Abstract
We have developed a polymer nanoparticle-based siRNA delivery system that exploits a cell-surface binding synergism between targeting ligands and cell-penetrating peptides. Nanoparticles were coated with folate and penetratin via a PEGylated phospholipid linker (DSPE-PEG): the combination of both of these ligands represents a strategy for enhancing intracellular delivery of attached polymer nanoparticles. Nanoparticles were characterized for size, morphology, density of surface modification, and ligand association and retention. The surface coverage achieved on DSPE-PEG-coated nanoparticles is as high as (or higher than) obtained with other ligand-modified nanoscale particulate systems (~0.5-5 pmol ligand/cm2). Additionally, these nanoparticles were loaded with a high density of siRNA (~130-140 pmol siRNA/mg nanoparticles), which is slowly released upon incubation in water. Synergies between the activity of surface binding and cell internalizing ligands on these siRNA-loaded nanoparticles impart delivery enhancements that improve their gene silencing efficacy both in culture and in tumor models. Traditionally, targeting ligands function by binding to cell surface receptors, while cell-penetrating peptides function by nonspecifically transporting across cell membranes. Interestingly, we have observed that improved delivery of these dual-functionalized nanoparticles was in part, a result of increased cell-surface avidity afforded by both ligands. This siRNA delivery system presents an approach to surface modification of nanovehicles, in which multiple ligands additively function in parallel to enhance cell binding and uptake.
Keywords: Nanoparticle, Copolymer, Gene therapy, Phospholipid, Drug delivery, Controlled drug release
1. Introduction
Polymer nanoparticles (NP) are effective drug delivery vehicles, which can be useful in treatment of tumors in a variety of animal models [1]. NP composed of poly(lactic-co-glycolic acid) (PLGA), which is the most well-studied degradable synthetic polymer in humans, are particularly attractive carriers for treating tumors in patients. Unmodified PLGA NP are internalized by many types of non-phagocytic cells in culture [2]; however, systemic delivery of these particles most often leads to substantial distribution to normal tissues and low uptake in tissues of interest. Potentially, NP uptake and selectivity can be improved through surface modification with ligands that promote cell binding and internalization [3-5]. NP formulated from PLGA are amenable to surface functionalization, using a variety of approaches [6].
Active targeting to cancer cells is a widely employed drug delivery strategy, in which ligand-conjugates directly interact with complementary moieties present on the surface of target cells [7-9]. Folate (FOL) is often used as a targeting ligand: it has a high specificity and affinity (Kd ~ 10-9 M) for cell-surface folate receptors, which are overexpressed on a variety of epithelial cancer cells, including KB cells [10,11]. When displayed by multivalent nanovehicles, FOL increases the binding avidity and subsequent cellular uptake of the attached cargo [12,13]. Cell-penetrating peptides (CPPs) are another commonly utilized class of ligands that can enhance NP uptake [14]. Penetratin (ANTP) is a well-studied CPP derived from the protein transduction domain of the antennapedia homeodomain; as with FOL, ANTP has been demonstrated to increase cell membrane translocation of attached high molecular weight cargo [15].
Many nanovehicle surface modification strategies support the attachment of multiple, different ligands. Most multi-functional modification strategies published so far employ a combination of ligands that potentially aid in overcoming sequential delivery barriers, such as targeting ligands (to elicit cell-surface binding and receptor-mediated endocytosis) and endosomal escape ligands (to promote delivery to the cytosol and avoid endolysosomal degradation) [16,17]. But no studies that we know of describe the combination of both targeting ligands and CPPs on the surface of polymer NP. There is some evidence that this strategy is valuable in other nano-scale delivery systems, particularly liposomes. For example, Zhao et al. recently reported that liposomes conjugated with folate and TAT (another CPP) were more effective in delivering paclitaxel than liposomes conjugated with folate or TAT alone [18]. The combination of FOL and ANTP as NP-bound ligands is uncommon, perhaps because their functions appear redundant, since both ligands purportedly enhance cell internalization. However, combinations of FOL and ANTP can exploit synergies in the inherent cellular affinities of each ligand. Folate binds its cognate cell-surface receptor and positively-charged ANTP has been purported to exhibit a nonspecific attraction to plasma membranes as well as specific interactions with cell surface glycosaminoglycans [19,20]. Therefore, while FOL and ANTP offer redundant functions at the cell-entry level, the combination of both ligands may increase NP avidity and enhance cell association.
Small interfering RNAs (siRNAs) are valuable tools in analyzing gene function and hold great therapeutic promise [21]. Since they are membrane-impermeant, siRNAs require a delivery vector to efficiently enter cells and localize at the site-of-action in the cytosol. Because of the specificity of siRNA-mediated gene knockdown, delivery of siRNA can be used as a sensitive method to assess the ability of ANTP/FOL-NP to deliver therapeutics intracellularly. To assess the drug delivery potential of polymer NP with both targeting and cell-penetrating ligands, siRNA was encapsulated into these dual-coated NP.
Many nano-scale drug delivery systems have demonstrated increased efficacy due to the presence of either targeting ligands or CPPs. The goals of this study were to (1) design, develop, and characterize NP coated with both FOL and ANTP; (2) comparatively evaluate single- and multi-ligand efficacy in enhancing cell uptake and siRNA delivery; and (3) decouple and understand the specific roles that FOL and ANTP have in increasing NP uptake.
2 Materials and Methods
2.1 Materials
Poly (D,L-lactide-co-glycolide) with terminal ester group (PLGA, 50:50 monomer ratio and 0.55–0.75 dL/g inherent viscosity) was purchased from Durect Corporation (Pelham, AL). The lipids purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) include 1,2-distearoyl-snglycero-3-phosphoethanolamine-N-[carboxy(polyethylene glycol)-2000] (DSPE-PEG), DSPE-PEG-FOL, DSPE-PEG-amine, and DSPE-PEG-maleimide. Bond-Breaker tris(2-carboxyethyl)phosphine (TCEP) and protein quantification reagents were purchased from Thermo Fisher Scientific (Rockford, IL). Folate, Coumarin 6 (C6), and polyvinyl alcohol (PVA) were purchased from Sigma-Aldrich (St. Louis, MO). Sepharose CL-2B and PD-10 gravity flow columns were from GE Healthcare Life Sciences (Piscataway, NJ). Luciferase Assay Reagent and lysis buffers were from Promega (Madison, WI). Fluorescein-5-isothiocyanate (FITC), Lipofectamine™ RNAiMAX, cell dissociation buffer (enzyme free, Hank's-based), Texas Red®-X phalloidin, Hoescht 33342, and Trypan blue stain were purchased from Invitrogen (Carlsbad, CA).
