4 was chosen based on its expression in brain regions involved in addiction-related behaviors (Bulchand et al. 2003; Hermanson et al. 1999). We generated a mouse strain, Lmo4Gt, from embryonic stem cells (line RR0142, http://baygenomics.ucsf.edu/) carrying a gene-trap insertion in the fourth intron of Lmo4 (Fig. 1a). Lmo4Gt/+ heterozygous mice express ~50% of wild-type Lmo4 transcript levels in the brain as determined by quantitative RT-PCR (qPCR), whereas expression of the closely related Lmo gene Lmo3 is unaffected (Fig. 1b). Lmo4Gt/+ mice are viable and fertile with no obvious behavioral defects. These data, together with our inability to recover homozygous Lmo4Gt mice, indicate that we have generated a null (or strongly hypomorphic) allele of Lmo4; null mutations in Lmo4 have been reported to die during development because of a failure in neural tube closure (Hahm et al. 2004; Lee et al. 2005; Tse et al. 2004). The Lmo4Gt line produces an LMO4–β-Galactosidase fusion protein, the expression of which is regulated by endogenous Lmo4 cis-regulatory elements. Intense expression of β-galactosidase in adult Lmo4Gt/+ mice was restricted to specific brain regions, including cortex, basolateral amygdala, Cpu, Acb and subregions of the hippocampus (CA3 and subiculum) (Fig. 1c–g), all brain regions that have been implicated in drug addiction (Nestler 2001).
Figure 1. Gene-trap insertion in Lmo4.
(a) Schematic representation of the mouse Lmo4 gene and position of the gene-trap vector insertion. Boxes show the location of the five exons, shaded boxes represent LMO4 protein-coding regions and the arrow indicates the direction of transcription. (b) Reduced Lmo4 expression in the brains of Lmo4Gt/+ mice. cDNA was synthesized from whole brains of three wild-type (+/+) or two Lmo4Gt/+(Gt/+) mice and Lmo4 levels were determined by qPCR. (c) Sagittal section from the brain of a 4-week-old male Lmo4Gt/+ mouse stained with X-Gal (blue). Intense β-galactosidase expression is observed in neocortex (Ctx; layers III and V), hippocampus (Hipp; CA3 regions and subiculum), Acb and Cpu; expression in most other brain regions is very low or not detectable. (d, e) Higher magnification view of β-galactosidase expression in the Acb (d) and Hipp (e). (f, g) Coronal sections showing expression in the prefrontal cortex (PFC, f) and basolateral amygdala (BLA, g). Scale bars: (c) 1 mm; (d–g) 500 μm. ac = anterior commissure.
Lmo4Gt mice exhibit enhanced responses to chronic cocaine
We next assessed the locomotor responses of Lmo4Gt/+ mice upon cocaine exposure. Previously, we found modest enhancements in the acute locomotor-stimulant response and stereotypes to a low dose of cocaine (5 mg/kg) in Lmo4Gt/+ mice compared with their wild-type littermates (Heberlein et al. 2009). Repeated cocaine exposure leads to sensitization of behavioral responsiveness, a long-lasting form of behavioral plasticity that has been proposed to model addictive liability (Robinson & Berridge 1993; Robinson & Kolb 2004). Lmo4Gt/+ mice were subjected to a sensitization protocol involving daily injections of 15 mg/kg of cocaine or saline as a control (Fig. 2a). To show that cocaine significantly increased locomotion in the sensitization protocol, data from days 3–7 (saline and cocaine) were analyzed by three-way ANOVA for genotype × drug × day. We found a significant effect of genotype (F1,210 = 14.62, P < 0.001), drug (F1,210 = 184.7, P < 0.001) and day (F4,210 = 3.15, P = 0.015) and significant genotype × drug (F1,210 = 9.35, P = 0.003) and drug × day interactions (F4,210 = 3.5, P = 0.009). These results show that cocaine significantly increases locomotor activity compared with saline, that the genotypes differ in their drug responses and that the locomotor response is dependent on the number of drug injections over the time course. We next performed a more detailed analysis of the saline (days 1–7) and cocaine (days 3–17) data separately by two-way repeated measures ANOVA. For the saline injections, there was no significant effect of genotype (F1,17 = 0.40, P = 0.53), but a significant effect of day (F6,102 = 9.77, P < 0.001) and a genotype × day interaction (F6,102 = 2.72, P = 0.017). For the cocaine treatments, Lmo4Gt/+ mice showed a significantly increased locomotor response to repeated 15 mg/kg doses of cocaine (genotype: F1,25 = 4.31, P = 0.048; day: F5,125 = 20.4, P < 0.001; genotype × day interaction: F5,125 = 1.54, P = 0.182). To independently examine the expression of cocaine sensitization 10 days after the last sensitizing drug treatment, mice from the saline group were given an acute cocaine injection on day 17 and compared