Abstract
This review details the molecular virology of the hepatitis E virus (HEV). While replicons and in vitro infection systems have recently become available, a lot of information on HEV has been generated through comparisons with better-studied positive-strand RNA viruses and through subgenomic expression of viral open reading frames. These models are now being verified with replicon and infection systems. We provide here the current knowledge on the HEV genome and its constituent proteins – ORF1, ORF2 and ORF3. Based on the available information, we also modify the existing model of the HEV life cycle.
Keywords: HEV, ORF1, ORF2, ORF3, RNA replication
1. Introduction
Hepatitis E virus (HEV) is the causative agent of hepatitis E, a form of acute viral hepatitis that is endemic to many resource-limited regions of the world (Emerson and Purcell, 2006). It is estimated that about 2 billion people, which is about a third of the world population, live in areas endemic for HEV and are at risk for infection. In these areas, hepatitis E is the major form of acute hepatitis; in India for example about 50% of acute hepatitis is due to HEV. It results in morbidity and increased mortality during pregnancy; these clinical aspects of hepatitis E are covered in detail in another review in this issue (R. Aggarwal).
The recognition that hepatitis E was caused by a distinct agent dates back to the 1970s from investigations undertaken by Khuroo and co-workers who studied jaundice outbreaks in Kashmir, India, and Purcell and co-workers who retrospectively studied the 1955–56 outbreak of jaundice in New Delhi, India. The history of hepatitis E and HEV is also covered in greater detail in another review in this issue (M.S. Khuroo). The discovery of viruses similar to HEV in pigs (swine HEV), chickens (avian HEV) and more recently in rabbits and rodents, as well as the successful transmission of swine HEV to macaques, which are a model for human HEV transmission, led to the idea of HEV as a zoonosis. This belief is further strengthened by studies from Japan that have used nucleotide sequence identity to link viruses recovered from uncooked meat (pigs and deer) and humans who became infected after consuming the meat. The discovery and analysis of animal HEVs (X.J. Meng) and the unique transmission situations in Japan (T. Miyamura) are also covered separately in this issue.
HEV is a small non-enveloped virus with a size of 27–34 nm and is classified as a Hepevirus in the family Hepeviridae (Emerson and Purcell, 2006). So far only one serotype of HEV has been identified, but there are four phylogenetically distinct genotypes, each dominant in a given geographic area. Genotype 1 includes strains from Asia and Africa, genotype 2 includes the Mexican strain and a few variants from Africa, and genotypes 3 and 4 include human and swine HEV strains from industrialized countries and Asia (particularly China), respectively. While genotypes 1 and 2 have only been found in humans, genotypes 3 and 4 have been recovered from humans as well as pigs and other animal species. Genotype 3 is evenly distributed across the world while genotype 4 is found more often in China and Japan.
Early studies on HEV transmission and pathogenesis as well as preclinical vaccine development studies have mostly been carried out in non-human primates such as cynomolgus, rhesus and owl monkeys, and chimpanzees (Uchida et al., 1991, Purdy et al., 1992, Ticehurst et al., 1992, McCaustland et al., 2000). More recently, pigs have also been used for transmission and molecular studies (Meng et al., 1998). However, a small animal model for HEV is still elusive. That and the lack of a suitable in vitro cell culture system have hampered virological studies on HEV. However, cell culture systems based on replicon RNA transfection and more recently those using the virus, have become available. These are covered in greater detail in another review in this issue (Okamoto).
2. The HEV genome
2.1. Cloning and genome organization
The HEV genome was first cloned from cDNA libraries prepared from the bile of macaques experimentally inoculated with stool suspensions from human patients (Reyes et al., 1990, Tam et al., 1991). Similar and polymerase chain reaction based strategies were later used to clone the genomes of multiple geographically distinct isolates of HEV (Huang et al., 1992, Panda et al., 2000, Emerson et al., 2001).
The HEV genome is a single-stranded RNA of ∼7.2 kb that is positive-sense, with a 5′-methylguanine cap and a 3′ poly(A) stretch, and contains three partially overlapping open reading frames (ORFs) – called orf1, orf2 and orf3 (Tam et al., 1991). The viral genome also has short 5′- and 3′-untranslated regions (UTRs) and a conserved 58-nucleotide region within orf1; these elements are likely to fold into conserved stem-loop and hairpin structures. These structures and a sequence closer to the 3′ end of orf1, which has homology to the alphavirus junction region, are proposed to be important for HEV RNA replication (Purdy et al., 1993). The region between the end of orf1 and start of orf3/orf2 appears to be complex and contains regulatory elements. These details are shown in Fig. 1 .
Fig. 1.

The hepatitis E virus genome. The ∼7.2 kb positive sense RNA genome of HEV has a 7-Me-G cap at its 5′ end and a poly A tail at its 3′ end. There are short stretches of untranslated regions at the 5′ and 3′ ends that fold into stem-loop structures (shown in blue). The three open reading frames are shown. ORF1 encodes a nonstructural polyprotein and contains a 58-nucleotide stretch near its 5′ end that folds into a stem-loop structure (shown in green). The ORF2 and ORF3 proteins are translated from a 2.2 kb subgenomic RNA generated during viral replication. The boxed region on the upper right shows the sequence alignment of the junction region between orf1 and orf3 in HEV isolates representative of genotypes 1–4. The nucleotide positions are shown with respect to HEV genotype 1 (Sar55). Dots indicate identity and dashes represent deletions. The orf1 stop codon is shown in red. There is a single nucleotide insertion (T, indicated with filled triangle) between positions 5116 and 5117 in HEV genotype 4. The four initiation codons within this junction region are shown in yellow boxes, and are at positions 5104, 5113, 5131 and 5145. This region has been predicted to fold into a double stem-loop structure shown in the boxed region on the left. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
2.2. Viral RNA species
In the liver tissue of macaques experimentally infected with HEV, Tam et al. (1991) detected three RNA species of ∼7.2, 3.7 and 2 kb, which were designated as the genomic and two subgenomic RNAs, respectively. In this model, the orf1 stop codon at position 5105 (nucleotide position according to the genotype 1 SAR-55 strain) overlaps with the orf3 start codon at position 5104. Two in-frame AUG codons at positions 5113 and 5131 were considered to code for methionine residues in the ORF3 protein. These are followed by another AUG codon in the −1 frame, which was proposed to be the orf2 start codon. Thus, in this model, the 3.7 and 2 kb subgenomic RNAs would be used to translate the ORF3 and ORF2 proteins, respectively.
Graff et al. (2006) have challenged this model. In stable Huh-7 cell lines made from functional HEV RNA replicons expressing the neomycin resistance gene from orf2 and orf3, the genomic RNA and only one subgenomic RNA were observed. The capped 2.2 kb subgenomic RNA initiated at nucleotide position 5122, which was downstream of the first two methionine codons in orf3, and was bicistronic, for the translation of ORF3 and ORF2 (Fig. 1). This model also rationalizes the reading frame differences observed in genotype 4 HEV isolates, which contain a T base insertion between nucleotides 5116/5117 (in SAR-55), thus taking AUG3 in a different translation frame. Translation of the ORF3 protein from AUG3 starting at nucleotide 5131 (in the SAR-55 strain) would be the same in all four genotypes of HEV. This model was confirmed by intrahepatic inoculation of wild type and mutant genotype 3 swine HEV replicons into pigs (Huang et al., 2007). Mutation of AUG1 or the insertion of a T base as in genotype 4 did not affect virus infectivity or rescue, but the mutation of AUG3 abolished virus infectivity.
The support for a single subgenomic RNA model also comes from PLC/PRF/5 cells that were either transfected with infectious genotype 3 RNA produced in vitro (from a cloned cDNA) or inoculated with fecal suspension containing genotype 4 HEV. The RNA isolated from these cells only showed the 2.2 kb subgenomic species, whose 5′ end mapped to nucleotide 5122 (Ichiyama et al., 2009).
A conserved double stem-loop RNA structure is predicted in the junction region (Huang et al., 2007). Its role in viral replication was recently determined in Huh-7 cells transfected with wild type or stem-loop mutant replicons containing reporter genes (Cao et al., 2010). The viral negative-strand RNA is proposed to be a template for the synthesis of positive-strand genomic and subgenomic RNAs, the latter from within the junction region in a primer-independent manner. The junction region negative-strand RNA is predicted to fold into a stable stem-loop structure. Mutation of the AGA motif on the loop or sequences that are part of the stem or the subgenomic RNA start site significantly reduced or abolished reporter activity. Thus, the sequence of the junction region as well as the stem-loop structure within it, play important roles in HEV replication.
2.3. Genetic variants
Comparative analyses of the nucleotide sequences of complete genomes of HEV isolates has revealed extensive genomic diversity leading to the identification of four major genotypes and several subtypes within each genotype (Lu et al., 2006). Genomic regions that have been used for phylogenetic purposes include a 301 nt sequence at the 5′ end of the orf2 region and a 306 nt sequence in the 3′ end of orf1, within the RdRp region.
A virus with about 50% nucleotide sequence identity with mammalian HEVs, called avian HEV, was isolated from chickens affected by the hepatitis-splenomegaly syndrome (Haqshenas et al., 2001, Huang et al., 2004). Though initially considered as HEV genotype 5, avian HEV is now proposed as a new species within the family Hepeviridae (International Committee for Taxonomy of Viruses; Ninth Report). A unique strain of HEV was identified from farmed rabbits in China, which shared 74–79% nucleotide sequence identity to existing HEV strains, and 46% identity to avian HEV (Zhao et al., 2009). The rabbit HEV appears to be a distant variant of HEV genotype 3. Two additional unique HEV sequences were isolated recently – one from rats (Johne et al., 2010) and the other from a wild boar (Takahashi et al., 2011).