ANTP peptide (RQIKIWFQNRRMKWKKGGC) was synthesized and RP-HPLC purified at the W.M. Keck Peptide Synthesis Facility at Yale University (New Haven, CT). Luciferase-targeted siRNA (siLUC, 5’-GCUAUGAAGCGCUAUGGGC-3’) with and without cholesterol (3’ sense strand modification) was purchased from Dharmacon (Lafayette, CO) in deprotected and desalted form with 3’dTdT overhangs on both strands. Negative control siRNA (SCR, Silencer® Negative Control No.1) was purchased from Invitrogen (Carlsbad, CA). Luciferase expressing plasmid pGL4.15 with SV40 promoter supplied by Promega (Madison, WI) was a generous gift from John Schmitz and Edward Chu (University of Pittsburgh, PA).
KB cells were provided by the American Type Culture Collection (ATCC) (Manassas, VA) and maintained in folate-free RPMI-1640 medium from Invitrogen (Carlsbad, CA) containing 10% (v/v) fetal bovine serum (FBS) from Atlanta Biologicals (Lawrenceville, GA).
2.2 Synthesis of DSPE-PEG constructs
DSPE-PEG-ANTP was synthesized by coupling DSPE-PEG-maleimide to the C-terminal cysteine of ANTP. ANTP (2.1 µmol) was dissolved in DMF, and added to HEPES-EDTA buffer (100mM HEPES, 10mM EDTA, pH 7.5) containing TCEP (50 mM). After reduction of ANTP for one hour, maleimide-PEG-DSPE (30.9 mg) was added and the reaction mixture was incubated overnight at room temperature. DSPE-PEG-ANTP was purified by gel filtration. DSPE-PEG-FITC was synthesized by coupling FITC to DSPE-PEG-amine. FITC (20 mg) was dissolved in DMF and incubated overnight with DSPE-PEG-amine (14.3 mg) in borate buffer (50 mM borate, pH 8.5) at room temperature. The reaction mixture was dialyzed in PBS. Conjugation was verified by RP-HPLC.
2.3 Nanoparticle fabrication
Ligand-coated and unmodified PLGA NP containing siRNA were prepared using a modified water-in-oil-in-water (w/o/w) double emulsion technique. Briefly, siRNA was complexed with spermidine in pH 7.4 TE buffer (10 mM Tris-HCl, 1 mM EDTA) at an 8:1 polyamine nitrogen to nucleotide phosphate (N-to-P) ratio. After 20 min, the self-assembled siRNA-spermidine complexes were added drop-wise under vortex to PLGA dissolved in methylene chloride (DCM). This mixture was sonicated using a TMX 400 sonic disruptor (Tekmar, Cincinnati, OH) to yield a primary water-in-oil emulsion. The resultant emulsion was added to TE-buffered 5% (w/v) PVA which contained the various DSPE-PEG constructs at a conjugate to PLGA ratio of 1.6-25.9 (μg/mg). Note that before fabrication, DSPE-PEG conjugates were suspended in either PBS or methanol (DSPE-PEG-FOL). This mixture was sonicated to yield a secondary water-in-oil-in-water emulsion. This emulsion was immediately poured into TE-buffered 0.3% (w/v) PVA and NP were allowed to harden under continuous stirring for 3-4 hours as the DCM evaporated. NP were collected by centrifugation at 16,100 g for 15 min at 4°C, washed three times with deionized water, freeze-dried, and stored desiccated at -20°C. C6-loaded NP were similarly prepared using an oil-in-water single emulsion technique.
2.4. Nanoparticle Characterization
2.4.1. Nanoparticle size and morphology
Samples were visualized using a XL-30 ESEM-FEG scanning electron microscope (SEM) (FEI Company, USA). Dry NP mounted on carbon tape were sputter-coated with a thin layer (25 nm-thick) of gold under vacuum in an argon atmosphere (Dynavac Mini Coater, Dynavac, USA). Average particle diameter and size distribution were measured by micrograph analysis of at least 1,000 particles per batch using image analysis software (ImageJ, National Institute of Health). The average hydrodynamic radius of NP in solution was determined by dynamic light scattering (DLS) using a ZetaPals ζ-potential and particle size analyzer (Brookhaven Instruments).
2.4.2. Nanoparticle loading
The amount of siRNA encapsulated in NP was determined using a modified organic extraction method, as described previously [22]. Briefly, NP were dissolved in DCM for one hour. Two volumes of TE buffer were added to the organic solution, vortexed and centrifuged (16,100 g for 10 min at 4°C) to extract siRNA into the aqueous phase. The amount of siRNA in the resultant aqueous fraction was measured using the QuantIT™ PicoGreen™ assay according the manufacturer's instructions (Invitrogen). Loading of C6 was determined by dissolving NP in DMSO for one hour; C6 content was quantified by spectrofluorescence using a Molecular Devices (Sunnyvale, CA) SpectraMax M5 (excitation/emission: 458/505 nm).
2.3.3. Protection of siRNA
The stability of encapsulated siRNA in the presence of serum was monitored by exposing NP loaded with siRNA (7 ug) to 95% (v/v) FBS at 37°C; an equivalent amount of siRNA was used for the free and cholesterol-conjugated controls. After incubating for 12 hours, RNA was recovered by phenol extraction and concentrated by isopropanol precipitation. RNA integrity was analyzed by 15% TBE-Urea PAGE.
2.3.4. Nanoparticle sustained release
NP were suspended in phosphate buffered saline (pH 7.4), and incubated at 37°C with gentle shaking. Release of siRNA was monitored at several time intervals over 12 days. At each sampling time, the NP suspension was centrifuged for 5 min at 16,100 g. The supernatant was removed for determination of siRNA content, and an equal volume of PBS was replaced for continued monitoring of siRNA release. The amount of siRNA remaining in the NP after 12 days was determined using the organic extraction previously described. Retention of attached DSPE-PEG-FITC was similarly measured; FITC-NP were incubated in saline for 10 days, and supernatant aliquots were periodically analyzed for DSPE-PEG-FITC content (i.e. released ligand).
2.4. Quantification of ligand surface coverage
The amount of FOL associated with NP was determined by incubating NP in methanol (5mg/ml) at 37°C to release DSPE-PEG-FOL. After incubating for 1 hr, PLGA polymer was pelleted by centrifugation at 20,000 g for 5 min. Extracted DSPE-PEG-FOL was analyzed by RP-HPLC on a Shimadzu SIL-10A system (Kyoto, Japan) with an Ascentis C18 column (15 cm × 4.6 mm, 5 μm). The mobile phase consisted of methanol and 10 mM phosphate buffer pH 7 (92:8, v/v) at a flow rate of 1.0 ml/min. DSPE-PEG-FOL was detected by absorbance at both 256 and 368 nm.
The amount of NP-associated ANTP peptide was measured by hydrolyzing NP in 1N NaOH (5mg/ml) to release FITC-labeled DSPE-PEG-ANTP. After incubating for 1 hr, ANTP content was quantified by spectrofluorescence (excitation/emission: 494/520 nm). For both FOL and ANTP, the approximations for ligand density and number of ligands per NP were determined by assuming that the NP were spherical with a density of 1.2 g/cm3 [23].
