Abstract
Opioid addiction is a chronic disease with high genetic contribution and a large inter-individual variability in therapeutic response. The goal of this study was to identify pharmacodynamic factors that modulate methadone dose requirement. The neurotrophin family is involved in neural plasticity, learning memory and behavior and deregulated neural plasticity may underlie the pathophysiology of drug addiction. BDNF was shown to affect the response to methadone maintenance treatment. This study explores the effects of polymorphisms in the nerve growth factor (beta polypeptide) gene, NGFB, on the methadone doses required for successful maintenance treatment for heroin addiction. Genotypes of 14 NGFB polymorphisms were analyzed for association with the stabilizing methadone dose in 72 former severe heroin addicts with no major co-medications. There was significant difference in methadone doses required by subjects with different genotypes of the NGFB intronic SNP rs2239622 (P = 0.0002). These results may have clinical importance.
Keywords: methadone, opioid addiction, nerve growth factor, NGFB, heroin addiction
Introduction
Heroin addiction is a chronic disease characterized by compulsive drug seeking, drug abuse, physical dependence and tolerance.1 The genetic contribution to vulnerability to develop heroin addiction is estimated at 40–60%.2–4 Methadone, the major pharmacotherapy of opiate addiction, is a mu-opioid receptor full agonist and a moderate noncompetitive N-methyl-D-aspartic acid (NMDA) receptor antagonist. Adequate doses of methadone are an important factor of successful treatment.5, 6 Methadone is a synthetic opioid that is administered as a racemic mixture of (R)- and (S)-methadone enantiomers; the (R)-methadone is an active enantiomer at the mu-opioid receptor. The half-life of the racemic mixture in humans ranges from 16–28 hours.7 Methadone is metabolized primarily by CYP3A4, CYP2B6, and CYP2D6.8 The large inter-individual variability in the therapeutic response to methadone may be influenced by genetic background (e.g. variants in the genes encoding metabolizing enzymes, drug transporters, drug targets and regulatory factors).9
The neutrophins family consists of nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF) and neurotrophins 3–7 that are involved in neuron development and differentiation as well as maintenance of neuronal systems, modulation of neurotransmission, and higher-order activities like learning memory and behavior.10, 11 They are synthesized as precursors that are proteolytically cleaved to active neurotrophins. The pathophysiology of drug addiction may be a result of a deregulation of synaptic plasticity that is caused by altered expression or binding affinity of neurotrophins.12 BDNF and its receptor, TrkB, induce glutamate release and also regulate GABAergic synapses.11 Recent studies have linked BDNF polymorphisms with memory impairments and susceptibility to psychiatric disorders and personality traits, as well as polysubstance abuse and heroin dependence in Asian males.10, 13–15 In addition, BDNF was suggested to confer differential susceptibility to MMT response in opioid addicts.16 Animal studies demonstrated that BDNF regulates the mesolimbic dopamine pathway and is involved in the response to aversive social experiences.17 There are only a few studies of the effect of NGFB genetic variability in humans, mostly related to insensitivity to pain.18–20 NGFB variations were shown to be associated with anxiety, in a gender-dependent manner.21 In addition, mice with CNS deletion of neutrophin-3 (NT-3) have attenuated morphine withdrawal reaction that was restored by transgene-derived overexpression of NT-3.22
The goal of this pharmacogenetics study is to identify pharmacodynamic genetic factors involved in response to MMT. To explore the potential effects of NGFB variation on response to methadone, we have analyzed methadone dose data from a well-characterized sample of former heroin addicts successfully stabilized in a MMT clinic in Israel. In addition, we have reexamined the results of our previous hypothesis-driven case-control association studies with heroin addiction that included NGFB SNPs.23, 24
Material and Methods
Subjects
The sample consisted of 72 (33 females) unrelated former severe heroin addicts in MMT from the Dr. Miriam and Sheldon G. Adelson Clinic for Drug Abuse, Treatment and Research, Tel Aviv, Israel. The ages ranged from 18–65 years (mean 38 years). All subjects had one or more years of daily multiple uses of heroin and at least one withdrawal or failure in a detoxification center and at least 6 months in MMT with stable methadone dose for at least 4 weeks. Patients underwent repeated random and observed urine tests and had negative urine for illicit opiates, cocaine or benzodiazepines for at least 4 weeks prior to obtaining blood specimens for methadone plasma level.25 Patients with major co-medications were excluded. Patients with specific medications that are not known to affect methadone metabolism or drugs metabolized in a related way (e.g., acetylsalicylic acid, metformin, statin) and patients on prescribed benzodiazepines were included. All subjects signed informed consent for genetic studies. The studies were approved by the Helsinki Committee of Tel-Aviv Sourasky Medical Center and the Institutional Review Board of The Rockefeller University Hospital.
The samples for the case-control association studies with heroin addiction are described in details elsewhere.23, 24 Briefly, the Caucasians sample consisted of 350 former severe heroin addicts in MMT and 184 controls, and the African American sample consisted of 207 cases and 167 controls. All subjects were from the USA.
SNPs and Ancestry informative markers (AIMs) genotyping
Genomic DNA was extracted from whole-blood samples using standard techniques. Genotyping was performed on a 1536-plex GoldenGate Custom Panel (GS0007064-OPA, Illumina, San Diego, CA, USA) as described23, 26 and analyzed by Genome Studio software genotyping Module Version 1.0.10 (Illumina). The SNPs selected for this custom array are tag SNPs (based on HapMap data), non-synonymous SNPs and SNPs with the potential to alter splicing efficiency.26 Genotype data were filtered based on call rates and cluster separation. Ten percent of the sample were genotyped in duplicate.
186 AIMs markers were genotyped as described.23, 26, 27 168 SNPs with adequate quality were selected for further analysis. Biographic Ancestry Scores (e.g., fractions of genetic affiliation of the individual in each of a predetermined number of clusters) were estimated by Structure 2.028 using 1051 CEPH subjects represented in the Human Genome Diversity Cell Line Panel (HGDP-CEPH) as reference with K=7.