Recombination in HEV genomes was recently reported (Wang et al., 2010). Phylogenetic and recombination analyses on 134 complete HEV genomes showed three potentially significant intra- and inter-genotype recombination events. These results suggest that recombination is rare but possible between human and swine HEV strains, and might also contribute to viral diversity.
3. HEV proteins
Three viral proteins – called the ORF1, ORF2 and ORF3 proteins are expressed from the viral genome. While the kinetics of expression of these proteins during the viral life cycle is not fully understood, their expression during HEV infection is confirmed by the presence of antibodies to these proteins in infected humans and experimental animals (Khudyakov et al., 1994, Panda et al., 1995).
3.1. The ORF1 protein
The orf1 of HEV encodes a protein of 1693 amino acids, which was proposed to encode the viral nonstructural functions (Koonin et al., 1992). This proposal was based on the detection of several functional motifs and domains present in the nonstructural proteins of other positive-stranded RNA viruses. The functional domains identified in the HEV nonstructural polyprotein include (starting from the N-terminal end) – methyltransferase (MeT), papain-like cysteine protease (PCP), RNA helicase (Hel) and RNA dependent RNA polymerase (RdRp). Besides these, some uncharacterized domains also found that are homologous to other animal and plant positive-strand RNA viruses. The ‘Y’ domain spans ∼200 amino acids downstream of the MeT domain and includes a subsequence that scored high on alignments with the nonstructural proteins of Rubella virus (RubV) and beet necrotic yellow vein virus (BNYVV). A conserved domain, earlier called the ‘X’ domain, but now known as the ‘macro’ domain flanks the PCP domains in the nonstructural polyproteins of many positive-stranded animal viruses. This domain in the HEV ORF1 polyprotein shows significant homology with similar domains in RubV, BNYVV, alphaviruses and some coronaviruses. In HEV and RubV a proline-rich region that might be a flexible hinge between the ‘X’ domain and upstream domains precedes the ‘X’ domain (Fig. 2 ). This early bioinformatic analysis of the HEV nonstructural protein (Koonin et al., 1992, Koonin and Dolja, 1993, Liu et al., 2009) proposed many putative biochemical activities, a few of which have now been characterized in greater detail (discussed below). It also suggested HEV, RubV and BNYVV to form a distinct monophyletic group within the alpha-like supergroup of positive-strand RNA viruses, but this proposal has not passed the test of time and subsequent investigations.
Fig. 2.

The ORF1 protein. The orf1 gene encodes a nonstructural polyprotein with four predicted functional domains, designated as methyltransferase (MeT), papain-like cysteine protease (PCP), helicase (Hel) and RNA-dependent RNA polymerase (RdRp). Besides these regions, two other regions designated X (macro domain) and Y share significant homology with nonstructural proteins of other positive-strand RNA viruses. A proline-rich region (V) upstream of the macro domain may act as a flexible hinge. Numbers indicate the predicted boundaries of the different regions; these are based on the genotype 1 Burmese isolate.
It is not entirely clear whether the ORF1 polyprotein is processed into biochemically distinct units, as is the case with other positive-strand RNA viruses. When expressed in mammalian cells using recombinant vaccinia viruses, ORF1 yielded processed products of 78 and 107 kDa (Ropp et al., 2000), whereas its expression in Escherichia coli or HepG2 hepatoma cells showed no processing (Ansari et al., 2000). However, in HepG2 cells transfected with HEV genomic RNA produced in vitro from a cDNA, metabolic labeling and immunoprecipitation showed distinct processed products. Anti-MeT, anti-helicase and anti-RdRp antibodies detected specific products of ∼35 kDa, ∼38 kDa and ∼36 kDa, respectively (Panda et al., 2000). Similarly, when a His6-ORF1-FLAG fusion protein was expressed in insect cells using recombinant baculoviruses, multiple processed fragments could be detected with anti-hexahistidine or anti-FLAG tag antibodies (Sehgal et al., 2006). Of these, a 35-kDa N-terminal fragment was characterized by mass spectrometry to be the methyltransferase protein. In the same study, E-64d, a cell-permeable cysteine protease inhibitor blocked ORF1 processing, but it could not be established whether the processing protease was of cellular or viral origin. So far no viral infection study has addressed expression and processing of the ORF1-derived nonstructural polyprotein.
3.1.1. Methyltransferase
Computer-assisted alignments of the ORF1 polyprotein suggested that the amino terminal residues 60–240 might serve as the viral methyltransferase (Koonin et al., 1992). The ORF1 sequence also suggested that HEV might belong to the large group of alphavirus-like superfamily of positive-strand RNA viruses. Members of this virus superfamily code a distinctive methyltransferase enzyme (Rozanov et al., 1992). Viral methyltransferases catalyze the capping of viral RNA, and since the HEV genomic and subgenomic RNAs are capped, a methyltransferase was expected to be encoded on the HEV genome. Accordingly, Magden et al. (2001) expressed a HEV cDNA fragment encoding amino acids 1–979 in insect cells using the baculovirus expression system. A 110-kDa protein (P110) expressed in this system was shown to contain guanine-7-methyltransferase as well as guanyltransferase activities.
The capping of mRNA is well characterized in mammalian, yeast and several viral systems, in which it follows a common mechanism (Ahola and Kaariainen, 1995).
-
1.
pppN1pN2pN3- - - - - → ppN1pN2pN3- - - - - + Pi
-
2.
GTP + enzyme → enzyme-GMP + PPi
-
3.
enzyme-GMP + ppN1pN2pN3- - - - - → GpppN1pN2pN3- - - - - + enzyme
-
4.
GpppN1pN2pN3- - - - - + AdoMet → m7GpppN1pN2pN3- - - - - + AdoHcy
In step 1 RNA triphosphatase removes the 5′ gamma-phosphate from the nascent RNA molecules. In steps 2 and 3 the mRNA gyanyltransferase donates a GMP moiety to this RNA to form a 5′-5′-triphosphate linkage in the cap structure. Finally, in step 4 the cap is methylated using the guanine-7-methyltransferase activity and S-adenosylmethionine (AdoMet) as the donor to give the 7-methylguanosine capped RNA and S-adenosylhomocysteine (AdoHcy).
While the HEV methyltransferase showed guanine-7-methyltransferase and guanyltransferase activities (Magden et al., 2001), the source of the RNA triphosphatase (step 1) was not clear. A recent report (Karpe and Lole, 2010b) suggests that this activity may reside in the HEV helicase. When a purified recombinant HEV helicase protein was incubated with either alpha-32P-RNA or gamma-32P-RNA, it removed 32P only from the latter, suggesting that it possesses a gamma-phosphatase activity, which might catalyze the first step in RNA cap formation.
Two reports have shown the presence of a 5′ m7G cap on the HEV genomic RNA. The HEV genomic RNA transcribed in vitro from viral cDNA is infectious for primates only when it is capped (Emerson et al., 2001). A 5′ RNA ligase-mediated rapid amplification of cDNA ends (RACE) method designed to select capped RNAs amplified the 5′ ends of the SAR-55 (genotype 1) and MEX-14 (genotype 2), confirming the HEV genomic RNA to be capped (Zhang et al., 2001a, Zhang et al., 2001b).
Why is it important for viral RNAs to be capped? Interferons provide the most important innate defense against viruses and viral double-stranded RNA (dsRNA) was found to be a potent trigger of the interferon response (Colby and Morgan, 1971). More recently, the 5′ triphosphate (5′ppp) group on an RNA molecule was also shown to be a potent activator of the interferon response and could be an additional or alternative trigger to dsRNA (Pichlmair et al., 2006). RNA polymerase-directed RNA synthesis initiates with a 5′ppp nucleotide and thus all RNA molecules initially carry an exposed 5′ppp moiety, which is removed by processing. Thus, capping of the viral RNA, besides aiding its translation through recognition by the ribosome, would also be important in protecting the virus from innate host responses.
3.1.2. Protease
The computer-assisted assignment of functional domains also predicted the presence of a papain-like cysteine protease (PCP) in ORF1 with moderate similarity to the Rubella virus protease (Koonin et al., 1992). Though the alignment score was low, the prediction was strengthened by conservation of the ‘X’ domain in HEV, which in other systems was found exclusively in association with the PCP. This alignment placed the putative PCP domain in the region of amino acids 433–592 of the ORF1 polyprotein. However, no functional activity for the HEV protease has so far been described. As mentioned above, extended incubations of the ORF1 polyprotein expressed using a recombinant vaccinia virus yielded products of 107 kDa and 78 kDa (Ropp et al., 2000). But, mutagenesis of Cys483 predicted by sequence alignments to be part of the catalytic dyad, had no effect on the generation of these products. Further, the other member of the proposed catalytic dyad, His590 is missing from the putative HEV protease. The presence of a functional protease within ORF1 and its role in polyprotein processing thus remains an unanswered question.