2.5. Association of ligands with nanoparticles
The association of DSPE-PEG conjugates with NP was confirmed using size exclusion chromatography with a CL-2B column. C6-loaded NP (1 mg) and micelles composed of a mixture of DSPE-PEG-FITC and DSPE-PEG (1:1, 55 nmol) served as size controls; NP (150 nm diameter) eluted in the void volume, while DSPE-PEG-FITC micelles (20-40nm diameter) were fractionated by the gel filtration resin. Fractions (0.1 ml) were collected using a phosphate buffered saline (pH 7.4) elution buffer at a linear flow rate of 15 cm/h, and analyzed by spectrofluorescence.
2.6. Nanoparticle cellular uptake
NP association with KB cells was visualized by confocal microscopy. Cells were grown to 60% confluency on Lab-Tek™ (Rochester, NY) 8-well chambered coverglass. All groups were incubated in folate-free RPMI and 10% FBS, except for the free folate binding control, which contained 2 mM folate. C6-loaded NP were added to cells at a NP to cell ratio of 6×105. After incubating for 16 hrs, cells were extensively washed with PBS to remove unassociated NP. Nuclei were stained with membrane-permeable Hoescht 33342 (2ug/ml), and washed with HBSS-FBS (9:1, v/v). Live cells were analyzed on a Leica (Bannockburn, IL) TCS SP5 Spectral Confocal Microscope.
Cell internalization of NP was monitored using flow cytometry. Cells were grown to 70% confluency, and then C6-loaded NP were added at a NP to cell ratio of 8×104. After incubating for 4 hrs at either 37°C or 4°C, cells were washed with PBS and harvested using cell dissociation buffer. To isolate internalized NP, cells were incubated with 0.1% (w/v) Trypan Blue for 5 min to quench any extracellular fluorescence of live cells (i.e. surface-associated NP). Cells were washed with 1% BSA in PBS and then analyzed on a BD Biosciences FACScan (San Jose, CA).
2.7. Nanoparticle delivery of siRNA
2.7.1. Delivery to cultured cells
Knockdown of luciferase was utilized to assess siRNA delivery in vitro. KB cells that stably expressed luciferase were generated as previously described [24]. Cells were grown to 50% confluency and then incubated with the various siRNA formulations at a final concentration of 100 nM. NP were added based on the determined loading of siRNA. The Lipofectamine™ RNAiMAX control group was prepared using the manufacturer's protocol. After treating for 48 hrs, the cells were washed with PBS and lysed with Passive Lysis Buffer for 30 min at 4°C. Lysates were clarified by centrifugation at 20,000 g for 10 min at 4°C. Luciferase activity was measured on a Promega (Madison, WI) GloMax® 20/20 Luminometer using Luciferase Assay Reagent. Total protein was determined using the Coomasie Plus (Bradford) Protein Assay.
2.7.2. Delivery to tumors
All animal studies were approved by Yale University's Institutional Animal Care and Use Committee. Xenograft tumors were established by subcutaneously injecting luciferase-expressing KB cells (5×106 cells) into the flank of NU/NU nude mice (female, Crl:NU-Foxn1nu, 6 weeks old, Charles River Laboratories). The various siRNA formulations were intratumorally administered after the average tumor volume (0.5×L×W2) reached approximately 100 mm3. Mice received 2 doses (10 μg/kg) that were spaced 2 days apart. Tumors were harvested 7 days after initial treatment. Tumors were homogenized in Cell Culture Lysis Reagent (100mg/ml) at 4°C. After clarifying the homogenates by centrifugation, luciferase activity was measured using Luciferase Assay Reagent. Total protein was determined using the BCA Protein Assay (Reducing Reagent Compatible).
2.10. Statistical analysis
At least three independent experiments were conducted in all studies. Data were analyzed using a two-sided Student's t-test with a p-value of 0.05 or less defined as the threshold for statistical significance.
RESULTS AND DISCUSSION
3.1. Fabrication of nanoparticles
PEGylated phospholipids have been utilized to functionalize colloidal nanoparticulate systems. The addition of amphipathic DSPE-PEG to pre-formed liposomes results in the partitioning of the acyl chains of the DSPE phospholipid into the hydrophobic lipid bilayer, and the outward display of hydrophilic PEG [25,26]. Analogously, amphipathic molecules tend to associate at the surface of the oil-in-water emulsions with the hydrophobic ends associated with the oil phase and the hydrophilic end exposed to the aqueous phase [27,28]. We exploited this amphipathic partitioning effect to functionalize the surface of PLGA NP that were prepared using a modified double emulsion solvent evaporation process. During fabrication, the aqueous phase of the second emulsion comprised DSPE-PEG that associated with the hydrophobic water-in-oil droplets of the primary emulsion. As the solvent diffused and evaporated, the acyl chains remained associated with the hardened polymer, effectively tethering PEG to the surface of the NP.
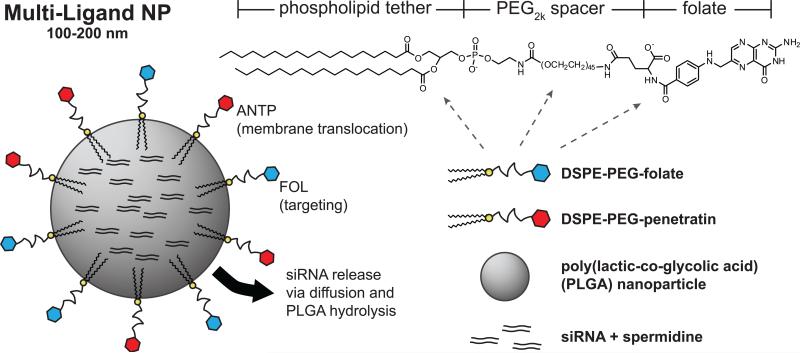
Ligand-coated particles (Fig. 1) were formulated by conjugating ligands to the distal end of DSPE-PEG before addition to the second emulsion. We postulated that the addition of these ligands would not drastically alter the association of DSPE-PEG with the NP because of the strong hydrophobic interactions between DSPE and PLGA dissolved in organic solvent.
1.
Schematic of multi-ligand-coated nanoparticles (ANTP/FOL-NPs). The siRNA-loaded NPs are comprised of a spherical PLGA core coated with phospholipid-linked PEGylated folate and penetratin ligands.