Statistical analyses
Linkage disequilibrium (D′ and r2) was estimated using R and Haploview version 4.2.29 Analysis of variance (ANOVA) was performed to determine if the mean levels of the daily methadone doses were significantly different among genotypes for each of the SNPs. It was also performed with ethnicity (Ashkenazi Jewish, non-Ashkenazi Jewish, Christian and Muslim), age and co-medication as covariates in the model. In a separate analysis, ANOVA was also performed with the homozygosity for the ABCB1 SNP rs1236 T allele, and the two CYP2B6 SNPs as covariates in the model.
Results
The NGFB gene (NM_002506.2) is located on chromosome 1p13.1 and consists of 3 exons encoding the preproNGF transcript. The complete proNGF protein is encoded by exon 3. Ten SNPs are described in the coding region (NCBI), of which 3 are synonymous and only one non-synonymous SNP (rs6330) is common. A total of 15 polymorphisms were genotyped for this study, of which three are non-synonymous, and the rest are intronic tag SNPs and SNPs from the 5′ region near the gene (Table 1, Supplementary Table S1). Four SNPs were excluded from analysis: two non-synonymous SNPs (rs11466110, rs11466112) were monomorphic, one SNP (rs6326) had very low minor allele frequency (MAF<0.01), and one SNP (rs10776799) had a low cluster separation score. Observed genotype distributions were consistent with Hardy-Weinberg equilibrium (HWE).
Table 1.
NGFB SNPs details
| # | SNP ID | Alleles | Position (build 37.1) |
Location | Protein |
|---|---|---|---|---|---|
| 1 | rs3811014 | A/G | 115882503 | 5' near gene | |
| 2 | rs4332358 | G/A | 115875645 | 5' near gene | |
| 3 | rs6537860 | C/T | 115856344 | Intron 1 | |
| 4 | rs4529705 | C/T | 115851491 | Intron 1 | |
| 5 | rs6678788 | G/A | 115839671 | Intron 1 | |
| 6 | rs2856813 | G/A | 115837919 | Intron 1 | |
| 7 | rs2239622 | C/T | 115837709 | Intron 1 | |
| 8 | rs910330 | C/A | 115835500 | Intron 2 | |
| 9 | rs2268793 | G/A | 115831783 | Intron 2 | |
| 10 | rs6326 | C/G | 115830461 | Intron 2 | |
| 11 | rs6328 | G/T | 115829943 | Intron 2 | |
| 12 | rs6330 | C/T | 115829313 | exon 3 | A35V |
| 13 | rs11466110 | G/A | 115829203 | exon 3 | V72M |
| 14 | rs11466112 | C/T | 115828756 | exon 3 | R221W |
The sample details for this study are described in Table 2 (see also Material and Methods). The stabilizing daily methadone dose ranges from 12.5–260 mg with a mean of 140±52 mg and normal distribution. There is no significant gender difference in the methadone daily dose. The mean trough plasma (R/S) methadone level is 498±269 ng/ml (range 100–1220 ng/ml) with a moderate correlation between trough plasma levels and methadone dose (r = 0.40). The sample consists of the 62 Jewish subjects and 10 subjects that are either Caucasians or Arabs. For simplicity, the Jewish subjects are divided into two main groups: Ashkenazi and “non-Ashkenazi”. The “non-Ashkenazi” group includes Moroccan, Yemenite, Iraqi, Turkish, Iranian, Libyan, Syrian, Greek subjects, and subjects with mixed origin or unknown origin. Five subjects are Jewish of mixed or unknown origin. Ancestry Biographic Scores (e.g., fractions of genetic affiliation of the individual in each of a predetermined number of clusters) were estimated based on genotypes of 168 ancestry informative markers (AIMs) with data from 1051 individuals representing 51 worldwide populations as a reference. 26, 27 There is a very low contribution of African, Far East Asian, Oceanian and/or Native American populations in this Israeli sample (data not shown). The Ashkenazi Jewish group has a mainly European contribution and some Middle Eastern contribution and the non-Ashkenazi group has a Middle Eastern or European contribution (Table 2).
Table 2.
Sample details
| # | Methadone Dose (mg/day) |
Trough Plasma levels (ng/ml) |
Ethnicity | European contributiona |
Middle Eastern contributiona |
|
|---|---|---|---|---|---|---|
| 1 | 12.5 | 100 | non Ashkenazi Jewish | Moroccan | 0.44 | 0.48 |
| 2 | 25 | 450 | non Ashkenazi Jewish | Moroccan | 0.04 | 0.94 |
| 3 | 50 | 230 | non Ashkenazi Jewish | Moroccan | 0.93 | 0.03 |
| 4 | 55 | 250 | Ashkenazi Jewish | 0.95 | 0.02 | |
| 5 | 55 | 280 | non Ashkenazi Jewish | Moroccan | 0.97 | 0.01 |
| 6 | 55 | 750 | non Ashkenazi Jewish | Syrian, Turkish | 0.06 | 0.85 |
| 7 | 60 | 180 | non Ashkenazi Jewish | Greek | 0.72 | 0.19 |
| 8 | 60 | 140 | non Ashkenazi Jewish | Moroccan, Spain | 0.03 | 0.92 |
| 9 | 70 | 280 | Ashkenazi Jewish | 0.90 | 0.08 | |
| 10 | 75 | 310 | Caucasian (non Jewish) | 0.02 | 0.82 | |
| 11 | 78 | 220 | non Ashkenazi Jewish | Yemenite | 0.02 | 0.89 |