3.1.3. Helicase
Many positive-stranded RNA viruses encode a RNA helicase, which is essential for replication of the genomes of these viruses (Kadare and Haenni, 1997). Helicases are divided into six superfamilies (SF-1 to SF-6) and further into subfamilies depending upon whether they unwind in the 3′ → 5′ (A) or 5′ → 3′ (B) direction. The SF-1 and SF-2 families of helicases are the most common and each has seven signature motifs that are involved in the binding and hydrolysis of nucleotide triphosphates (NTPs), and binding of the nucleic acids (DNA/RNA). The putative RNA helicase of HEV was shown to contain all the seven conserved motifs found in SF-1 helicases and proposed to have the NTPase as well as RNA-binding domains (Koonin et al., 1992). Experimental evidence for this was provided recently. Karpe and Lole (2010b) expressed the region corresponding to ORF1 amino acids 960–1204 in E. coli, and the purified recombinant protein showed both NTPase and RNA unwinding activities. The enzyme hydrolyzed all rNTPs but also dNTPs with a lower efficiency, and showed unwinding activity only on RNA duplexes with 5′ overhangs. Mutants in the nucleotide-binding motif I (GKS → GAS) or the Mg2+-binding motif II (DEAP → AAAP) showed reduced ATPase activity and had no RNA unwinding activity, indicating generation of energy as a critical requirement for helicase activity. The HEV helicase belongs to the 5′ → 3′ class of SF-1 family of helicases that are also found in other positive-strand RNA viruses such as alphaviruses, arteriviruses, coronaviruses and rubiviruses.
The HEV helicase also possesses a RNA 5′-triphosphatase activity, which is proposed to aid the HEV methyltransferase by catalyzing the first step of RNA capping (Karpe and Lole, 2010a).
3.1.4. RNA dependent RNA polymerase
The RdRp is an essential enzyme found in all RNA viruses; it is used to replicate the genomic RNA through an anti-genomic RNA intermediate. Comparison of the amino acid sequences of HEV and other positive-strand RNA viruses showed a strong homology of the ORF1 region encompassing amino acids 1207–1693 with the RdRp proteins of RubV and BNYVV (Koonin et al., 1992). The enzymes of these three viruses form a compact group within the “alpha-like” supergroup (III) of positive-strand RNA viruses. All eight conserved motifs, I-VIII described for positive-strand RNA virus RdRp proteins (Koonin, 1991) are also found in the HEV protein, including the GDD sequence that binds Mg2+, which is essential for replicase activity.
The RdRp region of HEV comprising nucleotides 3546–5106 was expressed as a 59 kDa His-tagged protein in E. coli (Agrawal et al., 2001). Using gel shift assays, this purified recombinant protein was shown to bind the 3′ end of HEV genomic RNA. Two stem-loop structures in the 3′ end of the HEV RNA as well as the poly(A) stretch were found to be required for this binding. Further, the recombinant RdRp was also able to use the 3′ end of the HEV RNA as a template to synthesize the complementary strand in vitro. Since anti-RdRp antibodies were earlier shown to immunoprecipitate an ∼37 kDa protein from cells transfected with an infectious replicon, a later study stably expressed an ∼40 kDa C-terminal region of the ORF1 polyprotein as a fusion with the enhanced green fluorescent protein (EGFP) (Rehman et al., 2008). This 68-kDa replicase-EGFP fusion protein localized to the endoplasmic reticulum (ER) and was a functional replicase due to its ability to copy HEV plus-strand RNA transfected into these cells. These studies also suggest that intracellular (ER) membranes are the likely sites for HEV replicase localization and possibly RNA replication too, as is the case with several other RNA viruses.
3.1.5. Macro domain
Macro domains are found in a variety of proteins of bacteria, archae and eukaryotes. These are domains with homology to the non-histone domain of the histone macroH2A (Pehrson and Fried, 1992). Their association with proteins involved in poly(ADP-ribose) polymerization, ADP-ribosylation and ATP-dependent chromatin remodeling, suggest that macro domains contribute to ADP-ribose metabolism and post-translational modifications (Aguiar et al., 2005). A few viral families, which include all members of Coronaviridae, rubella virus and alphaviruses (Togaviridae) and HEV, also encode macro domains (Snijder et al., 2003). All these viruses replicate their RNA in the cytoplasm of animal cells in membrane-associated replication complexes (Salonen et al., 2005). In these viruses, macro domains are closely associated with the typical replicase complexes that include RNA helicase and RdRp, and unlike some of their eukaryotic homologs, viral macro domain proteins do not enter the nucleus.
The first biochemical activity demonstrated in a macro domain protein was the hydrolysis of ADP-ribose 1″-phosphate (ADPR-1″P), a product of cellular pre-tRNA splicing (Snijder et al., 2003). Their ability to bind ADP-ribose and the association with poly(ADP-ribose) polymerase-1 (PARP-1) has also led to the suggestion that macro domain proteins might regulate apoptosis in eukaryotic cells. Though not experimentally demonstrated thus far, this property would be relevant for the viral macro domain proteins since viruses show broad effects on the apoptosis of infected cells.
Two reports describe the macro (or X) domain in HEV (Egloff et al., 2006, Neuvonen and Ahola, 2009). A region of the HEV ORF1 polyprotein including residues 775–960 (Burmese strain) was expressed with an N-terminal hexahistidine tag and purified from E. coli. This protein was found to contain ADPR-1″ phosphatase activity and bound in vitro to poly(ADP-ribose) and poly(A) (Egloff et al., 2006, Neuvonen and Ahola, 2009). While viral macro domain proteins from SARS coronavirus, SFV and HEV bound poly(ADP-ribose) with high affinity, they showed either poor binding (HEV) or no binding (SFV) to ADP-ribose; the reverse was observed with human macro domain proteins (Neuvonen and Ahola, 2009).
What might be the function of viral macro domains? These are present only in a small subset of RNA viruses that replicate in the cytosol, and form an essential part of the viral genome. Egloff et al. (2006) suggested that viral macro domains function as poly(ADP-ribose)-binding modules. Based on their in vitro binding and competition experiments, Neuvonen and Ahola (2009) suggest that the viral macro domains could have a role in viral RNA replication and/or transcription. Since the HEV and SFV macro domains can also bind poly(ADP-ribose) in the presence of poly(A), they could recruit poly(ADP-ribose)-modified cellular factors to the replication complex while bound to viral polyadenylated RNA. These possibilities await experimental confirmation.
3.2. The ORF2 protein
3.2.1. Expression and glycosylation
The ORF2 of HEV encodes the viral capsid protein of 660 amino acids (nt 5145–7125; SAR-55 isolate; Fig. 1), which was proposed to encapsidate the viral RNA genome (Purdy et al., 1993). When expressed in animal cells in culture, ORF2 proteins of ∼74 kDa and ∼88 kDa were observed, which were proven with tunicamycin and endoglysidase treatment studies to be the nonglycosylated and glycosylated forms, respectively (Jameel et al., 1996). Pulse-chase studies and the use of microsomal membranes showed the ORF2 protein to contain a N-terminal signal sequence that translocates the protein into the endoplasmic reticulum, where it acquires N-linked glycosylation (Jameel et al., 1996). The glycosylation sites on the ORF2 protein were mapped to conserved asparagine (Asn) residues 137, 310 and 562 (Zafrullah et al., 1999) (Fig. 1). Though the earlier studies were based on ectopic expression of the orf2 gene in cultured cells, replicon-based expression of the ORF2 protein has subsequently confirmed its N-linked glycosylation (Graff et al., 2008). Further, mutations in the glycosylation sites on the ORF2 protein prevented the formation of infectious virus particles from transfected replicons, and these particles had low infectivity in macaques (Graff et al., 2008). Recently, Yamada et al. (2009) constructed an infectious clone of HEV that propagates efficiently in cultured PLC/PRF/5 cells. Using this model they show the intracellular expression and secretion of an 83 kDa ORF2 protein. Though these authors did not directly test glycosylation of the ORF2 protein, a size significantly larger than the predicted size of ∼72 kDa, suggests this possibility. These observations are not in agreement with an earlier suggestion that the glycosylated form of the ORF2 protein is unstable and only the non-glycosylated protein forms the viral capsid (Torresi et al., 1999). Various observations suggest a role for cellular membranes in HEV replication and particle maturation. In such a scenario, glycosylation of the ORF2 capsid protein would be of advantage.
3.2.2. Self-assembly and virus-like particles
When expressed in insect cells using recombinant baculovirus, a N-terminal truncated ORF2 protein (aa 112–660) gave the expected product of 58 kDa, but also a 50 kDa protein that was efficiently secreted in the culture medium as virus-like particles of 23–24 nm diameter (Li et al., 1997). The full-length ORF2 protein when similarly expressed was processed into 63 kDa, 56 kDa and 53 kDa proteins, which correspond to truncated proteins spanning amino acid residues 112–660, 112–607 and 112–578, respectively (Robinson et al., 1998). Cryoelectron microscopy and image reconstruction methods showed the VLPs to have a T = 1 structure with protruding dimers at the icosahedral twofold symmetry axes such that the 60 monomers were organized as 30 morphological units (Xing et al., 1999). Subsequently, amino acids 126–601 in the ORF2 protein were found to be essential for the formation of secreted VLPs in this expression system (Li et al., 2005). The X-ray crystal structures obtained recently show the ORF2 protein monomers to contain three distinct domains – the shell (S), middle (M) and protruding (P) domains (Yamashita et al., 2009, Guu et al., 2009). Recently, Xing et al. (2010) have produced a large virion-sized and a small T = 1 particle by expressing the ORF2 protein in insect cells with or without the N-terminal 111 amino acids, respectively. The virion-sized particle showed a T = 3 icosahedral lattice and contained a ∼2 kb RNA derived from the orf2 gene; the T = 1 particle showed no encapsidated RNA. Though these data suggest that RNA encapsidation drives virion assembly, the presence of subgenomic RNA in the T = 3 particle is surprising, since RNA viruses package only the genomic RNA species. Further, a potential packaging signal was identified using a yeast three-hybrid approach towards the 5′ end of the genome, within the orf1 gene (Surjit et al., 2004). More structural details on the ORF2 capsid can be found in another article in this issue (Mori and Matsuura).