with mice treated on day 17 which had previously received cocaine during the sensitization protocol. Lmo4Gt/+ mice showed a trend toward enhanced sensitivity on day 17 by two-way ANOVA (genotype: F1,42 = 3.48, P = 0.069; treatment: F1,42 = 14.0, P < 0.001; genotype × treatment interaction: F1,42 = 0.77, P = 0.386). Taken together, these data show that global reduction in Lmo4 levels leads to increased sensitivity to cocaine and continued potentiation of the plastic changes underlying drug sensitization. Lmo4Gt/+ and control mice displayed similar levels of habituation to the locomotor chambers and response to a saline injection (Fig. 2b; genotype: F1,27 = 0.327, P = 0.572; time: F15,405 = 42.43, P < 0.001; genotype × time interaction: F15,405 = 1.26, P = 0.224), indicating that the mutant mice exhibit normal activity in the absence of cocaine. In summary, we show that LMO4 negatively regulates behavioral responsiveness to cocaine in mice, an effect that parallels previous findings with the Drosophila dLmo gene (Tsai et al. 2004).
RNAi targeting Lmo4 in Acb medium spiny neurons
The Acb is a key brain region involved in the psychomotor stimulant and rewarding effects of cocaine (Vanderschuren & Kalivas 2000) and a site of high Lmo4 expression in the adult mouse (Fig. 1d) (Hermanson et al. 1999). To test if Lmo4 expression in Acb regulates behavioral responses to cocaine, we utilized RNA interference to locally down-regulate Lmo4 mRNA levels in adult mice (Hommel et al. 2003). Three shRNAs were designed to target distinct and unique sequences in the Lmo4 mRNA (Fig. 3a) and cloned into a GFP-expressing lentiviral vector (Rubinson et al. 2003) for in vivo delivery. A virus expressing a scrambled shRNA (shScr), which should not target any gene encoded in the mouse genome, was generated as a negative control. Plasmids expressing all three Lmo4 shRNAs were efficient in downregulating endogenous Lmo4 levels when transfected into cultured Neuro-2a cells, as measured by qPCR (Fig. 3b). shLmo4.3 was chosen for in vivo experiments as it was most effective. To test knockdown of Lmo4 transcript in the brain, shScr or shLmo4.3 viruses were injected bilaterally into the Acb of wild-type mice. Twelve days after virus injection, infected Acb cells, visualized by GFP fluorescence, were isolated by laser-capture microdissection. qPCR of amplified mRNA isolated from infected cells showed that Lmo4 mRNA was reduced by 56% in cells infected with the shLmo4.3 virus compared with cells infected with control shScr virus (Fig. 3c). We also examined LMO4 protein expression in Acb neurons using immunohistochemistry. Nuclear LMO4 staining was observed in medium spiny neurons in the Acb, but not in astrocytes (Fig. 3d–g). We determined that LMO4 protein levels, as measured by immunohistochemistry, were reduced in Acb 1 month after infection with the shLmo4.3 virus (Fig. 3g,h), indicating that the shRNA is functional in vivo and that the effect persists for a long time period.
Figure 3. Lmo4 RNAi in cell culture and mouse Acb.
(a) Schematic representation of the Lmo4 transcript. The open box indicates the protein-coding region and the solid vertical lines the position of 19-nucleotide sequences used to design shRNAs that specifically target Lmo4. (b) qPCR of Lmo4 cDNA from Neuro-2a cells transfected with plasmids expressing one of three Lmo4 shRNAs (shLmo4.1–4.3) or a scrambled control shRNA (shScr). Total RNA was isolated 48 h after transfection. Data are presented as mean ± SD. (c) qPCR of cDNA from mouse Acb infected with lentivirus expressing shLmo4.3 or shScr. Infected cells were laser-capture microdissected from brain sections taken 12 days after stereotaxic injection of virus into Acb. Data are presented as mean ± SEM, n = 5 per group. (d–h) Infection of medium spiny neurons in the Acb with lentiviruses. (d) Partial three-dimensional reconstruction of a representative medium spiny neuron (MSN) in Acb that was infected with shScr virus (green). Boxed region shows an enlarged view of a dendrite. (e) Orthogonal plane of the same neuron. (f) MSN expressing nuclear LMO4 (red) as shown on a single optic section. (f′) The same cell, infected with shScr virus. (f″) Colocalization of LMO4 and GFP (yellow), as indicated by arrow. (g, h) Confocal images of Acb sections taken 1 month after stereotaxic injection of lentivirus expressing shScr (g) or shLmo4.3 (h). Colocalization of LMO4 and GFP staining is shown in yellow. Note lack of yellow staining in GFP-positive cells in (h) indicating reduced expression of LMO4. LMO4 expression is restricted to neurons (arrows); astrocytes (asterisk) do not express LMO4.