| 12 | 90 | 260 | Ashkenazi Jewish | 0.86 | 0.04 | |
| 13 | 90 | 650 | Ashkenazi Jewish | 0.91 | 0.05 | |
| 14 | 95 | 260 | non Ashkenazi Jewish | 0.14 | 0.83 | |
| 15 | 95 | 210 | non Ashkenazi Jewish | Turkish | 0.48 | 0.40 |
| 16 | 100 | 840 | non Ashkenazi Jewish | Turkish | 0.06 | 0.86 |
| 17 | 110 | 820 | non Ashkenazi Jewish | Iraqi | 0.01 | 0.89 |
| 18 | 110 | 300 | Caucasian (non Jewish) | 0.89 | 0.08 | |
| 19 | 115 | 890 | non Ashkenazi Jewish | Moroccan | 0.03 | 0.91 |
| 20 | 115 | 270 | Jewish | mix | 0.87 | 0.02 |
| 21 | 115 | 660 | Ashkenazi Jewish | 0.76 | 0.02 | |
| 22 | 120 | 500 | non Ashkenazi Jewish | 0.95 | 0.01 | |
| 23 | 120 | 330 | Arab | 0.01 | 0.96 | |
| 24 | 120 | 230 | Caucasian (non Jewish) | 0.93 | 0.04 | |
| 25 | 120 | 900 | non Ashkenazi Jewish | Moroccan | 0.07 | 0.92 |
| 26 | 125 | 230 | Arab | 0.04 | 0.81 | |
| 27 | 125 | 200 | non Ashkenazi Jewish | Yemenite | 0.19 | 0.77 |
| 28 | 130 | 580 | non Ashkenazi Jewish | Iranian | 0.76 | 0.19 |
| 29 | 130 | 390 | non Ashkenazi Jewish | 0.01 | 0.03 | |
| 30 | 130 | 340 | non Ashkenazi Jewish | 0.08 | 0.77 | |
| 31 | 130 | 290 | Caucasian (non Jewish) | 0.93 | 0.03 | |
| 32 | 135 | 330 | Ashkenazi Jewish | 0.92 | 0.05 | |
| 33 | 135 | 340 | non Ashkenazi Jewish | Yemenite | 0.73 | 0.26 |
| 34 | 135 | 100 | non Ashkenazi Jewish | Moroccan | 0.02 | 0.07 |
| 35 | 135 | 940 | non Ashkenazi Jewish | Libyan | 0.39 | 0.41 |
| 36 | 140 | 1220 | Ashkenazi Jewish | 0.37 | 0.56 | |
| 37 | 140 | 630 | non Ashkenazi Jewish | Iraqi | 0.97 | 0.02 |
| 38 | 140 | 630 | non Ashkenazi Jewish | Moroccan | 0.03 | 0.89 |
| 39 | 140 | 450 | Ashkenazi Jewish | 0.98 | 0.01 | |
| 40 | 140 | 230 | Jewish | mix | 0.28 | 0.17 |
| 41 | 140 | 600 | Ashkenazi Jewish | 0.96 | 0.02 | |
| 42 | 140 | 850 | Ashkenazi Jewish | 0.80 | 0.14 | |
| 43 | 145 | 780 | Jewish | mix | 0.93 | 0.04 |
| 44 | 150 | 250 | non Ashkenazi Jewish | Turkish, Iraqi | 0.01 | 0.96 |
| 45 | 150 | 450 | Arab | 0.58 | 0.33 | |
| 46 | 150 | 340 | Arab | 0.01 | 0.85 | |
| 47 | 160 | 110 | non Ashkenazi Jewish | Moroccan | 0.57 | 0.30 |
| 48 | 160 | 1000 | non Ashkenazi Jewish | Yemenite | 0.34 | 0.54 |
| 49 | 165 | 540 | non Ashkenazi Jewish | 0.08 | 0.66 | |
| 50 | 165 | 640 | Ashkenazi Jewish | 0.03 | 0.90 | |
| 51 | 170 | 400 | Caucasian (non Jewish) | 0.39 | 0.44 | |
| 52 | 170 | 500 | Ashkenazi Jewish | 0.91 | 0.02 | |
| 53 | 175 | 290 | non Ashkenazi Jewish | Turkish | 0.66 | 0.27 |
| 54 | 180 | 430 | Caucasian (non Jewish) | 0.22 | 0.68 | |
| 55 | 183 | 600 | Ashkenazi Jewish | 0.54 | 0.10 | |
| 56 | 185 | 490 | Jewish | unknown | 0.01 | 0.87 |
| 57 | 187.5 | 440 | Ashkenazi Jewish | 0.50 | 0.45 | |
| 58 | 190 | 820 | non Ashkenazi Jewish | Iraqi | 0.95 | 0.01 |
| 59 | 190 | 350 | non Ashkenazi Jewish | Syrian | 0.02 | 0.92 |
| 60 | 190 | 1210 | non Ashkenazi Jewish | 0.96 | 0.01 | |
| 61 | 190 | 380 | non Ashkenazi Jewish | 0.88 | 0.09 | |
| 62 | 190 | 940 | Ashkenazi Jewish | 0.37 | 0.56 | |
| 63 | 190 | 520 | non Ashkenazi Jewish | Yemenite | 0.09 | 0.85 |
| 64 | 195 | 490 | non Ashkenazi Jewish | Moroccan | 0.96 | 0.02 |
| 65 | 200 | 750 | non Ashkenazi Jewish | Yemenite | 0.05 | 0.93 |
| 66 | 205 | 390 | Ashkenazi Jewish | 0.43 | 0.29 | |
| 67 | 220 | 600 | Ashkenazi Jewish | 0.94 | 0.01 | |
| 68 | 220 | 700 | Ashkenazi Jewish | 0.96 | 0.03 | |
| 69 | 225 | 670 | non Ashkenazi Jewish | Moroccan | 0.95 | 0.01 |
| 70 | 230 | 580 | non Ashkenazi Jewish | 0.12 | 0.78 | |
| 71 | 240 | 680 | non Ashkenazi Jewish | Iraqi | 0.11 | 0.80 |
| 72 | 260 | 1070 | Jewish | mix | 0.67 | 0.25 |
The list is sorted by ascending methadone dose.
The proportion of ancestral contribution was calculated with AIMs data (see methods).
Only the two major contributors (European and Middle East), out of 7 calculated, are shown.
Listed in Table 3 are the minor allele frequencies (MAF) of the SNPs genotyped in this study, our previous association study in Caucasians,23 and in three HapMap populations (www.hapmap.org). HapMap data reveal major differences in the MAF of most of the SNPs between Caucasian, Asian and African populations, out of which, for six SNPs, the minor allele in Caucasians is the major allele in the African and/or the Chinese population. The non-synonymous SNP rs6330 is less frequent in Africans, SNP rs6328 is more common in Chinese, SNP rs6326 is African-specific, and rs2268793 is much more common in the Chinese population compared to the Caucasian and African populations. In general, the allele frequencies in our previous study of Caucasians are compatible with the HapMap data.