The recombinant ORF2 proteins expressed in E. coli have also been shown to assemble into higher order structures. The p239 (aa 368–606), E2 (aa 394–606) and E2a (459–660) proteins (Fig. 3 ) predominantly occur as homodimers under mildly dissociating conditions, which model the dominant antigenic determinants and neutralizing site of HEV (Zhang et al., 2001a, Zhang et al., 2001b, Li et al., 2005a). Under native conditions, the E2 and E2a proteins form hexamers, and the p239 protein forms a particle of 23 nm (Li et al., 2005a). Comparison of these three proteins and mutational analysis showed the dimeric interface to include a cluster of six hydrophobic residues (aa 597–602) and the domain involved in particle formation (aa 430–458) to also include a high proportion of hydrophobic residues (Li et al., 2005b). The crystal structure of the E2s protein (aa 455–602) shows it to form a tight homodimer that is essential for HEV interaction with the host cell, and contains the HEV neutralizing antibody recognition site (Li et al., 2009).
Fig. 3.

The ORF2 protein. The orf2 gene encodes the HEV capsid protein. The full-length protein is 660 amino acids long. In animal cells, pORF2 is glycosylated and three N-linked glycosylation sites have been mapped to the indicated asparagine (Asn) residues. A truncated protein of 56 kDa (amino acids 112–607) can self-assemble in insect cells to form virus-like particles (VLPs), which possess the same dominant antigenic epitopes as the virion. Various shorter constructs have been expressed in E. coli. Of these, p239 forms particles via dimeric interactions while E2 and E2a form hexameric surface protrusions. The E2s is proposed as the dimerization domain. Two of these proteins (yellow highlight) have undergone clinical testing in humans as HEV vaccine candidates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2.3. Neutralization epitopes and HEV vaccines
Various studies have looked at the neutralizing domains of the ORF2 capsid protein. Using phage display methods, Schofield et al. (2000) identified two neutralizing monoclonal antibodies from chimpanzees that had recovered from experimental HEV infection, and these mapped to overlapping epitopes in the region of amino acids 578–607. Meng et al. (2001) generated antibodies to multiple overlapping peptides and truncated ORF2 proteins and tested these in an in vitro cell culture infectivity assay to locate the minimal neutralizing domain to amino acids 452–617. The minimal neutralizing domain has now been mapped to residues 458–607 (Zhou et al., 2004). When combined with structural details, all of these mapping studies place the neutralizing epitope(s) in the protruding (P) domain of the ORF2 protein (Yamashita et al., 2009, Guu et al., 2009, Xing et al., 2011).
The immunological and structural studies on the ORF2 protein provide a basis for HEV vaccine development. Two recombinant vaccines against HEV have undergone successful clinical testing in humans (Shrestha et al., 2007, Zhu et al., 2010). These are based on recombinant ORF2 proteins either expressed as T = 1 VLPs in insect cells (Shrestha et al., 2007) or as p239 particles in E. coli (Zhu et al., 2010). Further details on these and other recombinant vaccine approaches can be found in another review in this issue (Kamili). The structural studies also provide a basis for using the ORF2 particles as an epitope presentation system (Xing et al., 2011), which when combined with the ability of these VLPs to render successful oral immunization (Li et al., 2004), can provide a powerful vaccine platform.
3.2.4. Interactions with target cells
The biochemical and structural characterization of the ORF2 capsid protein also offers an opportunity to study the interactions of HEV with its target cells. The purified p239 protein, which structurally and antigenically resembles the HEV capsomere, bound and penetrated different cell lines susceptible in vitro to HEV infection (He et al., 2008). In pull-down experiments, p239 showed binding to Grp78/BiP, alpha-tubulin and heat shock protein 90 (HSP90), and inhibitor studies showed HSP90 to be important for the intracellular trafficking of p239 (Zheng et al., 2010). The ORF2 VLPs produced in insect cells were also shown to bind Huh7 liver cells and this binding was dependent upon heparan surface proteoglycans (HSPGs) present on cell surface Syndecans (Kalia et al., 2009). Analogous to binding studies with p239 (He et al., 2008), our studies show that the VLPs also bind to cell lines susceptible to HEV infection; these are further taken up into Huh7 cells through fluid endosomes (M. Kalia, R. Holla, S. Jameel; unpublished results). More studies along these lines are likely to identify the cellular receptor for HEV and characterize the virus uptake and uncoating pathway in infected cells.
Viral capsid proteins are involved in extensive interactions with cellular proteins for the purpose of capsid assembly and virus egress. For non-enveloped viruses, the capsid protein interactions are also important for virus entry, intracellular trafficking and signaling (Tsai, 2007, Kalia and Jameel, 2009). Very little is known about the interactions of the ORF2 protein with cellular components and pathways, besides that with HSP90 (He et al., 2008) and HSPGs (Kalia et al., 2009) summarized above. The ORF2 protein expressed in animal cells was observed to localize to the ER, where it causes ER stress and some of the protein is retrotranslocated into the cytoplasm through a canonical ER-associated degradation (ERAD) pathway (Surjit et al., 2007). Whether these effects of the ORF2 protein have a bearing on HEV assembly and egress or on virus entry and intracellular trafficking remains to be seen.
3.3. The ORF3 protein
3.3.1. Expression and subcellular localization
The orf3 of HEV was initially predicted to express a protein of 123 amino acids. It was recently suggested that the ORF3 protein is translated from a bicistronic subgenomic RNA from an AUG codon at position 5131 (in SAR-55 isolate), which would result in a 114 amino acid protein; that would be 9 amino acids shorter at its N-terminus than the earlier proposal (Graff et al., 2006). Using a replicon system based on genomic RNA produced in vitro, orf3 was found to be dispensable for replication in vitro in Huh7 and other cell lines (Emerson et al., 2006). However, it was required for infection in monkeys that received intrahepatic inoculation of HEV genomic RNA (Graff et al., 2005). These observations suggest that the ORF3 protein functions as a viral accessory protein, and is likely to affect the host response to infection.
The ORF3 protein has been expressed using various systems that include E. coli (Panda et al., 1995), the yeast Pichia pastoris (Lal et al., 1997) and mammalian cell lines (Jameel et al., 1996). In mammalian cells orf3 expressed a protein of ∼13 kDa, which was phosphorylated at a single Serine residue by cellular mitogen-activated protein kinase (MAPK) (Zafrullah et al., 1997). Cells transfected with HEV genomic RNA prepared in vitro were also found to express a protein of the same size, which could be immunoprecipitated with anti-ORF3 antibodies (Panda et al., 2000, Graff et al., 2005, Emerson et al., 2006).
Subcellular fractionation showed the ORF3 protein to associate with the cytoskeleton (Zafrullah et al., 1997). Confocal microscopy has revealed its punctate and filamentous intracellular distribution; the latter is attributed to its interaction with microtubules (Kannan et al., 2009). The punctate distribution of the ORF3 protein localized it to early and recycling endosomes (Chandra et al., 2008a).
3.3.2. Protein domains and effects on cellular functions
Primary sequence analysis of the ORF3 protein does not show domains homologous to any other protein or any other distinguishing features, except the proline-rich nature of the protein. It contains two large N-terminal hydrophobic domains – D1 (aa 7–23) and D2 (aa 28–53), and two proline-rich domains – P1 (aa 66–77) and P2 (aa 95–111) (Fig. 4 ).
Fig. 4.
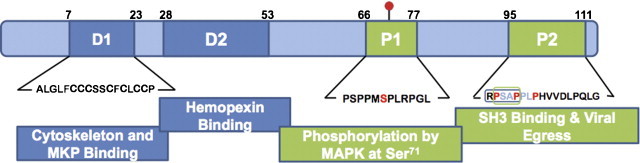
The ORF3 protein. The orf3 gene encodes a 114-amino acid multi-functional protein, which has two hydrophobic domains (D1 and D2) and two proline rich domains (P1 and P2). In mammalian cells, the ORF3 protein is phosphorylated at Serine 71 (red dot) by cellular MAPK. The proline-rich P2 domain contains a PXXP motif (RPSAP, in box) that is implicated in binding to SH3 domains in cellular proteins. An intact PSAP motif is also required for viral egress. Regions of the ORF3 protein experimentally shown to be involved in various interactions are indicated. Numbers represent the proposed boundaries of the various domains. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Of these, D1 is extremely rich in cysteine residues and is required for the ORF3 protein to associate with the cytoskeleton (Zafrullah et al., 1997), to bind microtubules (Kannan et al., 2009) and a MAPK phosphatase (Kar-Roy et al., 2004); however, none of these activities have been linked conclusively to one or more cysteine residues present in this domain. Domain P1 contains two overlapping motifs that have homology to kinase substrates – PMSP that is phosphorylated by MAPK and SPLR that has a loose homology to the SPKK motif that is phosphorylated by cdc2 kinase. Of these, Ser-80 (now Ser-71 in the 114 aa ORF3 protein) is phosphorylated by MAPK, both in transfected cells as well as in vitro (Zafrullah et al., 1997); whether the ORF3 protein expressed during viral infection is also phosphorylated has not been tested. The P2 domain has two overlapping PXXP motifs, which have been described in many viral and cellular proteins involved in signal transduction, and bind the Src homology 3 (SH3) domains found in other signal-transducing molecules (Pawson, 1995). Indeed, this domain was shown to be responsible for the interaction of the ORF3 protein with many cellular SH3 domain-containing proteins (Korkaya et al., 2001).