Lmo4 RNAi in Acb enhances sensitivity to cocaine
The effect of Lmo4 downregulation on cocaine-induced locomotor behaviors was analyzed in mice injected with shLmo4.3 lentivirus bilaterally into Acb; mice with bilateral shScr virus injections were used as controls. Nine days after surgery, mice were tested for cocaine-induced locomotor activity using the same sensitization protocol described for Lmo4Gt/+ mice (Fig. 4a). Three-way ANOVA was used to analyze days 3–7 of saline and cocaine treatments (shRNA × drug × day) and indicated a significant effect of shRNA (F1,155 = 5.87, P = 0.017), drug (F1,155 = 193.22, P < 0.001) and a trend toward an effect of day (F4,155 = 2.13, P = 0.08). A focused analysis of the saline data (days 1–7) showed no effect of shRNA (F1,7 = 1.11, P = 0.327), an effect of day (F6,42 = 15.6, P < 0.001) and no shRNA × day interaction (F6,42 = 1.09, P = 0.386). For the cocaine data, there was a statistically significant effect of shRNA (F1,24 = 5.082, P = 0.034) and day (F5,120 = 17.79, P < 0.001), but no significant interaction of shRNA × day (F5,120 = 0.690, P = 0.632). Taken together, these data indicate that mice with local downregulation of Lmo4 in the Acb exhibit enhanced sensitivity to cocaine throughout the sensitization protocol and a normal response to saline. We next examined the expression of sensitization by injecting all mice with cocaine on day 17. Animals that were previously injected with saline during the sensitization protocol were compared with mice previously treated with cocaine. We found no effect of shRNA (F1,31 = 1.02, P = 0.320) but an effect of prior treatment (F1,31 = 17.79, P < 0.001) and no shRNA × treatment interaction (F1,31 = 2.5, P = 0.124). The accuracy and degree of infection were analyzed after the completion of behavioral tests by immunohistochemistry with an antibody recognizing virally encoded GFP. Viral infection, spreading in general ~500 μm from the site of injection, was still visible in Acb 26 days after virus injection (Fig. 4b, Fig. S1). As a control, shScr and shLmo4.3 viruses were injected bilaterally into Cpu (where Lmo4 is also expressed at high levels; Fig. 1c) and mice subjected to the cocaine sensitization protocol. No differences in cocaine sensitization between shScr-and shLmo4.3-injected mice were observed (Fig. 4c). Thus, downregulation of Lmo4 in the Acb of adult mice, using a shRNA delivered by lentiviral infection, resulted in enhanced locomotor responses to repeated cocaine. This effect is qualitatively similar to what was observed upon global reduction in Lmo4 (as in Lmo4Gt/+mice). In summary, our data show that LMO4 functions continuously in the mature nervous system to negatively regulate the behavioral responses to cocaine administration and identifies the Acb as an important site of LMO4 function.
Figure 4. Acb injections of Lmo4 shRNA enhance locomotor responses to cocaine.
(a) Cocaine sensitization in mice expressing shLmo4.3 (triangles) or shScr (circles) in the Acb. Mice received either cocaine (open symbols) or saline (closed symbols) on days 3–7; all animals received cocaine on day 17. (b) Lentivirus infection in Acb as visualized by immunohistochemistry with an anti-GFP antibody (brown). Sections were also stained with cresyl violet to show cytoarchitecture. Infection generally spread 0.5 mm from the injection site. Lower panel shows a higher magnification view of GFP-positive neurons near the edge of the infection site. ac = anterior commissure; LV = lateral ventricle (c) Cocaine sensitization in mice expressing shLmo4.3 (open circles) and shScr (closed circles) in Cpu. No differences were observed between mice expressing shLmo4.3 or shScr.
Discussion
Lmo4 has been characterized primarily for its role in breast cancer and neural tube development (Hahm et al. 2004; Sum et al. 2005; Visvader et al. 2001) and shown to interact, chiefly in epithelial cells, with a number of transcriptional regulators (Manetopoulos et al. 2003; Sugihara et al. 1998; Sum et al. 2002). We provide evidence that Lmo4 functions in the adult brain, specifically the Acb, to regulate long-lasting behavioral sensitization obtained upon repeated cocaine administration.