3.2. Characterization of nanoparticles
3.2.1. Size and morphology of nanoparticles
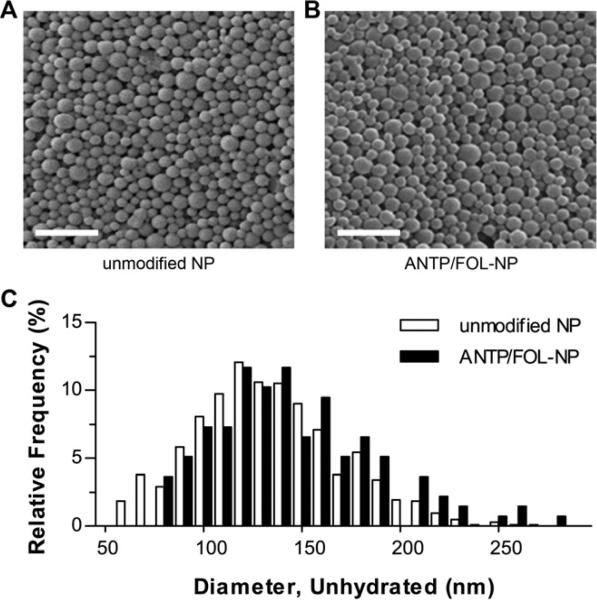
The incorporation of DSPE-PEG onto PLGA NP did not noticeably alter particle size and morphology (Fig. 2). Image analysis of representative SEM micrographs was used to calculate the respective mean diameters of unmodified NP and NP with surface attached ANTP and FOL (ANTP/FOL-NP) as 133 ± 37 nm and 144 ± 62 nm. The size distribution of ANTP/FOL-NP was slightly shifted toward larger particles, relative to unmodified NP (Fig. 2C). While the size difference was not statistically significant, the slightly larger diameter of the ligand-coated NP could be caused by destabilization of the emulsified droplets due to interactions between DSPE-PEG and the water-in-oil interface or the polyvinyl alcohol emulsion stabilizer. Alternately, the addition of PEG2000 to the NP surface could account for a 7-9 nm increase in diameter, depending on whether the PEG is in the “mushroom” or “brush” conformation [29,30]. The tunable PEG surface densities achievable by this method (Table 1), suggests that PEG can coat NP in either conformation [31].
2.
Size and morphology of nanoparticles. Scanning electron micrograph of (A) uncoated nanoparticles and (B) nanoparticles coated with both penetratin (ANTP) and folate (FOL) via coupling to DSPE-PEG. Scale bar represents 1μm. (C) Size distributions of unhydrated nanoparticles, n > 600.
1.
Quantification of nanoparticle-bound ligands. Surface density was measured for nanoparticles coated with only FOL, only ANTP, and both FOL and ANTP; data represent mean ± S.D.; n=3. Surface coverage and molecules per NP were determined using surface area and density approximations for spherical PLGA nanoparticles.
| Ligand | Surface Density (pmol/mg NP) | Surface Coverage (pmol/cm2 NP) | Molecules per NP |
|---|---|---|---|
| FOL (only) | 205 ± 45 | 0.6 ± 0.1 | 260 |
| ANTP (only) | 1734 ± 99 | 5.2 ± 0.3 | 2210 |
| FOL (mixed) | 211 ± 0 | 0.6 ± 0.0 | 270 |
| ANTP (mixed) | 1531 ± 51 | 4.6 ± 0.2 | 1950 |
Dynamic light scattering was used to measure the hydrodynamic diameter of unmodified NPs and ANTP/FOL-NPs, yielding 160 ± 47 nm and 152 ± 35 nm, respectively. An overall size increase upon hydration of the polymer was expected. Contrary to the results from the SEM analysis, the hydrated diameter of the unmodified NP was greater than that of the ANTP/FOL-NP. This disproportionate size increase was likely due to slight aggregation of the unmodified NP; PEG present on the surface of NP can function as both a steric barrier and solubilizing element that can decrease the occurrence of nonspecific interactions and subsequent NP agglomeration [32]. Both NP formulations were small enough to be internalized by cells via endocytosis [33,34].
3.2.2. Quantification of ligand surface coverage
We anticipate that the density of ligands on the surface of NP is directly related to the function and performance of the vehicle [35,36]. Two methods were employed to determine the degree of ligand attachment (Table 1). Spectrophotometric quantitation of FOL on the surface of NP is confounded by high background due to turbidity and light scattering; therefore, HPLC was used to isolate and quantify the extracted analyte. The amount of ANTP coverage was determined using spectrofluorimetry, by quantifying the FITC-labeled peptide after dissolving the NP.
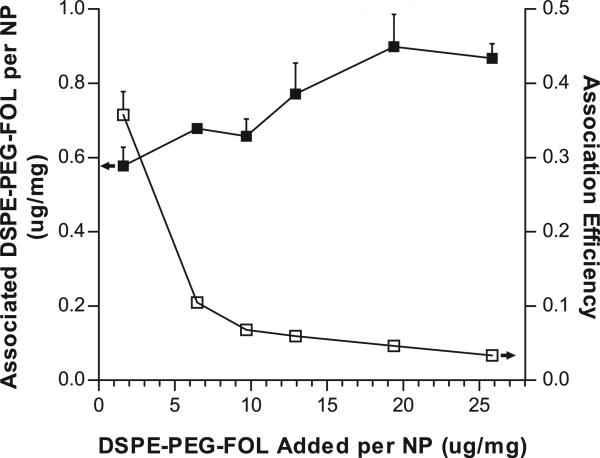
The degree of ligand attachment was proportional to the amount of ligand added during NP fabrication. The initial amount of DSPE-PEG-FOL was titrated from 1.6-25.9 μg ligand/mg NP; the respective association of DSPE-PEG-FOL ranged from 0.6-0.9 μg ligand/mg NP (Fig. 3, left y-axis). However, the association efficiency decreased (ranging from 38.9%-3.0%) as more DSPE-PEG-FOL was added (Fig. 3, right y-axis). Due to these diminishing returns, for this study, the maximum amount of FOL surface decoration was not investigated. Additionally, the amount of ANTP (7.2 μg/mg) used in this work was sufficient to enhance NP delivery.
3.
Degree and efficiency of DSPE-PEG-FOL association with nanoparticles. DSPE-PEG-FOL was titrated during NP fabrication to achieve different surface densities (filled squares, left y-axis). Association efficiency was expressed as the ratio between the amount of DSPE-PEG-FOL associated with NPs and the amount initially added to NPs (empty squares, right y-axis). Data represent mean ± S.D.; n=3.
To compare the efficacies of the different ligand-modifications, it was important to ensure that the ligand densities between the mixed and individual formulations were similar. Respectively, the amounts of DSPE-PEG-FOL and DSPE-PEG-ANTP initially added were 1.3% and 0.7% (w/w, ligand/polymer). The densities of the individual ligands, 205 ± 49 pmol/mg NP for FOL only and 1734 ± 99 pmol/mg NP for ANTP only, were similar to the corresponding densities of the cognate ligands for the mixed formulation, 211 ± 0 pmol FOL/mg NP and 1531 ± 51 pmol ANTP/mg NP for ANTP/FOL-NP. For both ligands, ligand deposition was proportional to the starting amount added; however, the efficiency of association was ~5% for DSPE-PEG-FOL and ~80% for DSPE-PEG-ANTP. The greater efficiency of DSPE-PEG-ANTP association could be a result of electrostatic interactions between the arginine- and lysine-rich peptide with the negatively charged PLGA NP.