Such a domain structure and the ability of the ORF3 protein to interact with multiple cellular proteins suggested a potential role for it in optimizing the cellular environment for viral infection and replication. Various studies have been carried out in this direction, but most of these are based on ORF3 over-expression in hepatic cell lines and remain to be confirmed in an efficient viral infection system. Nevertheless, these studies provide some insights into the possible multi-pronged functions of this HEV protein.
The ORF3 protein was found to activate the extracellularly regulated kinase (Erk), a member of the MAPK family of signal transducing molecules (Korkaya et al., 2001). This Erk activation was independent of the traditional Raf/MEK pathway, but depended upon the ability of the ORF3 protein to bind and inactivate Pyst1, a prototypic member of the Erk-specific MAPK phosphatase (Kar-Roy et al., 2004). By binding to the central linker region of Pyst1 through its N-terminal D1 domain, the ORF3 protein was proposed to block conformational changes in the phosphatase, which are required for its Erk-mediated activation (Kar-Roy et al., 2004). Prolonged Erk activation was proposed to generate a survival and proliferative signal. Cells expressing the ORF3 protein also displayed higher levels of hexokinase and oligomeric forms of the voltage-dependent anion channel (VDAC), which lead to attenuation of mitochondrial death signaling (Moin et al., 2007). Subcellular localization of the ORF3 protein to early and recycling endosomes delayed post-internalization trafficking of the activated epidermal growth factor receptor (EGFR) (Chandra et al., 2008a). This was recently found to be true for the hepatocyte growth factor receptor (c-Met) as well, and depended upon the ability of the ORF3 protein to interact with CIN85, a multidomain adaptor protein implicated in the downregulation of receptor tyrosine kinases (Chandra et al., 2010). In the suggested model, the ORF3 protein binds to CIN85, possibly through the proline-rich P2 domain in the former and SH3 domains in the latter. This competes with formation of the growth factor receptor–Cbl–CIN85 complex, resulting in reduced trafficking of activated growth factors towards the late endosomal/lysosomal compartment where the receptor is degraded (Chandra et al., 2010). Such a scenario would prolong endomembrane growth factor signaling and again promote cell survival and proliferation.
Growth factor receptor endocytosis and intracellular trafficking are also required for the nucleo-cytoplasmic transfer of phosphorylated forms of the signal transducer and activator of transcription 3 (STAT3) protein (Bild et al., 2002). As a consequence of its effects on EGFR trafficking, reduced levels of phospho-STAT3 (pSTAT3) are found in the nuclei of ORF3-expressing cells (Chandra et al., 2008a). This leads to decreased transcription of genes involved in the acute phase response, a major determinant of inflammation in the host. Again, this would create an environment conducive to viral replication. A proteomic study recently showed reduced levels of a number of acute phase proteins, such as serum amyloid P precursor, haptoglobin, hemopexin, alpha-1-acid glycoprotein, glycosylated fibrinogen, reinol binding protein, transthyretin and albumin, in the plasma of hepatitis E patients compared to healthy controls (Taneja et al., 2009, Taneja et al., 2011).
In a yeast two-hybrid screen for ORF3-interacting cellular proteins, the alpha-1-microglobulin and bikunin precursor protein (AMBP) and its constituents, alpha-1-microglobulin and bikunin (Tyagi et al., 2004, Tyagi et al., 2005) were identified. Increased secretion of alpha-1-microglobulin was also observed from ORF3-expressing cells (Surjit et al., 2006). This was mediated by the tumor susceptibility gene 101 (Tsg101) protein, a member of the endosomal-sorting complex, which bound the ORF3 protein through a conserved PSAP motif in its P2 domain. Since alpha-1-microglobulin is an immunosuppressive protein, this was also proposed to protect virus-infected cells in the liver (Surjit et al., 2006). In support of these in vitro results, alpha-1-microglobulin levels were significantly higher in the urine of hepatitis E patients compared to acute hepatitis B patients and healthy controls (Taneja et al., 2009, Taneja et al., 2011).
Fibrinogen beta chain and hemopexin were also identified from the yeast two-hybrid screen. Its interaction with the ORF3 protein led to reduced fibrinogen beta secretion from cells; the transcription of alpha, beta and gamma fibrinogen genes, was also reduced in ORF3-expressing cells (Ratra et al., 2009). This is likely to be due to the effects of the ORF3 protein on intracellular trafficking since fibrinogen is also an acute phase protein whose expression is induced by interleukin-6 (IL-6) through the STAT3 transcription factor. Hemopexin is a heme-binding 60 kDa acute phase plasma glycoprotein, which protects cells from hemoglobin-mediated oxidative damage during intravascular hemolysis. It also plays an important role in iron metabolism, and together with transferrin and haptoglobin is responsible for iron homeostasis. The ORF3 protein binds hemopexin through its D2 domain, and this is proposed to aid in viral infection by affecting cellular iron homeostasis (Ratra et al., 2008). Reduced levels of hemopexin and haptoglobin have also been observed in the plasma of hepatitis E patients compared to healthy controls (Taneja et al., 2009, Taneja et al., 2011).
A proteomic and transcriptional study of cells stably expressing the ORF3 protein revealed increased expression of many glycolytic enzymes (Moin et al., 2009). This was regulated by increased levels of the hypoxia inducible factor-1 (HIF-1) transcription factor, which in turn was brought about by increased stability of the alpha subunit of HIF-1 in ORF3-expressing cells (Moin et al., 2009). The HIF-1 complex recruits phosphorylated p300/CBP to target gene promoters and the ORF3 protein increased p300/CBP phosphorylation through Erk activation. Thus, the ORF3 protein appears to use a dual strategy to modulate energy homoestasis.
Two broad roles were predicted for the ORF3 protein in HEV pathogenesis (Chandra et al., 2008b). It promotes cell survival and proliferation through Erk activation, extended endomembrane signaling from activated growth factor receptors, and attenuation of mitochondrial death signals. The ORF3 protein also dampens innate host responses through an attenuated acute phase response and increased secretion of immunosuppressive factors such as alpha-1-microglobulin. A third possible role of regulating cellular energy homeostasis can be added to these. It must again be emphasized that almost all of these results are based on over-expression of the ORF3 protein in cell lines. They await confirmation in virus infection systems.
3.3.3. Role in virion morphogenesis and release
Some recent studies also suggest a role for the ORF3 protein in virus egress and its association with virions. An ORF3-deficient mutant of the infectious cDNA clone pJE03-1760F/wt replicated efficiently in PLC/PRF/5 and A549 cells, but compared to the wild type clone produced less than 1% detectable virus in the culture supernatant (Yamada et al., 2009). Using an immunocapture PCR assay, the ORF3 protein was found on the surface of cell culture generated HEV, which also showed a lower density than the ORF3-deficient virus. These observations indicate that the ORF3 protein is important for HEV egress and that it is present on the virion surface, possibly in association with lipids (Yamada et al., 2009). An earlier study had shown that anti-ORF3 antibodies could capture HEV particles from the serum, but not the feces of hepatitis E patients (Takahashi et al., 2008). In this study as well, the serum HEV banded at a lower density compared to fecal HEV, suggesting that the former is associated with lipids, and that this coat is possibly shed during its passage through the enteric system.
A genotype 1 HEV was similarly found to replicate in Caco-2 intestinal and Huh-7 hepatoma cells and its egress from these cells depended on the presence of a functional ORF3 protein (Emerson et al., 2010). In the polarized Caco-2 cells, the ORF3 protein accumulated at the apical membrane and virus egress also took place from this surface. The PXXP motif in the P2 domain of the ORF3 protein was found to be important for virus egress, but not for its accumulation at the apical end. One of the mutant viruses showed no infection following intrahepatic inoculation in macaques, but another was infectious and the recovered virus was found to have completely reverted to a wild type ORF3 sequence. These results support previous observations of the ORF3 requirement for infection (Graff et al., 2005). In this study also, the ORF3 protein, in association with lipids, was found to coat the virion surface.
A recent study shows the requirement of an intact PSAP motif within the P2 domain for the formation of membrane associated HEV particles with the ORF3 protein on their surface (Nagashima et al., 2011). Though neither this nor the Emerson et al. (2010) study implicated the role of any host protein in ORF3-mediated HEV release, the requirement of the PSAP motif suggests a possible role for Tsg101, which was earlier shown to bind the ORF3 protein (Surjit et al., 2006). Several RNA viruses bear proline-rich sequences in their ‘late domains’, which are required for viral budding. These include P(S/T)AP and PPXY (X, any amino acid) motifs, which hijack host proteins in the vacuolar protein sorting (VPS) pathway. This pathway gives rise to multivesicular bodies (MVBs), which is topologically identical to virus budding (Chen and Lamb, 2008). Viral ‘late domains’ interact with members of the VPS pathway, and redirect the complexes to the site of virus budding on the plasma membrane (Chen and Lamb, 2008). Though HEV is a non-enveloped virus, its association with lipids, subcellular localization of the ORF3 protein to endosomes (Chandra et al., 2008a) and a requirement for its PSAP motif in viral egress suggests that HEV follows the VPS pathway for its release from infected cells (Fig. 5 ).
Fig. 5.