Table 3.
Allele frequencies of the NGFB SNPs in different populations
| Israeli samplea |
European Americans23 | HapMap | |||||
|---|---|---|---|---|---|---|---|
| Cases | Controls | CEUc | YRId | CHBe | |||
| # | SNP ID | n =72 | n=350b | n=184 | |||
| 1 | rs3811014 | 0.16 | 0.18 | 0.21 | 0.16 | 0.60f | 0.16 |
| 2 | rs4332358 | 0.45 | 0.28 | 0.36 | 0.23 | 0.70f | 0.57f |
| 3 | rs6537860 | 0.41 | 0.31 | 0.34 | 0.30 | 0.65f | 0.81f |
| 4 | rs4529705 | 0.42 | 0.31 | 0.34 | 0.40g | 0.63f | 0.80f |
| 5 | rs6678788 | 0.39 | 0.30 | 0.31 | 0.29 | 0.43 | 0.49 |
| 6 | rs2856813 | 0.42 | 0.46 | 0.48 | 0.48 | 0.89f | 0.70f |
| 7 | rs2239622 | 0.38 | 0.29 | 0.30 | 0.28 | 0.19 | 0.46 |
| 8 | rs910330 | 0.39 | 0.30 | 0.31 | 0.29 | 0.25 | 0.49 |
| 9 | rs2268793 | 0.12 | 0.07 | 0.10 | 0.07 | 0.02 | 0.30 |
| 10 | rs6326 | 0.01 | 0.01 | 0.01 | 0.01 | 0.31h | 0.00 |
| 11 | rs6328 | 0.42 | 0.37 | 0.36 | 0.34 | 0.29 | 0.53f |
| 12 | rs6330 | 0.37 | 0.43 | 0.40 | 0.44 | 0.16 | 0.13 |
| 13 | rs11466110 | 0.00 | 0.00 | 0.00 | 0.01 | 0.04 | 0.00 |
| 14 | rs11466112 | 0.00 | 0.00 | 0.00 | 0.00 | 0.003 | 0.00 |
Numbers in bold are significantly different than those of HapMap Caucasians.
This study.
Sample does not include the Israeli subjects.
Caucasians.
Africans from Yoruba.
Han Chinese.
The minor allele in CEU is the major allele in YRI or CHB.
Other NCBI Caucasian population (no data in HapMap).
African-specific.
Interestingly, the MAF of several SNPs in the Israeli sample are higher than those found in the European American sample from our previous study and from those reported in the HapMap Caucasian population but are not as high as those for the African and Asian populations (Table 3). Interestingly, the MAF of SNPs rs2239622 and rs910330 is higher in the Israeli sample than in both Caucasian and African HapMap samples and is closer to the MAF in the Chinese HapMap sample. No data is publicly available on the frequency of these SNPs in Middle Eastern populations and the significance of these findings is still in question because the sample is small and may not represent the general population because of the non-random selection.
Association of NGFB genetic variability with methadone dose
Individual SNPs were analyzed for association with stabilizing methadone dose (Table 4, Supplementary Table S1). There were significant differences in the mean daily methadone dose in subjects with different genotype groups of SNP rs2239622 (Figure 1). The mean daily methadone doses in subjects homozygous for the variant T allele (81.7 mg) were lower than those of the heterozygotes and the non–carriers (140 and 153 mg, respectively) (P = 0.0002 for genotype test with recessive mode). The range of the daily methadone doses in this group was 25–135 mg. Analysis of the data for SNP rs2239622 by 3 dose groups (<80 mg, 80–149, >149) resulted in similar results (P = 0.0015). The subjects that are homozygous (TT) for SNP rs2239622 include two Ashkenazi Jews, a non-Jewish Caucasian, and six non-Ashkenazi Jews, so this genotype group is not limited to a specific subgroup. The results were comparable when ethnicity, age and co-medication were used as covariates in the model (P = 0.0004). No significant differences in the mean trough plasma levels between the genotype groups were found (data not shown).
Table 4.
Association of individual SNPs with daily methadone dose
| Genotypea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/B | B/B | |||||||||
| SNP ID | n | Mean Methadone dose |
SE | n | Mean Methadone dose |
SE | n | Mean Methadone dose |
SE | P-valueb | |
| 1 | rs3811014 | 49 | 138.3 | 8.3 | 23 | 141.5 | 7.6 | 0 | - | - | 0.8087 |
| 2 | rs4332358 | 19 | 115.5 | 12.3 | 40 | 151.3 | 7.2 | 12 | 128.8 | 15.6 | 0.0311 |
| 3 | rs6537860 | 23 | 127.7 | 11.3 | 39 | 151.9 | 7.6 | 10 | 117.0 | 18.0 | 0.0702 |
| 4 | rs4529705 | 22 | 127.1 | 11.9 | 40 | 151.6 | 7.4 | 10 | 117.0 | 18.0 | 0.0699 |
| 5 | rs6678788 | 24 | 138.5 | 10.7 | 40 | 147.1 | 7.6 | 8 | 103.1 | 22.1 | 0.0920 |
| 6 | rs2856813 | 22 | 123.0 | 11.6 | 39 | 154.3 | 6.7 | 11 | 118.9 | 20.4 | 0.0268 |
| 7 | rs2239622 | 26 | 139.7 | 9.9 | 37 | 153.1 | 7.9 | 9 | 81.7 | 11.8 | 0.0007c |
| 8 | rs910330 | 25 | 139.3 | 10.3 | 38 | 145.6 | 7.9 | 9 | 112.8 | 21.7 | 0.2392 |
| 9 | rs2268793 | 57 | 137.4 | 6.9 | 13 | 149.6 | 16.2 | 2 | 127.5 | 7.5 | 0.7152 |
| 10 | rs6326 | 70 | 2 | 0 | NA | ||||||
| 11 | rs6328 | 25 | 134.9 | 12.0 | 34 | 153.4 | 6.4 | 13 | 111.0 | 16.7 | 0.0367 |
| 12 | rs6330 | 29 | 134.8 | 9.3 | 33 | 144.1 | 8.5 | 10 | 136.8 | 22.3 | 0.7750 |
| 13 | rs11466110 | 0 | 0 | 0 | NA | ||||||
| 14 | rs11466112 | 0 | 0 | 0 | NA | ||||||
A represents the more common allele and B represents the less common variant
Genotype test with co-dominant mode.