Surface density was used to calculate ligand coverage per surface area and the number of ligand molecules per NP. For the ANTP/FOL-NP, the maximal amount of attached DSPE-PEG was 2,220 molecules per NP. The cross-sectional headgroup area of DSPE-PEG is 0.58 or 1.24 nm2, depending on the conformational state of PEG [37]. Assuming a surface monolayer with hexagonal close packing, a spherical NP with a diameter of 150 nm can accommodate approximately 60,000-120,000 DSPE-PEG molecules. Based on these calculations, if the ligand distribution is homogeneous, then these formulations do not form a tightly packed surface coating.
We postulate that residual polyvinyl alcohol stabilizes DSPE-PEG on the NP surface, but also provides a steric barrier that can impede DSPE-PEG association. To test this hypothesis, the attachment of DSPE-PEG-FITC to NP was investigated under different emulsion conditions. NP formulated with both 5% (w/v) PVA and 1.6% (w/w) DSPE-PEG-FITC were coated with 1160 ± 160 pmol FITC/mg NP, while NP synthesized using the same amount of DSPE-PEG-FITC but no PVA were coated with 2960 ± 250 pmol/mg. Therefore, the presence of PVA may impede DSPE-PEG attachment. Due to differences in HLB values, DSPE-PEG and PVA have distinct surfactant properties. PVA is an effective emulsion stabilizer that can yield 150 nm NP in the presence of DSPE-PEG-FITC (Fig. 4A); NP formulated using only DSPE-PEG-FITC (Fig. 4B) appeared to fuse together and not form distinct NP. In the absence of PVA, the addition of DSPE-PEG-FITC with additional 5% (w/v) DSPE-PEG produced NP around 300-500 nm (Fig. 4C) that were coated with 2080 ± 160 pmol FITC/mg NP. These formulations illustrate that certain dynamics between PVA and DSPE-PEG dictate ligand attachment and NP size. In similar DSPE-PEG NP systems, helper lipids like soybean lecithin and 1,2-dilauroylphosphatidylocholine have been added to produce a lipid monolayer on the surface of NP that may aid in the association of DSPE-PEG [38,39]. Such lipid-polymer hybrids have not been extensively characterized, and it is unclear how the lipid coat alters the well-characterized physicochemical and pharmacokinetic properties of PLGA NP. Regardless, the ligand densities achieved with our formulations compare favorably to the densities reported for other effective functionalized NP [40].
4.
Evaluation of emulsion stabilizing properties of DSPE-PEG and PVA. Scanning electron micrograph of NPs stabilized with DSPE-PEG-FITC and (A) 5% (w/v) PVA, (B) no additional stabilizer, and (C) 5% (w/v) DSPE-PEG. Insets represent 10-fold decrease in magnification of corresponding micrograph.
3.2.3. Association of ligands with nanoparticles
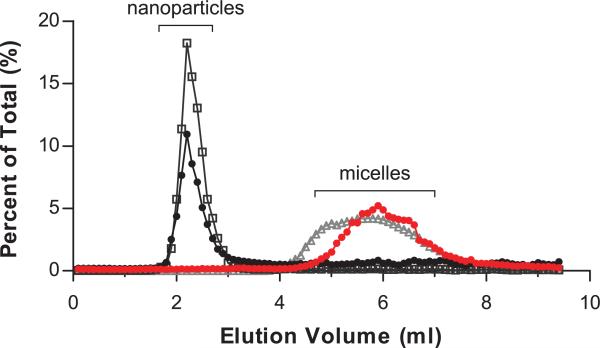
All of the ligand concentrations used during NP synthesis were above the critical micelle concentration (CMC) of DSPE-PEG in water (0.5 μM) [41]. Therefore, during NP fabrication, DSPE-PEG could exist in two energetically favorable states: in micelles and at the interface of the oil-in-water emulsion droplets. (We note that the aqueous phase of the double emulsion consisted of polyvinyl alcohol and trace amounts of methanol, which likely altered the effective CMC of DSPE-PEG [42,43].) Transfer of DSPE-PEG from micelles to liposomes is known to occur spontaneously [25,44]; therefore, we postulated that DSPE-PEG would similarly transfer from micelles to emulsified NP. Although any residual micelles should be washed away during the centrifugation steps of NP fabrication, we used size exclusion chromatography to ensure that the ligands were attached to NP (Fig 5). NP coated with FITC-DSPE-PEG or loaded with coumarin 6 both eluted in the column void volume (~2.3 ml), while FITC-DSPE-PEG micelles and FITC-NP dissolved in NaOH (to release micellar FITC-PEG-DSPE) shared similar elution profiles (~6 ml), which indicated an association between DSPE-PEG and NP.
5.
Size exclusion chromatography confirmed the association of ligands with nanoparticles. Nanoparticles coated with DSPE-PEG-FITC that were suspended in PBS pH 7.4 (black closed circles) eluted similarly to NPs loaded with fluorescent dye (gray open boxes). Nanoparticles coated with DSPE-PEG-FITC that were hydrolyzed in 0.1N NaOH (red closed circles) had a similar elution profile as free micellar DSPE-PEG-FITC (gray open triangles). Shown are representative data from three separate experiments.
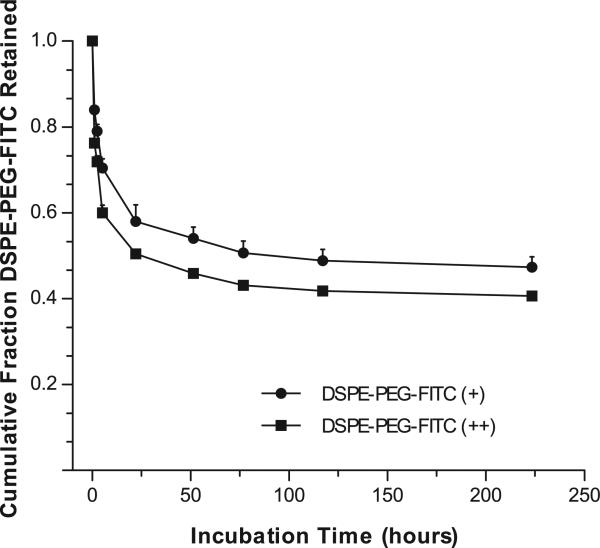
DSPE-PEG exhibits a dynamic association with NP in which the non-covalently attached ligand is released over time (Fig. 6). DSPE-PEG-FITC was attached to NP at surface densities similar to those used for FOL- and ANTP-NP; DSPE-PEG-FITC (+) NP were coated with 232 ± 58 pmol/mg and DSPE-PEG-FITC (++) NP were coated with 1160 ± 160 pmol/mg. At both surface densities, when suspended in saline at 37°C, FITC-NPs exhibited a loss of DSPE-PEG in the first 24 hours, which was followed by sustained ligand retention over the next week. After 10 days, both formulations retained around 50% of the total associated ligand, which implied that DSPE-PEG attaches to NP with different levels of stability. We have developed different methods of surface modification that retain near 100% of ligand over days to weeks [6,27]; however the ligand loss inherent to the DSPE-PEG attachment strategy has certain advantages. For example, targeting ligands and CPPs are only needed for delivering NP to target cells; once the NP are localized, the ligands may serve as a hindrance to payload release. The impact of dynamic DSPE-PEG ligand attachment and drug release may contribute to the efficacy of these NP in delivering siRNA.