Proposed replication cycle of hepatitis E virus. The viral particles are concentrated on the surface of target cells through heparin sulfate proteoglycans acting as attachment factors (red wavy lines) (step 1) and subsequently bind a specific yet uncharacterized receptor (step 2), following which the particles are internalized (step 3). The virus then uncoats (step 4) to release genomic RNA (red noodles) that is translated in the cytoplasm into nonstructural proteins (step 5). These nonstructural proteins include the RNA dependent RNA polymerase that replicates the positive sense genomic RNA into negative sense transcripts (purple noodles) (step 6); the latter then act as templates for the synthesis of a 2.2 kb subgenomic RNA (step 7a) as well as full-length positive sense transcripts (step 7b). The positive sense subgenomic RNA is translated into ORF2 (blue) and ORF3 (crimson) proteins (step 8). The ORF2 protein packages the genomic RNA to assemble new virions (step 9) while the ORF3 protein may optimize the host cell environment for viral replication. The ORF3 protein is also associated with endomembranes (step 10a) or plasma membranes (step 10b) and may aid in viral egress. Recent studies suggest that mature virions are associated with the ORF3 protein and lipids (step 11), which are subsequently removed through a process that is not understood at present, to resume a fresh infection cycle. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
4. HEV life cycle and genome replication
The life cycle of HEV is poorly understood, largely because of the non-availability of efficient in vitro culture methods or small animal models of infection. The original view of the HEV life cycle (Reyes et al., 1993), which was based primarily on its genome analysis and analogy to other positive-strand RNA viruses, is still broadly relevant. However, based on in vitro expression and replicon studies, some details have now begun to emerge. The basic model and additions to it are shown in Fig. 5.
Various transformed cell lines are permissive for HEV infection and virion production. These include hepatic cell lines such as Huh7, PLC/PRF/5 and HepG2, the lung carcinoma A549 cell line and the colon carcinoma Caco-2 cell line. The binding and entry of HEV is poorly understood. No cellular receptor for HEV has been identified, but HSPGs appear to be required as attachment factors (Kalia et al., 2009). The intracellular trafficking following entry is also poorly understood; HSP90 and tubulin appear to be involved in this process (Zheng et al., 2010). The location and mechanism of uncoating to release viral RNA is also not known.
Once viral RNA is released in the cytosol it is translated directly into the ORF1 polyprotein, as is the case with all positive-sense RNA viruses. It is not clear whether the polyprotein is processed into individual functional units (see Section 3.1). However, the regions predicted to encode viral methyltransferase, helicase and replicase produce functionally active proteins when expressed in heterologous systems (see Sections 3.1.1, 3.1.2, 3.1.3, 3.1.4). The genomic RNA would replicate into negative-sense RNA intermediates, which have been detected in replicon-transfected cells (Panda et al., 2000) as well as in the livers of experimentally infected macaques (Nanda et al., 1994) and pigs (Meng et al., 1998). These would serve as templates for the synthesis of genomic as well as subgenomic positive-sense RNAs, of which the latter are translated into the ORF2 (capsid) and ORF3 proteins (Graff et al., 2006, Huang et al., 2007, Ichiyama et al., 2009). The ORF2 protein packages the genomic positive-sense RNA into progeny virions (see Section 3.2.2), and recent evidence suggests that the ORF3 protein together with lipids coats this particle, possibly during the budding process (see Section 3.3.3).
It is believed that the primary site of HEV replication is the liver, with hepatocytes being the most likely cell type. However, in vitro results also support infection and replication in non-hepatic cell types such as A549 lung carcinoma cells and in Caco-2 colon carcinoma cells. In pigs experimentally infected with swine HEV, while positive-sense viral RNA was detected in almost all tissues at some point during the infection, negative-sense replicative RNA intermediates were detected primarily in the small intestine, lymph node, colon and liver (Williams et al., 2001). In a recent report from hepatitis E patients, HEV RNA was detected in peripheral blood mononuclear cells, but there was no evidence for viral replication in this compartment (Ippagunta et al., 2010).
5. Potential targets and antiviral agents
Various steps in the HEV life cycle can be potential targets for the development of antiviral drugs. The methyltransferase and guanyltransferase activities in the ORF1 protein (see Section 3.1.1) are strictly virus-specific and thus good targets for antiviral development (Magden et al., 2001). The RNA helicase of HEV has been biochemically characterized and is essential for replication of the viral RNA genome (Karpe and Lole, 2010a), but it is not clear how distinct it is from human helicases to be a potential drug target. This would have to await elucidation of its structural details. The HEV RdRp expressed in E. coli was shown to bind the 3′ end of the viral RNA genome (Agrawal et al., 2001), but its biochemical activity has so far not been characterized. Since the RdRp is unique to RNA viruses, it would again be a good drug target, and perhaps pan-viral inhibitors can be explored against this target.
Interference with HEV RNA replication has been attempted using ribozymes and small interfering RNAs. Mono- and di-hammerhead ribozymes designed against the 3′ end of the HEV genomic RNA were shown to inhibit expression from a reporter construct in HepG2 cells (Sriram et al., 2003). In A549 cells infected with HEV, small interfering RNAs (siRNAs) against the orf2 region were shown to offer protection (Huang et al., 2010). While such approaches are feasible in vitro, the delivery and targeting of such inhibitors in vivo would be the real challenge. At least one study in immunocompromised transplant patients with chronic HEV infection has also shown the efficacy of Ribavarin monotherapy (Kamar et al., 2010). Again, the utility of this approach among the vast majority of HEV infections that are acute remains questionable.
6. Summary
Since the discovery of HEV in the early 1970s and the molecular cloning of its genome two decades later, research on the molecular virology of HEV has been slow due to the lack of in vitro culture systems or small animal models of viral infection. Much of the early information and models of the HEV life cycle and genome replication were based on comparative information from other better-characterized positive-strand RNA viruses. Viral ORFs have been expressed in various heterologous systems and the proteins characterized biochemically. With the recent availability of replicon- and infection-based in vitro cell culture models, the molecular virology of HEV is becoming increasingly clear. While there are a few surprises, such as the association of lipids with a non-enveloped virus, many earlier observations have also been confirmed in recent experiments.
This review summarizes the current knowledge in the area and presents a modified view of the HEV life cycle.
Acknowledgements
The HEV work from the authors’ laboratory has been funded by grants from the Wellcome Trust, UK, National Institutes of Health, USA, Department of Biotechnology, India, and from ICGEB internal resources. All are gratefully acknowledged. Support in the form of Junior Research Fellowships (to IA and RPH) from the Council for Scientific and Industrial Research, India, is also acknowledged.
References
- Agrawal S., Gupta D., Panda S.K. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp) Virology. 2001;282:87–101. doi: 10.1006/viro.2000.0819. [DOI] [PubMed] [Google Scholar]
- Aguiar R.C., Takeyama K., He C., Kreinbrink K., Shipp M.A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- Ahola T., Kaariainen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U.S.A. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari I.H., Nanda S.K., Durgapal H., Agrawal S., Mohanty S.K., Gupta D., Jameel S., Panda S.K. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1) J. Med. Virol. 2000;60:275–283. [PubMed] [Google Scholar]
- Bild A.H., Turkson J., Jove R. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 2002;21:3255–3263. doi: 10.1093/emboj/cdf351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Huang Y.W., Meng X.J. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J. Virol. 2010;84:13040–13044. doi: 10.1128/JVI.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Kalia M., Hajela K., Jameel S. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl–CIN85 complex. J. Virol. 2010;84:3857–3867. doi: 10.1128/JVI.01994-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Kar-Roy A., Kumari S., Mayor S., Jameel S. The hepatitis E virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the acute-phase response. J. Virol. 2008;82:7100–7110. doi: 10.1128/JVI.00403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Taneja S., Kalia M., Jameel S. Molecular biology and pathogenesis of hepatitis E virus. J. Biosci. 2008;33:451–464. doi: 10.1007/s12038-008-0064-1. [DOI] [PubMed] [Google Scholar]
- Chen B.J., Lamb R.A. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT. Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby C., Morgan M.J. Interferon induction and action. Annu. Rev. Microbiol. 1971;25:333–360. doi: 10.1146/annurev.mi.25.100171.002001. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S.U., Nguyen H.T., Torian U., Burke D., Engle R., Purcell R.H. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J. Virol. 2010;84:9059–9069. doi: 10.1128/JVI.00593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S.U., Purcell R.H. Hepatitis E virus. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. 5th edition. Lippincott Williams & Wilkins; Philadelphia: 2006. pp. 3047–3058. [Google Scholar]