P =0.0002 for genotype test with recessive mode.
Abbreviations: n, number of subjects; SE, standard error; NA, not applicable.
Figure 1.
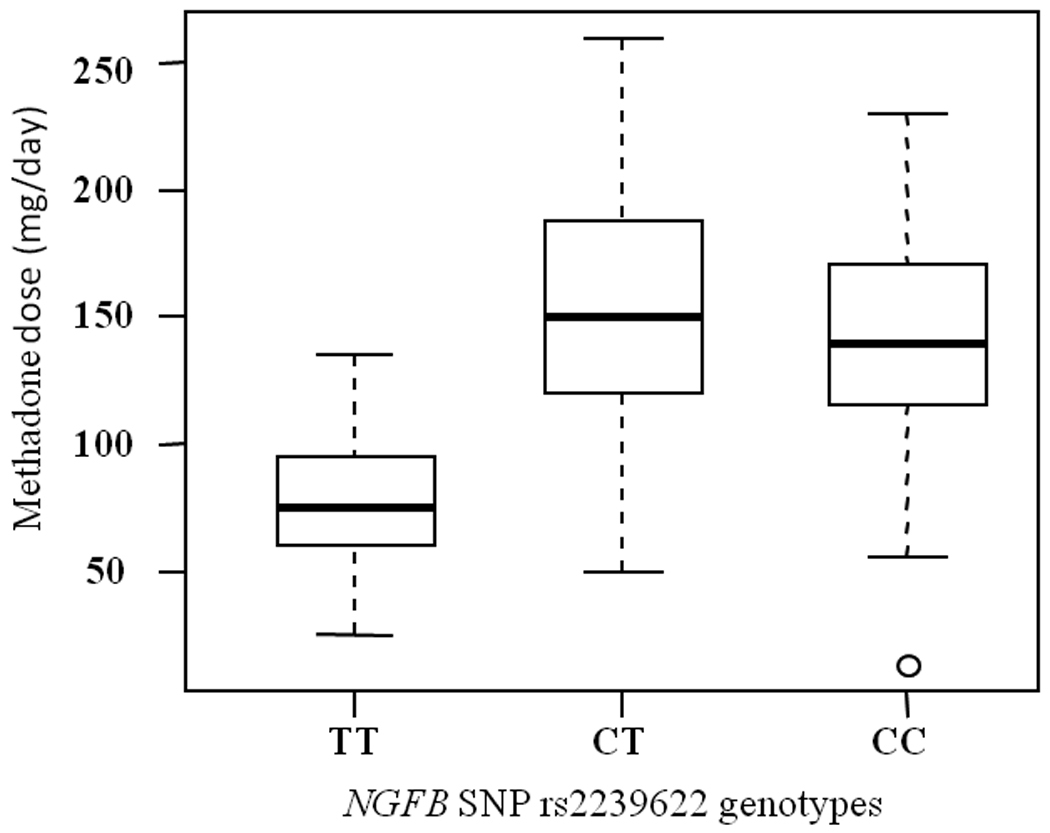
Association of NGFB SNP rs2239622 with daily methadone doses required for successful treatment. The results are shown as box plots where the box represents the middle 50% of the data and the whiskers represent the spread of the remaining data. The median is represented by the line in the center. Stable methadone doses are plotted against the genotypes.
We have previously reported an association of homozygosity T/T for SNP 1236C>T (rs1128503) in the p-glycoprotein encoding gene, ABCB1, with high methadone doses (>150 mg/day).30 In addition, we have recently found that carriers of two CYP2B6 SNPs require relatively lower methadone doses (Levran et al., manuscript in preparation). To account for a potential multiple gene effect we have analyzed the combined data of NGFB SNP rs2239622, CYP2B6 SNPs and ABCB1 SNP rs1128503, and this analysis substantiated the original result (P = 0.0001).
Revisiting the hypothesis-driven case-control association study with heroin addiction
We have previously reported a hypothesis-driven case-control association study of 130 genes.23, 24 The finding of association of the NGFB variant with stabilizing methadone dose prompted us to reexamine the results of these studies, since only a limited number of SNPs that gave the lowest P values in the association test were originally reported. These studies were performed on the same array as the current study, so the same NGFB SNPs were genotyped. Intriguingly, the result for NGFB SNP rs4332358 was just slightly above the cutoff chosen in the original study. The frequency of subjects with the G/G genotype among the heroin addicts was nominally significantly higher than that of controls (P = 0.003, odds ratio = 1.73, 95%CI (1.18, 2.53)) for a genotype test with a recessive mode; see also Table 3 for the differences in allele frequency between cases and controls). The minor allele ‘A’ may be considered a protective allele. Notably, this allele is the major allele in the African population (Table 3). The results for the rest of the NGFB SNPs were not significant. No significant associations of any NGFB SNPs were found in African Americans. Of importance, AIMs data revealed that only 2% of the Caucasian subjects and none of the African American subjects in the two previous studies have a major Middle Eastern contribution (>.75) (Levran et al., manuscript in preparation).
Linkage Disequilibrium (LD) analysis
Most of the SNPs genotyped in this study were chosen as tagging SNPs26 and are expected to show low LD and represent different haplotypes. Analysis of the genotype data revealed high LD between the following SNPs: rs910330 and rs6678788 (D′ = .97, r2 = .94), rs2239622 and rs6678788 (D′ = .81, r2 = .64), rs910330 and rs2239622 (D′ = .81, r2 = .64), and rs4529705 and rs6537860 (D′ = .10, r2 = .97). These results are compatible with those of the HapMap Caucasian population. Listed in Supplementary Table S2 are HapMap SNPs that are tagged by SNPs from this study (D′ > .8, r2 ≥ .6) in the HapMap Caucasian population. There is no data available for SNPs rs4529705 and rs6326 in HapMap. Notably, the non-synonymous SNP rs6330 is in complete LD with SNP rs6327 located at a distance of 673 bp in intron 2 that was not analyzed in this study. SNPs rs2239622 and rs4332358, indicated in this study for association with methadone dose or heroin addiction, respectively, are in high LD with several SNPs in intron 1 and 2.