6.
Retention of DSPE-PEG-FITC by NPs. In solution, FITC-NPs coated with DSPE-PEG-FITC at different surface densities retain a fraction of the ligand over time; data represent mean ± S.D.; n=3.
3.3. Encapsulation and release of siRNA
3.3.1. Nanoparticle Loading
We have previously reported the encapsulation of a high density of siRNA into PLGA NP [22]; we used similar methods to load siRNA into particles coated with DSPE-PEG conjugates. The loading of siRNA was 132.3 ± 10.9 pmol siRNA/mg NP for unmodified NP and 142.7 ± 23.8 pmol siRNA/mg NP for ANTP/FOL-NP; similar values were measured for FOL-NPs and ANTP-NP. There was no statistically significant difference between the loading values of the different NP formulations, so the presence of the ligand did not appear to affect the encapsulation of siRNA. Note that during NP fabrication, siRNA is incorporated during the primary emulsion, while DSPE-PEG ligands are incorporated during the second emulsion.
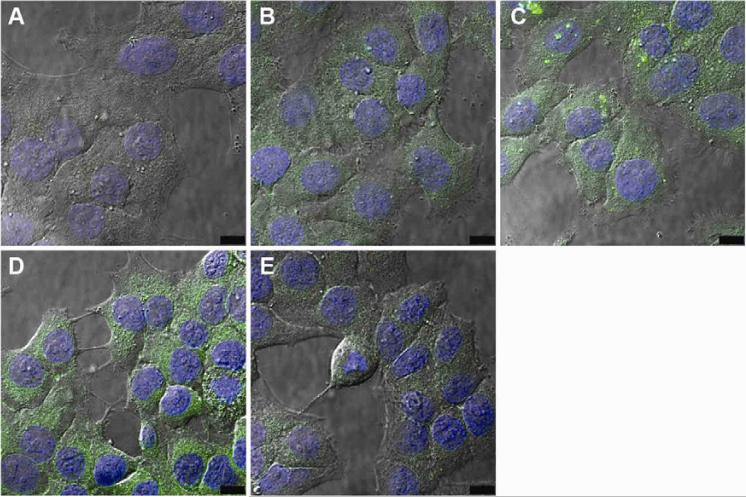
We have also observed that adding DSPE-PEG to NP that were fabricated using a single emulsion technique did not alter drug encapsulation. Coumarin 6 is a hydrophobic dye, so fluorescent NP were formulated by an oil-in-water single emulsion method. All NP formulations encapsulated around 1.7 μg C6/mg NP, which corresponded to 85% encapsulation efficiency. Loading consistency across different NP formulations is essential for comparative cell uptake studies of fluorescently-tagged NP (Fig. 8 and 9).
8.
Confocal microscopy showing the effect of ligand coating on nanoparticle uptake. KB cells were incubated with (A) uncoated NPs, (B) FOL-NPs, (C) ANTP-NPs, (D) ANTP/FOL-NPs, and (E) ANTP/FOL-NPs with excess folate. Each panel represents an experiment of three, in which a single confocal slice with nuclei stained with Hoechst 33342 (blue) and nanoparticles loaded with coumarin 6 (green) is overlaid onto a DIC image.
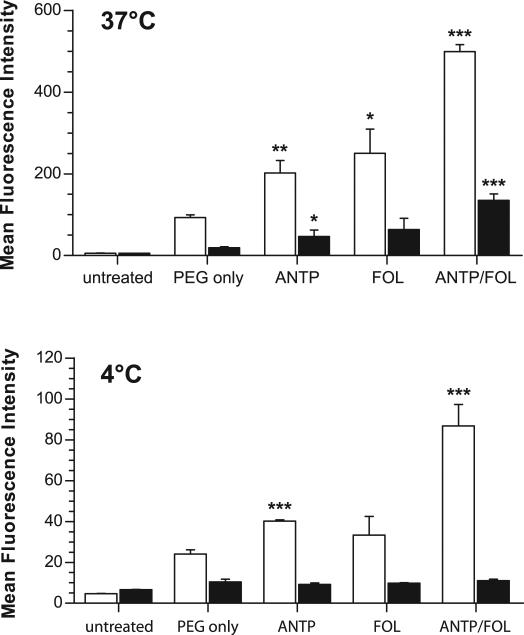
9.
FACS analysis showing the effect of ligand coating on NP-cell surface association and internalization. KB cells were incubated with the indicated nanoparticle formulations at (top) 37°C or (bottom) 4°C to prevent NP uptake. Internalized nanoparticles (black bars) were isolated from total cell-associated nanoparticles (surface and internalized, white bars) by quenching of extracellular fluorescence using trypan blue. Data represent mean ± S.D.; n=3 of 10,000 live-cell gated counts per sample; * p<0.05, ** p<0.005, *** p<0.0005 compared to PEG only control.
3.3.2. Protection of siRNA
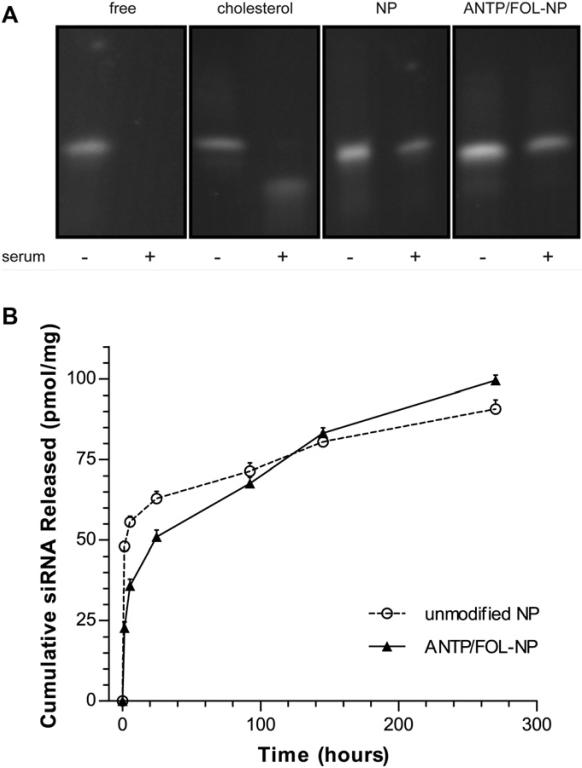
Instability is one of the main limitations in utilizing siRNA as a therapeutic agent; we observed that siRNA encapsulated in NP was protected from serum degradation. NP loaded with siRNA were suspended in 95% FBS for 12 hours, RNase was deactivated, and siRNA was extracted by phenol extraction. Isolated nucleic acids were analyzed by denaturing PAGE (Fig. 7A). Both NP formulations effectively protected encapsulated siRNA. The presence of surface modifications may be more effective than unmodified NP because the presence of DSPE-PEG produces an attenuated burst release phase as well as a steric barrier to RNases. Cholesterol-conjugated siRNA was used as a positive control because it has a greater serum half-life than unmodified siRNA [45]. Both NP formulations were more effective than cholesterol conjugation in preventing siRNA degradation.