- Emerson S.U., Nguyen H., Torian U., Purcell R.H. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 2006;80:10457–10464. doi: 10.1128/JVI.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S.U., Zhang M., Meng X.J., Nguyen H., St. Claire M., Govindarajan S., Huang Y.K., Purcell R.H. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15270–15275. doi: 10.1073/pnas.251555098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Nguyen H., Yu C., Elkins W.R., St. Claire M., Purcell R.H., Emerson S.U. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J. Virol. 2005;79:6680–6689. doi: 10.1128/JVI.79.11.6680-6689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Torian U., Nguyen H., Emerson S.U. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 2006;80:5919–5926. doi: 10.1128/JVI.00046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Zhou Y.H., Torian U., Nguyen H., St. Claire M., Yu C., Purcell R.H., Emerson S.U. Mutations within potential glycosylation sites in the capsid protein of hepatitis E virus prevent the formation of infectious virus particles. J. Virol. 2008;82:1185–1194. doi: 10.1128/JVI.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guu T.S., Liu Z., Ye Q., Mata D.A., Li K., Yin C., Zhang J., Tao Y.J. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqshenas G., Shivaprasad H.L., Woolcock P.R., Read D.H., Meng X.J. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 2001;82:2449–2462. doi: 10.1099/0022-1317-82-10-2449. [DOI] [PubMed] [Google Scholar]
- He S., Miao J., Zheng Z., Wu T., Xie M., Tang M., Zhang J., Ng M.H., Xia N. Putative receptor-binding sites of hepatitis E virus. J. Gen. Virol. 2008;89:245–249. doi: 10.1099/vir.0.83308-0. [DOI] [PubMed] [Google Scholar]
- Huang C.C., Nguyen D., Fernandez J., Yun K.Y., Fry K.E., Bradley D.W., Tam A.W., Reyes G.R. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virol. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- Huang F.F., Sun Z.F., Emerson S.U., Purcell R.H., Shivaprasad H.L., Pierson F.W., Toth T.E., Meng X.J. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J. Gen. Virol. 2004;85:1609–1618. doi: 10.1099/vir.0.79841-0. [DOI] [PubMed] [Google Scholar]
- Huang Y.W., Opriessnig T., Halbur P.G., Meng X.J. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 2007;81:3018–3026. doi: 10.1128/JVI.02259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Zhou J., Yang Z., Cui L., Zhang W., Yuan C., Yang S., Zhu J., Hua X. RNA interference inhibits hepatitis E virus mRNA accumulation and protein synthesis in vitro. Vet. Microbiol. 2010;142:261–267. doi: 10.1016/j.vetmic.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama K., Yamada K., Tanaka T., Nagashima S., Jirintai D., Takahashi M., Okamoto H. Determination of the 50-terminal sequence of subgenomic RNA of hepatitis E virus strains in cultured cells. Arch. Virol. 2009;154:1945–1951. doi: 10.1007/s00705-009-0538-y. [DOI] [PubMed] [Google Scholar]
- Ippagunta, S.K., Naik, S., Jameel, S., Ramana, K.N., Aggarwal, R., 2010. Viral RNA but no evidence of replication can be detected in the peripheral blood mononuclear cells of hepatitis E virus-infected patients. J. Viral Hepat., doi:10.1111/j.1365-2893.2010.01351.x. [DOI] [PMC free article] [PubMed]
- Jameel S., Zafrullah M., Ozdener M.H., Panda S.K. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 1996;70:207–216. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Plenge-Boenig A., Hess M., Ulrich R.G., Rettz J., Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 2010;91:750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- Kadare G., Haenni A.L. Virus-encoded RNA helicases. J. Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia M., Chandra V., Rahman S.A., Sehgal D., Jameel S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 2009;83:12714–12724. doi: 10.1128/JVI.00717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia M., Jameel S. Virus entry paradigms. Amino Acids. 2009 doi: 10.1007/s00726-009-0363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N., Rostaing L., Abravanel F., Garrouste C., Lhomme S., Esposito L., Basse G., Cointault O., Ribes D., Nogier M.B. Ribavirin therapy inhibits viral replication on patients with chronic hepatitis E virus infection. Gastroenterology. 2010;139:1612–1618. doi: 10.1053/j.gastro.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Kannan H., Fan S., Patel D., Bossis I., Zhang Y.J. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J. Virol. 2009;83:6375–6382. doi: 10.1128/JVI.02571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 2010;84:3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J. Virol. 2010;84:9637–9641. doi: 10.1128/JVI.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar-Roy A., Korkaya H., Oberoi R., Lal S.K., Jameel S. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J. Biol. Chem. 2004;279:28345–28357. doi: 10.1074/jbc.M400457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov Y.E., Favorov M.O., Jue D.L., Hine T.K., Fields H.A. Immunodominant antigenic regions in a structural protein of the hepatitis E virus. Virology. 1994;198:390–393. doi: 10.1006/viro.1994.1048. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya A.E., Purdy M.A., Rozanov M.N., Reyes G.R., Bradley D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H., Jameel S., Gupta D., Tyagi S., Kumar R., Zafrullah M., Mazumdar M., Lal S.K. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 2001;276:42389–42400. doi: 10.1074/jbc.M101546200. [DOI] [PubMed] [Google Scholar]
- Lal S.K., Tulasiram P., Jameel S. Expression and characterization of the hepatitis E virus ORF3 protein in the methylotrophic yeast, Pichia pastoris. Gene. 1997;190:63–67. doi: 10.1016/s0378-1119(96)00698-1. [DOI] [PubMed] [Google Scholar]
- Li S., Tang X., Seetharaman J., Yang C., Gu Y., Zhang J., Du H., Shih J.W., Hew C.L., Sivaraman J. Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus–host interaction. PLoS Pathog. 2009;5:e1000537. doi: 10.1371/journal.ppat.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.W., Zhang J., Li Y.M., Ou S.H., Huang G.Y., He Z.Q., Ge S.X., Xian Y.L., Pang S.Q., Ng M.H. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine. 2005;23:2893–2901. doi: 10.1016/j.vaccine.2004.11.064. [DOI] [PubMed] [Google Scholar]
- Li S.W., Zhang J., He Z.Q., Gu Y., Liu R.S., Lin J., Chen Y.X., Ng M.H., Xia N.S. Mutational analysis of essential interactions involved in the assembly of hepatitis E virus capsid. J. Biol. Chem. 2005;280:3400–3406. doi: 10.1074/jbc.M410361200. [DOI] [PubMed] [Google Scholar]
- Li T.C., Suzuki Y., Ami Y., Dhole T.N., Miyamura T., Takeda N. Protection of cynomolgus monkeys against HEV infection by oral administration of recombinant hepatitis E virus-like particles. Vaccine. 2004;22:370–377. doi: 10.1016/j.vaccine.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Li T.C., Takeda N., Miyamura T., Matsuura Y., Wang J.C., Engvall H., Hammar L., Xing L., Cheng R.H. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 2005;79:12999–13006. doi: 10.1128/JVI.79.20.12999-13006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M.A., Uchida T., Takeda N., Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fu Y., Jiang D., Li G., Xie J., Peng Y., Yi X., Ghabrial S.A. A novel mycovirus that is related to the human pathogen hepatitis E virus and rubi-like viruses. J. Virol. 2009;83:1981–1991. doi: 10.1128/JVI.01897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Li C., Hagedorn C.H. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- Magden J., Takeda N., Li T., Auvinen P., Ahola T., Miyamura T., Merits A., Kaariainen L. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 2001;75:6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaustland K.A., Krawczynski K., Ebert J.W., Balayan M.S., Andjaparidze A.G., Spelbring J.E., Cook E.H., Humphrey C. Hepatitis E virus infection in chimpanzees: a retrospective analysis. Arch. Virol. 2000;145:1909–1918. doi: 10.1007/s007050070065. [DOI] [PubMed] [Google Scholar]
- Meng J., Dai X., Chang J.C., Lopareva E., Pillot J., Fields H.A., Khudyakov Y.E. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology. 2001;288:203–211. doi: 10.1006/viro.2001.1093. [DOI] [PubMed] [Google Scholar]
- Meng X.J., Halbur P.G., Haynes J.S., Tsareva T.S., Bruna J.D., Royer R.L., Purcell R.H., Emerson S.U. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch. Virol. 1998;143:1405–1415. doi: 10.1007/s007050050384. [DOI] [PubMed] [Google Scholar]
- Moin S.M., Chandra V., Arya R., Jameel S. The hepatitis E virus ORF3 protein stabilizes HIF-1alpha and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell. Microbiol. 2009;11:1409–1421. doi: 10.1111/j.1462-5822.2009.01340.x. [DOI] [PubMed] [Google Scholar]
- Moin S.M., Panteva M., Jameel S. The hepatitis E virus Orf3 protein protects cells from mitochondrial depolarization and death. J. Biol. Chem. 2007;282:21124–21133. doi: 10.1074/jbc.M701696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S., Takahashi M., Jirintai D., Tanaka T., Yamada K., Nishizawa T., Okamoto H. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J. Gen. Virol. 2011;92:269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- Nanda S.K., Panda S.K., Durgapal H., Jameel S. Detection of the negative strand of hepatitis E virus RNA in the livers of experimentally infected rhesus monkeys: evidence for viral replication. J. Med. Virol. 1994;42:237–240. doi: 10.1002/jmv.1890420306. [DOI] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S.K., Ansari I.H., Durgapal H., Agrawal S., Jameel S. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 2000;74:2430–2437. doi: 10.1128/jvi.74.5.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S.K., Nanda S.K., Zafrullah M., Ansari I.H., Ozdener M.H., Jameel S. An Indian strain of hepatitis E virus (HEV): cloning, sequence, and expression of structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J. Clin. Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Purdy M.A., McCaustland K.A., Krawczynski K., Tam A., Beach M.J., Tassopoulos N.C., Reyes G.R., Bradley D.W. Expression of a hepatitis E virus (HEV) trpE fusion protein containing epitopes recognized by antibodies in sera from human cases and experimentally infected primates. Arch. Virol. 1992;123(335):349. doi: 10.1007/BF01317268. [DOI] [PubMed] [Google Scholar]