Discussion
One of the mechanisms underlying drug addiction may be due to synaptic plasticity changes which may be a result of alterations in the expression of genes encoding neurotrophins and their receptors, among other genes.31 The NGFB gene consists of 3 exons encoding the preproNGF transcript. After removal of the signal peptide, the precursor generates proNGF. The complete proNGF protein is encoded by exon 3 and undergoes further post-translational processing to generate a mature product. It was demonstrated that proNGF is the predominant form of NGF in human and rodent brain tissue, suggesting that proNGF may have independent biological activity.32, 33 The effect of NGFB is mediated by tyrosine kinase receptor NTRK1 (TrkA) and the cytokine p75 neurotrophin receptor through activation of intracellular signaling cascades that regulate several processes including gene expression. Numerous stimuli including stress cause dynamic modulation of NGF and NGF receptor expression.34 NGF also modulates the neuronal and inflammatory component of pain.33 NGF was suggested to function as a molecular switch that redirects the delta opioid receptor (OPRD1) to the surface membrane of central synaptic terminals under chronic opioid conditions.35 OPRD1 plays an important role in pain control 36 and is also interacting with OPRM1.37 An NGF- responsive region was identified in the rodent oprd1 promoter.38, 39 Perinatal methadone exposure was shown to reduce rat striatal NGF content, but not mRNA levels, suggesting regulation on other levels (e.g., processing, stability or release).40 Chronic alcohol exposure has been shown to down-regulate human plasma NGF with a greater decrease in patients with family history of alcohol dependency.41
Individual methadone dosage optimization is one of the key factors in effective MMT, since methadone has large inter-individual variability in response and a narrow therapeutic index. Several pharmacogenetics studies aimed to identify pharmacodynamic genetic factors that modulate response to methadone maintenance were reported.30, 42–50 BDNF was suggested to confer differential susceptibility to MMT response in opioid addicts.16 The main findings of these earlier studies are that variants in ABCB1, OPRM1, dopamine D2 receptor (DRD2), BDNF, and potassium inwardly-rectifying channel KCNJ6 may be related to MMT response.
The major finding of this study is that the NGFB intronic variant rs2239622 is associated with relatively low methadone doses required in some patients for successful treatment of opiate addiction, in an Israeli population with Caucasian and Middle Eastern ancestry. The mean daily methadone doses in subjects homozygous for the variant T allele was 81.7 mg (range 25–135 mg) compared to 148 mg in heterozygotes and non–carriers. This is the first study associating NGFB variation with methadone dose requirement. The effect of NGFB on MMT response is not known and may be related to alteration in NGFB expression and/or function that leads to alteration in neural plasticity. The biological effect of the specific intronic variant is not currently known and it may be a marker for another functional SNP or a haplotype. This is the first step toward identifying specific gene variants associated with dose requirements that may possibly allow prediction of individual response to MMT, which is of clinical significance.
NGFB variants are unlikely to be acting alone in modulating the response to methadone, and multivariate analysis reflects more realistically the potential contribution of several genetic factors to dose requirement. For example, a NGFB variant was shown to modify the risk for eating disorders conferred by the risk genotype of a SNP in the neurotrophin receptor gene NTRK3, suggesting epistatic interaction.51 Methadone is a substrate of the efflux transporter P-glycoprotein that is encoded by the ABCB1 gene. We have previously reported that homozygosity to the T allele of ABCB1 SNP 1236C>T is associated with higher methadone doses.30 In addition, we have recently found that carriers of two variants in the gene encoding cytochrome P450 methadone metabolizing enzyme CYP2B6 require relatively lower methadone doses (Levran et al., manuscript in preparation). An analysis of NGFB SNP rs2239622 with the three SNPs mentioned above as co-variates substantiated the significance of the results obtained in the single SNP analysis.
The efficacy of methadone is significantly altered by several medications that are often consumed by MMT patients (e.g., treatments for HIV/AIDS and affective disorders).52, 53 To eliminate the effect of other drugs on the results, subjects selected for this study were, for the most part, not receiving other prescribed medication and none had evidence of ongoing drug abuse.
The significant variation in allele frequency of NGFB SNPs among different populations, as demonstrated in HapMap, and elucidated to a limited extent in this study, may have clinical relevance and may reflect an evolutionary dynamic process of selective advantage or genetic drift. It is especially relevant for admixed populations such as these in Israel. There is high genetic similarity between the Jewish and the Caucasian populations, but also some differences that may be explained by the complex demographic history of the Jewish people.54, 55 If the results from this small non-randomly selected sample reflect the allele frequencies of this population, the NGFB gene is one of the genes in which Jewish people and/or Middle Eastern populations differ from Caucasians.
This study has several limitations: a) small sample size; b) a relatively large number of SNPs have been analyzed, increasing the risk for type I error; c) all the patients are recruited from one clinic in one country; and d) the sample ethnicity is a quite unique admixture of Caucasians and Middle Eastern. To address these limitations, larger studies in other populations and clinics are warranted.
In summary, the NGFB SNP rs2239622 is shown to be associated with relatively low stabilizing methadone doses in patients in MMT in Israel, and NGFB SNP rs4332358 has been associated with heroin addiction in European Americans. Further studies are necessary to confirm these results, to determine the functionality of these SNPs (or linked SNPs) and the mechanism by which they may affect methadone response. An improved understanding of the role of neutrophins in drug addiction and its treatment may facilitate the search for effective treatment and prevention.
Supplementary Material
Acknowledgments
We would like to thank and acknowledge the contributions of Pei-Hong Shen, Dr. David Goldman, Dr. Ann Ho, Susan Russo and Anat Sason.
This work was supported in part by NIDA-P60-05130 (M.J.K.) and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Footnotes
Conflict of interest statement. None declared.