The functional and structural neuroadaptations underlying sensitization to the psychostimulant effects of abused drugs have been proposed to contribute to addictive behaviors (Kalivas et al. 1998; Robinson & Berridge 2000). Chronic drug exposure leads to molecular and morphological alteration of the Acb (Robinson & Kolb 2004; Thomas et al. 2001; Wolf 1998), resulting, at least in part, from drug-induced changes in gene expression. LMO4, through its ability to modulate activity of LIM homeodomain and other transcription factors, is well suited to regulate such events. Indeed, both the levels and activity of Lmo4 are dynamically regulated in the nervous system. For example, Lmo4 expression is under circadian regulation (Panda et al. 2002) and induced by sleep deprivation (Cirelli & Tononi 2000). In addition, LMO4 was recently identified as a calcium-regulated transcriptional activator (Aizawa et al. 2004) required for proper patterning of somatosensory barrels in the mouse cortex (Kashani et al. 2006). We did not observe changes in the levels of Lmo4 in striatum upon acute or chronic cocaine exposure (data not shown), suggesting that the function rather than the levels of LMO4 may be regulated by cocaine. Because LMO4 activity is regulated by calcium (Aizawa et al. 2004; Kashani et al. 2006) and dopamine affects intracellular calcium levels (Bergson et al. 2003), it is tempting to speculate that cocaine-induced changes in dopamine (and calcium) levels may affect LMO4 activity, which in turn would produce transcriptional changes regulating drug-induced plasticity. These molecular events would act normally to curb the development of the sensitized state, because loss of Lmo4 function leads to an enhanced response to repeated cocaine exposure. A potential partner for LMO4 in such process may be the transcription factor CREB, which has been shown recently to interact physically with LMO4 in cultured cells (Kashani et al. 2006). The role of CREB in the biochemical and behavioral responses to abused drugs, particularly psychostimulants, has been well documented (Carlezon et al. 2005; Levine et al. 2005). It is therefore possible that the activity of CREB may be modulated by LMO4, an event that would, in turn, affect cocaine-induced neural plasticity in the striatum. Future studies will examine the transcriptional targets and binding partners of LMO4 that mediate behavioral responses to cocaine.
Supplementary Material
Figure S1: Schematic representation of sequential coronal brain sections showing the main site of infection in Acb for mice that were used in the behavioral experiments in Fig. 4a. Red dots illustrate infection foci, shaded gray areas the Acb core. Numbers show the distance in mm anterior to bregma.
Table S1: Primer and shRNA sequences.
Acknowledgments
We thank Lorien Hollfelder and Stacy Taylor for technical assistance. This work was supported by funds provided to UH by the State of California for medical research on alcohol and substance abuse through the University of California at San Francisco, the Sandler Family Foundation and NIH (DA14809).
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Amara SG, Kuhar MJ. Neurotransmitter transporters: recent progress. Annu Rev Neurosci. 1993;16:73–93. doi: 10.1146/annurev.ne.16.030193.000445. [DOI] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Bergson C, Levenson R, Goldman-Rakic PS, Lidow MS. Dopamine receptor-interacting proteins: the Ca(2+) connection in dopamine signaling. Trends Pharmacol Sci. 2003;24:486–492. doi: 10.1016/S0165-6147(03)00232-3. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–321. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH. Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol Cell Biol. 2004;24:2074–2082. doi: 10.1128/MCB.24.5.2074-2082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U, Tsai LT, Kapfhamer D, Lasek AW. Drosophila, a genetic model system to study cocaine-related behaviors: a review with focus on LIM-only proteins. Neuropharmacology. 2009;56 (Suppl 1):97–106. doi: 10.1016/j.neuropharm.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O, Sugihara TM, Andersen B. Expression of LMO-4 in the central nervous system of the embryonic and adult mouse. Cell Mol Biol (Noisyle-grand) 1999;45:677–686. [PubMed] [Google Scholar]
- Hogan B. Manipulating the Mouse Embryo: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Plainview, NY: 1994. [Google Scholar]
- Hommel JD, Sears RM, Georgescu D, Simmons DL, DiLeone RJ. Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med. 2003;9:1539–1544. doi: 10.1038/nm964. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry. 2005;162:1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Pierce RC, Cornish J, Sorg BA. A role for sensitization in craving and relapse in cocaine addiction. J Psychopharmacol. 1998;12:49–53. doi: 10.1177/026988119801200107. [DOI] [PubMed] [Google Scholar]