- Purdy M., Tam A., Huang C., Yarbough P., Reyes G. Hepatitis E virus: a non-enveloped member of the ‘alpha-like’ RNA virus supergroup. Semin. Virol. 1993;4:319–326. [Google Scholar]
- Ratra R., Kar-Roy A., Lal S.K. The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry. 2008;47:1957–1969. doi: 10.1021/bi7016552. [DOI] [PubMed] [Google Scholar]
- Ratra R., Kar-Roy A., Lal S.K. ORF3 protein of hepatitis E virus interacts with the Bβ chain of fibrinogen resulting in decreased fibrinogen secretion from HuH-7 cells. J. Gen. Virol. 2009;90:1359–1370. doi: 10.1099/vir.0.009274-0. [DOI] [PubMed] [Google Scholar]
- Reyes G.R., Huang C.C., Tam A.W., Purdy M.A. Molecular organization and replication of hepatitis E virus (HEV) Arch. Virol. Suppl. 1993;7:15–25. doi: 10.1007/978-3-7091-9300-6_2. [DOI] [PubMed] [Google Scholar]
- Reyes G.R., Purdy M.A., Kim J.P., Luk K.C., Young L.M., Fry K.E., Bradley D.W. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- Rehman S., Kapur N., Durgapal H., Panda S.K. Subcellular localization of hepatitis E virus (HEV) replicase. Virology. 2008;370:77–92. doi: 10.1016/j.virol.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Robinson R.A., Burgess W.H., Emerson S.U., Leibowitz R.S., Sosnovtseva S.A., Tsarev S., Purcell R.H. Structural characterization of recombinant hepatitis E virus ORF2 proteins in baculovirus-infected insect cells. Prot. Exp. Purif. 1998;12:75–84. doi: 10.1006/prep.1997.0817. [DOI] [PubMed] [Google Scholar]
- Ropp S.L., Tam A.W., Beames B., Purdy M., Frey T.K. Expression of the hepatitis E virus ORF1. Arch. Virol. 2000;145:1321–1337. doi: 10.1007/s007050070093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov M.N., Koonin E.V., Gorbalenya A.E. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kaariainen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield D.J., Glamann J., Emerson S.U., Purcell R.H. Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J. Virol. 2000;74:5548–5555. doi: 10.1128/jvi.74.12.5548-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal D., Thomas S., Chakraborty M., Jameel S. Expression and processing of the hepatitis E virus ORF1 nonstructural polyprotein. Virol. J. 2006;3:38. doi: 10.1186/1743-422X-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha M.P., Scott R.M., Joshi D.M., Mammen M.P., Jr., Thapa G.B., Thapa N., Myint K.S., Fourneau M., Kuschner R.A., Shrestha S.K. N. Engl. J. Med. 2007;356:895–903. doi: 10.1056/NEJMoa061847. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram B., Thakral D., Panda S.K. Targeted cleavage of hepatitis E virus 3′ end RNA mediated by hammerhead ribozymes inhibits viral RNA replication. Virology. 2003;312:350–358. doi: 10.1016/s0042-6822(03)00259-9. [DOI] [PubMed] [Google Scholar]
- Surjit M., Jameel S., Lal S.K. The ORF2 protein of hepatitis E virus binds the 5′ region of viral RNA. J. Virol. 2004;78:320–328. doi: 10.1128/JVI.78.1.320-328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Jameel S., Lal S.K. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J. Virol. 2007;81:3339–3345. doi: 10.1128/JVI.02039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Oberoi R., Kumar R., Lal S.K. Enhanced alpha1 microglobulin secretion from Hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J. Biol. Chem. 2006;281:8135–8142. doi: 10.1074/jbc.M509568200. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Nishizawa T., Sato H., Sato Y., Jirintai D., Nagashima S., Okamoto H. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 2011 doi: 10.1099/vir.0.029470-0. (Out ahead of print) [DOI] [PubMed] [Google Scholar]
- Takahashi M., Yamada K., Hoshino Y., Takahashi H., Ichiyama K., Tanaka T., Okamoto H. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 2008;153:1703–1713. doi: 10.1007/s00705-008-0179-6. [DOI] [PubMed] [Google Scholar]
- Tam A.W., Smith M.M., Guerra M.E., Huang C.C., Bradley D.W., Fry K.E., Reyes G.R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja S., Sen S., Gupta V.K., Aggarwal R., Jameel S. Plasma and urine biomarkers in acute viral hepatitis E. Proteome Sci. 2009;7:39. doi: 10.1186/1477-5956-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja S., Ahmad I., Sen S., Kumar S., Arora R., Gupta V.K., Aggarwal R., Narayanasamy K., Reddy V.S., Jameel S. Plasma peptidome profiling of acute hepatitis E patients by MALDI-TOF/TOF. Proteome Sci. 2011;9:5. doi: 10.1186/1477-5956-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticehurst J., Rhodes L.L., Jr., Krawczynski K., Asher L.V., Engler W.F., Mensing T.L., Caudill J.D., Sjogren M.H., Hoke C.H., Jr., LeDuc J.W. Infection of owl monkeys (Aotus trivirgatus) and cynomolgus monkeys (Macaca fascicularis) with hepatitis E virus from Mexico. J. Infect. Dis. 1992;165:835–845. doi: 10.1093/infdis/165.5.835. [DOI] [PubMed] [Google Scholar]
- Torresi J., Li F., Locarnini S.A., Anderson D.A. Only the non-glycosylated fraction of hepatitis E virus capsid (open reading frame 2) protein is stable in mammalian cells. J. Gen. Virol. 1999;80:1185–1188. doi: 10.1099/0022-1317-80-5-1185. [DOI] [PubMed] [Google Scholar]
- Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- Tyagi S., Surjit M., Lal S.K. The 41-amino-acid C-terminal region of the hepatitis E virus ORF3 protein interacts with bikunin, a kunitz-type serine protease inhibitor. J. Virol. 2005;79:12081–12087. doi: 10.1128/JVI.79.18.12081-12087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Surjit M., Roy A.K., Jameel S., Lal S.K. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 2004;279:29308–29319. doi: 10.1074/jbc.M402017200. [DOI] [PubMed] [Google Scholar]
- Uchida T., Suzuki K., Iida F., Shikata T., Ichikawa M., Rikihisa T., Mizuno K., Win K.M. Animal model, virology and gene cloning of hepatitis E. Gastroenterol. Jpn. 1991;26(Suppl. 3):148–151. doi: 10.1007/BF02779286. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang W., Ni B., Shen H., Song Y., Wang X., Shao S., Hua X., Cui L. Recombination analysis reveals a double recombination event in hepatitis E virus. Virol. J. 2010;7:129. doi: 10.1186/1743-422X-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.P., Kasorndorkbua C., Halbur P.G., Haqshenas G., Guenette D.K., Toth T.E., Meng X.J. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J. Clin. Microbiol. 2001;39:3040–3046. doi: 10.1128/JCM.39.9.3040-3046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Kato K., Li T., Takeda N., Miyamura T., Hammar L., Cheng R.H. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology. 1999;265:35–45. doi: 10.1006/viro.1999.0005. [DOI] [PubMed] [Google Scholar]
- Xing L., Li T.C., Mayazaki N., Simon M.N., Wall J.S., Moore M., Wang C.Y., Takeda N., Wakita T. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Wang J.C., Li T.C., Yasutomi Y., Lara J., Khudyakov Y., Schofield D., Emerson S.U., Purcell R.H., Takeda N., Miyamura T., Cheng R.H. Spatial configuration of hepatitis E virus antigenic domain. J. Virol. 2011;85:1117–1124. doi: 10.1128/JVI.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Takahashi M., Hoshino Y., Takahashi H., Ichiyama K., Tanaka T., Okamoto H. Construction of an infectious cDNA clone of hepatitis E virus strain JE03-1760F that can propagate efficiently in cultured cells. J. Gen. Virol. 2009;90:457–462. doi: 10.1099/vir.0.007559-0. [DOI] [PubMed] [Google Scholar]
- Yamashita T., Mori Y., Miyazaki N., Cheng R.H., Yoshimura M., Unno H., Shima R., Moriishi K., Tsukihara T., Li T.C. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12986–12991. doi: 10.1073/pnas.0903699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M., Ozdener M.H., Kumar R., Panda S.K., Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 1999;73:4074–4082. doi: 10.1128/jvi.73.5.4074-4082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M., Ozdener M.H., Panda S.K., Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Ng M.H., Xia N.S., Lau S.H., Che X.Y., Chau T.N., Lai S.T., Im S.W. Conformational antigenic determinants generated by interactions between a bacterially expressed recombinant peptide of the hepatitis E virus structural protein. J. Med. Virol. 2001;64:125–132. doi: 10.1002/jmv.1027. [DOI] [PubMed] [Google Scholar]
- Zhang M., Purcell R.H., Emerson S.U. Identification of the 5′ terminal sequence of the SAR-55 and MEX-14 strains of hepatitis E virus and confirmation that the genome is capped. J. Med. Virol. 2001;65:293–295. doi: 10.1002/jmv.2032. [DOI] [PubMed] [Google Scholar]
- Zhao C., Ma Z., Harrison T.J., Feng R., Zhang C., Qiao Z., Fan J., Ma H., Li M., Song A., Wang Y. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 2009;81:1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- Zheng Z.Z., Miao J., Zhao M., Tang M., Yeo A.E., Yu H., Zhang J., Xia N.S. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J. Gen. Virol. 2010;91:1728–1736. doi: 10.1099/vir.0.019323-0. [DOI] [PubMed] [Google Scholar]
- Zhou Y.H., Purcell R.H., Emerson S.U. An ELISA for putative neutralizing antibodies to hepatitis E virus detects antibodies to genotypes 1, 2, 3, and 4. Vaccine. 2004;22:2578–2585. doi: 10.1016/j.vaccine.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Zhu F.C., Zhang J., Zhang X.F., Zhou C., Wang Z.Z., Huang S.J., Wang H., Yang C.L., Jiang H.M., Cai J.P. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet. 2010;376:895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]