References
- 1.Kreek MJ. Methadone-related opioid agonist pharmacotherapy for heroin addiction. History, recent molecular and neurochemical research and future in mainstream medicine. Ann N Y Acad Sci. 2000;909:186–216. doi: 10.1111/j.1749-6632.2000.tb06683.x. [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- 3.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 4.Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, et al. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.Amato L, Davoli M, Perucci CA, Ferri M, Faggiano F, Mattick RP. An overview of systematic reviews of the effectiveness of opiate maintenance therapies: available evidence to inform clinical practice and research. J Subst Abuse Treat. 2005;28:321–329. doi: 10.1016/j.jsat.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Kreek MJ, LaForge KS, Butelman E. Pharmacotherapy of addictions. Nat Rev Drug Discov. 2002;1:710–726. doi: 10.1038/nrd897. [DOI] [PubMed] [Google Scholar]
- 7.Kreek MJ, Vocci FJ. History and current status of opioid maintenance treatments: blending conference session. J Subst Abuse Treat. 2002;23:93–105. doi: 10.1016/s0740-5472(02)00259-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Kantelip JP, Gerritsen-van Schieveen P, Davani S. Interindividual variability of methadone response: impact of genetic polymorphism. Mol Diagn Ther. 2008;12:109–124. doi: 10.1007/BF03256276. [DOI] [PubMed] [Google Scholar]
- 10.Chao MV, Rajagopal R, Lee FS. Neurotrophin signalling in health and disease. Clin Sci (Lond) 2006;110:167–173. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho AL, Caldeira MV, Santos SD, Duarte CB. Role of the brain-derived neurotrophic factor at glutamatergic synapses. Br J Pharmacol. 2008;153(Suppl 1):S310–S324. doi: 10.1038/sj.bjp.0707509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolanos CA, Nestler EJ. Neurotrophic mechanisms in drug addiction. Neuromolecular Med. 2004;5:69–83. doi: 10.1385/NMM:5:1:069. [DOI] [PubMed] [Google Scholar]
- 13.Lang UE, Hellweg R, Kalus P, Bajbouj M, Lenzen KP, Sander T, et al. Association of a functional BDNF polymorphism and anxiety-related personality traits. Psychopharmacology (Berl) 2005;180:95–99. doi: 10.1007/s00213-004-2137-7. [DOI] [PubMed] [Google Scholar]
- 14.Uhl GR, Liu QR, Walther D, Hess J, Naiman D. Polysubstance abuse-vulnerability genes: genome scans for association, using 1,004 subjects and 1,494 single-nucleotide polymorphisms. Am J Hum Genet. 2001;69:1290–1300. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng CY, Hong CJ, Yu YW, Chen TJ, Wu HC, Tsai SJ. Brain-derived neurotrophic factor (Val66Met) genetic polymorphism is associated with substance abuse in males. Brain Res Mol Brain Res. 2005;140:86–90. doi: 10.1016/j.molbrainres.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 16.de Cid R, Fonseca F, Gratacos M, Gutierrez F, Martin-Santos R, Estivill X, et al. BDNF variability in opioid addicts and response to methadone treatment: preliminary findings. Genes Brain Behav. 2008;7:515–522. doi: 10.1111/j.1601-183X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 17.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 18.Einarsdottir E, Carlsson A, Minde J, Toolanen G, Svensson O, Solders G, et al. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet. 2004;13:799–805. doi: 10.1093/hmg/ddh096. [DOI] [PubMed] [Google Scholar]
- 19.Larsson E, Kuma R, Norberg A, Minde J, Holmberg M. Nerve growth factor R221W responsible for insensitivity to pain is defectively processed and accumulates as proNGF. Neurobiol Dis. 2009;33:221–228. doi: 10.1016/j.nbd.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgibbon GJ, Kingston H, Needham M, Gaunt L. Haploinsufficiency of the nerve growth factor beta gene in a 1p13 deleted female child with an insensitivity to pain. Dev Med Child Neurol. 2009;51:833–837. doi: 10.1111/j.1469-8749.2008.03173.x. [DOI] [PubMed] [Google Scholar]
- 21.Lang UE, Hellweg R, Bajbouj M, Gaus V, Sander T, Gallinat J. Gender-dependent association of a functional NGF polymorphism with anxiety-related personality traits. Pharmacopsychiatry. 2008;41:196–199. doi: 10.1055/s-0028-1082070. [DOI] [PubMed] [Google Scholar]
- 22.Akbarian S, Bates B, Liu RJ, Skirboll SL, Pejchal T, Coppola V, et al. Neurotrophin-3 modulates noradrenergic neuron function and opiate withdrawal. Mol Psychiatry. 2001;6:593–604. doi: 10.1038/sj.mp.4000897. [DOI] [PubMed] [Google Scholar]
- 23.Levran O, Londono D, O'Hara K, Nielsen DA, Peles E, Rotrosen J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levran O, Londono D, O'Hara K, Randesi M, Rotrosen J, Casadonte P, et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8:531–540. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelson M, Peles E, Bodner G, Kreek MJ. Correlation between high methadone doses and methadone serum levels in methadone maintenance treatment (MMT) patients. J Addict Dis. 2007;26:15–26. doi: 10.1300/J069v26n01_03. [DOI] [PubMed] [Google Scholar]
- 26.Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Levran O, O'Hara K, Peles E, Li D, Barral S, Ray B, et al. ABCB1 (MDR1) genetic variants are associated with methadone doses required for effective treatment of heroin dependence. Hum Mol Genet. 2008;17:2219–2227. doi: 10.1093/hmg/ddn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 32.Fahnestock M, Michalski B, Xu B, Coughlin MD. The precursor pro-nerve growth factor is the predominant form of nerve growth factor in brain and is increased in Alzheimer's disease. Mol Cell Neurosci. 2001;18:210–220. doi: 10.1006/mcne.2001.1016. [DOI] [PubMed] [Google Scholar]