7.
PLGA NPs encapsulate and release siRNA. (A) NPs protect siRNA from degradation in serum; siRNA was extracted from NPs, and analyzed by denaturing PAGE. (B) Controlled release profiles of siRNA for ANTP/FOL-NPs and unmodified NPs; data represent mean ± S.D.; n=3.
3.3.3. Sustained release of siRNA
All NP preparations provided a slow release of siRNA molecules during incubation in buffered saline. Unmodified and ligand-coated NP both exhibited a substantial burst release; after 48 hours, unmodified NP released 48% of the loaded siRNA and ANTP/FOL-NP released 36% (Fig. 7B). After the burst phase, both formulations showed sustained release; however, ANTP/FOL-NP released siRNA at a faster rate than unmodified NP. After 11 days, unmodified NP released an additional 17% and ANTP/FOL-NP released an additional 35% of the total amount of loaded siRNA per formulation. Of note, FOL-NP and ANTP-NP had similar release profiles as ANTP/FOL-NPs (data not shown). DSPE-PEG present on the surface of NP may function as a steric and hydrophobic barrier that limits diffusion and release of siRNA from the NP, which would explain the lower burst with particles coated with DSPE-PEG and its derivatives.
The increased siRNA release rate after the attenuated burst phase (after around 6 hrs of incubation) is likely due to the concomitant release of the DSPE-PEG (Fig. 6). Within 6 hrs, 30-40% of the attached DSPE-PEG has been released. (We note that properties of the attached ligand may impact NP retention, so FITC may exhibit different release kinetics than FOL and ANTP.) This loss of DSPE-PEG would relieve the diffusion constraint on the encapsulated siRNA. Alternately, the higher long-term release rate for particles coated with DSPE-PEG molecules might be simply due to the lower burst, which results in a higher internal siRNA concentration at later time and, therefore, a higher siRNA gradient to support release.
As shown below, significant NP uptake by target cells occurred within the first hour of administration. Although release from NP persists for long periods of times, a substantial portion of the siRNA may be released before sufficient intracellular localization occurs; this effect is particularly important for unmodified particles which released 48.1 ± 1.5 pmol siRNA/mg NP during the first hour in solution (Fig. 7B). The reduction of the extent of burst, which we have observed in all ligand-coated NP (e.g. ANTP/FOL-NP released 22.8 ± 1.8 pmol/mg over the first hour), may be important for the successful delivery of sufficient quantities of siRNA to the cytosol.
3.4. Cellular uptake of targeted nanoparticles
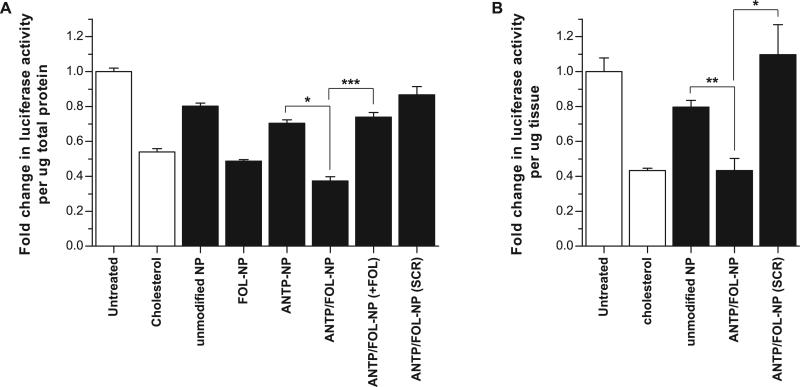
We used confocal microscopy to investigate the impact of ligand modification on cellular uptake of NPs loaded with fluorescent dye. All NP formulations were internalized by KB cells, as indicated by nuclear exclusion and occupation of a substantial proportion of the intracellular milieu (Fig. 8). Relative to the cell-associated fluorescence observed for unmodified NPs (Fig. 8A), ANTP-NP (Fig. 8B) and FOL-NP (Fig. 8C) exhibited more total fluorescence and thus greater uptake. Flow cytometry was used to quantify these differences in total cell-associated fluorescence (Fig. 9; unmodified NP = 92.8 ± 6.7 RFU, ANTP-NP = 202.3 ± 30.0 RFU, and FOL-NPs = 250.5 ± 58.7 RFU). Compared to all other formulations ANTP/FOL-NP (Fig. 8D) produced the maximal amount of cellular accumulation (499.3 ± 17.0 RFU). The addition of excess folate to compete out folate-mediated binding resulted in a decrease in the amount of ANTP/FOL-NP uptake (Fig. 8E) that was similar to the level of ANTP-NP, yet greater than that of unmodified NP. This observation indicates that FOL and ANTP function additively to increase cellular uptake of NP.
3.5. Inhibition of gene expression using nanoparticles
3.5.1. Gene silencing in vitro
We evaluated the ability of ligand-modified NP to deliver siRNA and knockdown luciferase gene expression in vitro. NP were added to culture medium and suspended above KB cells that had been stably transfected with luciferase. As expected based on the measurements of cellular uptake (Fig. 8 and 9), the extent knockdown was dependent on ligand functionalization. Relative to untreated samples, unmodified NP produced 20% knockdown, while ANTP-NP and FOL-NP reduced luciferase expression by 30% and 51%, respectively (Fig. 10A). Although FOL-NP and ANTP-NP exhibited similar levels of cell uptake, FOL-NP generated almost 2-fold greater knockdown than ANTP-NP, suggesting that FOL-NP and ANTP-NP, which are equally effective at entering cells, have different intracellular fates. Folate is an ideal targeting ligand because, after folate receptor-mediated endocytosis, FOL is released into the cytosol for use in metabolic processes [46]. Exploitation of this entry pathway should promote the localization of attached cargo in the cytosol; in effect, avoiding endo-lysosomal degradation. On the contrary, the mechanism of penetratin uptake and localization is unclear, so vesicular sequestration of attached cargo is a possibility [47].
10.
Luciferase gene silencing effect of different ligand-coated NP formulations (black bars) compared to untreated and cholesterol-conjugated siRNA controls (white bars) delivered (A) in vitro and (B) in vivo. Luciferase signal was normalized to total protein level. (+FOL) indicates addition of 2mM free folate, (SCR) indicates negative control siRNA. Data represent mean ± S.D.; (A) n=3, (B) n=5; * p<0.05, ** p<0.005, *** p<0.0005.