- Kashani AH, Qiu Z, Jurata L, Lee SK, Pfaff S, Goebbels S, Nave KA, Ghosh A. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–8408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Jr, Whisler K, Gilden L, Beckmann AM, Steffen C, Zhang YJ, Marotti L, Self DW, Tkatch T, Baranauskas G, Surmeier DJ, Neve RL, Duman RS, Picciotto MR, Nestler EJ. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Janak PH, He L, Whistler JL, Heberlein U. Downregulation of mu opioid receptor by RNA interference in the ventral tegmental area reduces ethanol consumption in mice. Genes Brain Behav. 2007;6:728–735. doi: 10.1111/j.1601-183X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Nowak R, Lettieri K, Kenny DA, Pfaff SL, Gill GN. The LIM domain-only protein LMO4 is required for neural tube closure. Mol Cell Neurosci. 2005;28:205–214. doi: 10.1016/j.mcn.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci U S A. 2005;102:19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetopoulos C, Hansson A, Karlsson J, Jonsson JI, Axelson H. The LIM-only protein LMO4 modulates the transcriptional activity of HEN1. Biochem Biophys Res Commun. 2003;307:891–899. doi: 10.1016/s0006-291x(03)01298-1. [DOI] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW. DeltaFosB: a sustained molecular switch for addiction. Proc Natl Acad Sci U S A. 2001;98:11042–11046. doi: 10.1073/pnas.191352698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2. Academic Press; San Diego, CA: 2001. [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, III, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Res Brain Res Rev. 1993;18:247–291. doi: 10.1016/0165-0173(93)90013-p. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. The psychology and neurobiology of addiction: an incentive-sensitization view. Addiction. 2000;95(Suppl 2):S91–S117. doi: 10.1080/09652140050111681. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. Incentive-sensitization and addiction. Addiction. 2001;96:103–114. doi: 10.1046/j.1360-0443.2001.9611038.x. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47(Suppl 1):33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Sugihara TM, Bach I, Kioussi C, Rosenfeld MG, Andersen B. Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc Natl Acad Sci U S A. 1998;95:15418–15423. doi: 10.1073/pnas.95.26.15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sum EY, Peng B, Yu X, Chen J, Byrne J, Lindeman GJ, Visvader JE. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. doi: 10.1074/jbc.M110603200. [DOI] [PubMed] [Google Scholar]
- Sum EY, Segara D, Duscio B, Bath ML, Field AS, Sutherland RL, Lindeman GJ, Visvader JE. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci U S A. 2005;102:7659–7664. doi: 10.1073/pnas.0502990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MJ, Beurrier C, Bonci A, Malenka RC. Long-term depression in the nucleus accumbens: a neural correlate of behavioral sensitization to cocaine. Nat Neurosci. 2001;4:1217–1223. doi: 10.1038/nn757. [DOI] [PubMed] [Google Scholar]
- Tsai LT, Bainton RJ, Blau J, Heberlein U. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2004;2:e408. doi: 10.1371/journal.pbio.0020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH, Boehm T, Rabbitts TH. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 2004;24:2063–2073. doi: 10.1128/MCB.24.5.2063-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Hall FS, Sora I. Cocaine, reward, movement and monoamine transporters. Mol Psychiatry. 2002;7:21–26. doi: 10.1038/sj.mp.4000964. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, Kalivas PW. Alterations in dopaminergic and glutamatergic transmission in the induction and expression of behavioral sensitization: a critical review of preclinical studies. Psychopharmacology (Berl) 2000;151:99–120. doi: 10.1007/s002130000493. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O’Reilly L, White D, Williams R, Armes J, Lindeman GJ. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FJ, Kalivas PW. Neuroadaptations involved in amphetamine and cocaine addiction. Drug Alcohol Depend. 1998;51:141–153. doi: 10.1016/s0376-8716(98)00072-6. [DOI] [PubMed] [Google Scholar]
- Wolf ME. The role of excitatory amino acids in behavioral sensitization to psychomotor stimulants. Prog Neurobiol. 1998;54:679–720. doi: 10.1016/s0301-0082(97)00090-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Schematic representation of sequential coronal brain sections showing the main site of infection in Acb for mice that were used in the behavioral experiments in Fig. 4a. Red dots illustrate infection foci, shaded gray areas the Acb core. Numbers show the distance in mm anterior to bregma.
Table S1: Primer and shRNA sequences.