- 33.Pezet S, McMahon SB. Neurotrophins: mediators and modulators of pain. Annu Rev Neurosci. 2006;29:507–538. doi: 10.1146/annurev.neuro.29.051605.112929. [DOI] [PubMed] [Google Scholar]
- 34.Fiore M, Chaldakov GN, Aloe L. Nerve growth factor as a signaling molecule for nerve cells and also for the neuroendocrine-immune systems. Rev Neurosci. 2009;20:133–145. doi: 10.1515/revneuro.2009.20.2.133. [DOI] [PubMed] [Google Scholar]
- 35.Bie B, Zhang Z, Cai YQ, Zhu W, Zhang Y, Dai J, et al. Nerve growth factor-regulated emergence of functional delta-opioid receptors. J Neurosci. 2010;30:5617–5628. doi: 10.1523/JNEUROSCI.5296-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narita M, Kuzumaki N, Miyatake M, Sato F, Wachi H, Seyama Y, et al. Role of delta-opioid receptor function in neurogenesis and neuroprotection. J Neurochem. 2006;97:1494–1505. doi: 10.1111/j.1471-4159.2006.03849.x. [DOI] [PubMed] [Google Scholar]
- 37.Kieffer BL, Gaveriaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 38.Chen YL, Monteith N, Law PY, Loh HH. Dynamic association of p300 with the promoter of the G protein-coupled rat delta opioid receptor gene during NGF-induced neuronal differentiation. Biochem Biophys Res Commun. 2010;396:294–298. doi: 10.1016/j.bbrc.2010.04.083. [DOI] [PubMed] [Google Scholar]
- 39.Chen YL, Law PY, Loh HH. Action of NF-kappaB on the delta opioid receptor gene promoter. Biochem Biophys Res Commun. 2007;352:818–822. doi: 10.1016/j.bbrc.2006.11.103. [DOI] [PubMed] [Google Scholar]
- 40.Wu VW, Mo Q, Yabe T, Schwartz JP, Robinson SE. Perinatal opioids reduce striatal nerve growth factor content in rat striatum. Eur J Pharmacol. 2001;414:211–214. doi: 10.1016/s0014-2999(01)00807-x. [DOI] [PubMed] [Google Scholar]
- 41.Yoon SJ, Roh S, Lee H, Lee JY, Lee BH, Kim YK, et al. Possible role of nerve growth factor in the pathogenesis of alcohol dependence. Alcohol Clin Exp Res. 2006;30:1060–1065. doi: 10.1111/j.1530-0277.2006.00120.x. [DOI] [PubMed] [Google Scholar]
- 42.Doehring A, Hentig N, Graff J, Salamat S, Schmidt M, Geisslinger G, et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenet Genomics. 2009;19:407–414. doi: 10.1097/FPC.0b013e328320a3fd. [DOI] [PubMed] [Google Scholar]
- 43.Crettol S, Besson J, Croquette-Krokar M, Hammig R, Gothuey I, Monnat M, et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1722–1727. doi: 10.1016/j.pnpbp.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Lawford BR, Young RM, Noble EP, Sargent J, Rowell J, Shadforth S, et al. The D(2) dopamine receptor A(1) allele and opioid dependence: association with heroin use and response to methadone treatment. Am J Med Genet. 2000;96:592–598. doi: 10.1002/1096-8628(20001009)96:5<592::aid-ajmg3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 45.Lotsch J, Skarke C, Wieting J, Oertel BG, Schmidt H, Brockmoller J, et al. Modulation of the central nervous effects of levomethadone by genetic polymorphisms potentially affecting its metabolism, distribution, and drug action. Clin Pharmacol Ther. 2006;79:72–89. doi: 10.1016/j.clpt.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Crettol S, Deglon JJ, Besson J, Croquette-Krokar M, Hammig R, Gothuey I, et al. ABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatment. Clin Pharmacol Ther. 2006;80:668–681. doi: 10.1016/j.clpt.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Coller JK, Barratt DT, Dahlen K, Loennechen MH, Somogyi AA. ABCB1 genetic variability and methadone dosage requirements in opioid-dependent individuals. Clin Pharmacol Ther. 2006;80:682–690. doi: 10.1016/j.clpt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Fonseca F, Gratacos M, Escaramis G, De Cid R, Martin-Santos R, Fernandez-Espejo E, et al. Response to methadone maintenance treatment is associated with the MYOCD and GRM6 genes. Mol Diagn Ther. 2010;14:171–178. doi: 10.1007/BF03256370. [DOI] [PubMed] [Google Scholar]
- 49.Barratt DT, Coller JK, Somogyi AA. Association between the DRD2 A1 allele and response to methadone and buprenorphine maintenance treatments. Am J Med Genet B Neuropsychiatr Genet. 2006;141:323–331. doi: 10.1002/ajmg.b.30319. [DOI] [PubMed] [Google Scholar]
- 50.Lotsch J, Pruss H, Veh RW, Doehring A. A KCNJ6 (Kir3.2, GIRK2) gene polymorphism modulates opioid effects on analgesia and addiction but not on pupil size. Pharmacogenet Genomics. 2010;20:291–297. doi: 10.1097/FPC.0b013e3283386bda. [DOI] [PubMed] [Google Scholar]
- 51.Mercader JM, Saus E, Aguera Z, Bayes M, Boni C, Carreras A, et al. Association of NTRK3 and its interaction with NGF suggest an altered cross-regulation of the neurotrophin signaling pathway in eating disorders. Hum Mol Genet. 2008;17:1234–1244. doi: 10.1093/hmg/ddn013. [DOI] [PubMed] [Google Scholar]
- 52.Kharasch ED, Bedynek PS, Walker A, Whittington D, Hoffer C. Mechanism of ritonavir changes in methadone pharmacokinetics and pharmacodynamics: II. Ritonavir effects on CYP3A and P-glycoprotein activities. Clin Pharmacol Ther. 2008;84:506–512. doi: 10.1038/clpt.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCance-Katz EF, Sullivan LE, Nallani S. Drug interactions of clinical importance among the opioids, methadone and buprenorphine, and other frequently prescribed medications: a review. Am J Addict. 2010;19:4–16. doi: 10.1111/j.1521-0391.2009.00005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Need AC, Kasperaviciute D, Cirulli ET, Goldstein DB. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 2009;10:R7. doi: 10.1186/gb-2009-10-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