ANTP/FOL NP were the most effective gene silencing formulation; they produced 63% knockdown (Fig. 10A). This decrease in gene expression was statistically significant compared to FOL-NP (p<0.05). The addition of excess folate lowered the knockdown efficacy of ANTP/FOL-NP to the level of ANTP-NP (p<0.0005), which further suggested an additive relationship between the two ligands. ANTP/FOL-NP loaded with a negative control siRNA did not produce statistically significant knockdown relative to untreated cells.
Cholesterol-conjugated siRNA has been used extensively for in vitro and in vivo gene silencing [45]. We used chol-siRNA as a positive control, and observed that after 48 hours, FOL-NPs and ANTP/FOL-NP elicited greater knockdown than chol-siRNA (46%) in culture. Typical siRNA-mediated knockdown lasts between 3-7 days; however, we have recently shown that siRNA-loaded PLGA NP can achieve sustained gene expression inhibition for at least 14 days [22]. In summary, ligand-coated NP achieve greater knockdown levels than cholesterol-conjugated siRNA, and they show promise for sustained knockdown due to controlled release of siRNA over time.
3.5.2. Gene silencing in vivo
We used a xenograft tumor model to assess the in vivo efficacy of ANTP/FOL-NP. Tumors were established in nude mice using luciferase-expressing KB cells. Direct intratumoral injection of ANTP/FOL-NP resulted in 60% knockdown, which was comparable to that achieved by the cholesterol-conjugated siRNA control, as well as statistically significant relative to unmodified NP and ANTP/FOL-NP loaded with an untargeted control siRNA (Fig. 10B). Because ANTP/FOL-NP were more effective than unmodified NP, the presence of both ligands enhanced the delivery efficacy, presumably through improved targeting and cell penetration. PLGA NP are effective in vivo drug delivery vehicles [32]; this tumor model demonstrates the utility of modifying NP with both FOL and ANTP.
3.6. Enhanced cell-surface association of ligand-coated NPs
To understand why ANTP/FOL-NP were more effective than the individually modified FOL-NP and ANTP-NP, we investigated the specific roles of the ligands. Folate increases NP avidity by binding to extracellular folate receptors, and it also enhances cell uptake by triggering receptor-mediated endocytosis [48]. ANTP enhances cell uptake by translocation across the plasma membrane, but it may also increase NP avidity through electrostatic interactions of basic peptide residues with the negatively charged cell surface [47]. Furthermore, there is evidence that CPPs, such as ANTP, bind to cell surfaces via interactions with glycosaminoglycans [15]. We hypothesize that one advantage of ANTP/FOL-NP is an increase in overall NP avidity due to the presence of both FOL and ANTP ligands.
To test this hypothesis, we used FACS to monitor cellular association and internalization of NP. NP uptake via receptor-mediated endocytosis and CPP-mediated entry is temperature dependent—although the mechanism and conditions for CPP-mediated uptake are still unclear [47]. At both 37°C (Fig. 9, top, white bars) and 4°C (Fig. 9, bottom, white bars), all ligand-coated NP formulations showed enhanced cell association compared to control NP coated with only PEG-DSPE. At both temperatures, ANTP/FOL-NP showed an increase in cell-association relative to the both ANTP-NP and FOL-NP. If ANTP only functioned at the cell-entry level, the amount of cell-associated ANTP/FOL-NP would be similar to the amount of cell-associated FOL-NP at 4°C, in the absence of cell entry. However, at 4°C, ANTP/FOL-NP exhibited a more than 2-fold increase in cell-association than FOL-NP. Therefore, ANTP has an active role in enhancing the binding of ANTP/FOL-NP to cell surfaces.
Some of the earliest reports on the mechanisms of CPPs purport that cell entry is temperature-independent; however, more recent studies refute those claims [40,47]. To ensure that cell entry of CPP-bound NP was inhibited at low temperatures, we used trypan blue to quench the fluorescence of surface-bound NP (Fig. 9, black bars). Trypan blue is impermeable to the membranes of live cells, and has been used as a tool to quench extracellular fluorescence, particularly in phagocytosis assays [49]. (We note that acid-washing is commonly used to remove cell surface-bound materials [50] and could have been used as an alternate method here; however, the acidic pH required for this technique may affect the release of the lipophilic C6 dye from the surface-bound PLGA NP, and result in false-positive cellular fluorescence [51].) At 37°C, the amount of internalized NP was proportional to the amount of total cell-associated fluorescence for each formulation. At 4°C, all of the internalized NP were near background levels, indicating that few NP were internalized and most were surface-bound. These results indicate that cell entry of ANTP-coated NP is temperature-dependent, and support the assertion that ANTP functions primarily to enhance NP avidity.
Conclusion
We have developed a siRNA delivery vehicle comprising a PEGylated PLGA NP that exhibits enhanced knockdown efficacy by exploiting the synergistic activity of two different ligands: folate and penetratin. ANTP/FOL-NP display a high density of surface ligands, encapsulate and protect large amounts of siRNA, and demonstrate favorable siRNA release kinetics. Furthermore, the ligands function in concert to enhance cellular uptake and subsequent gene knockdown. Further investigation is necessary to understand the exact functions of FOL and ANTP in enhancing the efficacy of ANTP/FOL-NP, but our findings suggest that at the level of cell-association, both ligands increase the avidity of multi-functional NP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tong R, Cheng J. Anticancer polymeric nanomedicines. Polymer Reviews. 2007;47:345–381. [Google Scholar]
- 2.Cartiera MS, Johnson KM, Rajendran V, Caplan MJ, Saltzman WM. The uptake and intracellular fate of PLGA nanoparticles in epithelial cells. Biomaterials. 2009;30:2790–2798. doi: 10.1016/j.biomaterials.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen MØ, Lichawska A, Arpanaei A, Rask Jensen SM, Kaur H, Oupicky D, et al. Surface functionalisation of PLGA nanoparticles for gene silencing. Biomaterials. 2010;31:5671–5677. doi: 10.1016/j.biomaterials.2010.03.069. [DOI] [PubMed] [Google Scholar]
- 4.Fahmy TM, Fong PM, Goyal A, Saltzman WM. Targeted for drug delivery. Materials Today. 2005;8:18–26. [Google Scholar]
- 5.Cu Y, LeMoëllic C, Caplan MJ, Saltzman WM. Ligand-modified gene carriers increased uptake in target cells but reduced DNA release and transfection efficiency. Nanomedicine. 2010;6:334–343. doi: 10.1016/j.nano.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keegan ME, Saltzman WM. Surface-modified biodegradable microspheres for DNA vaccine delivery. Methods Mol Med. 2006;127:107–113. doi: 10.1385/1-59745-168-1:107. [DOI] [PubMed] [Google Scholar]
- 7.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 8.Gu F, Karnik R, Wang A, Alexis F, Levy-Nissenbaum E, Hong S, et al. Targeted nanoparticles for cancer therapy. Nano Today. 2007;2:14–21. [Google Scholar]
- 9.Zureikat AH, McKee MD. Targeted therapy for solid tumors: current status. Surg Oncol Clin N Am. 2008;17:279–301. doi: 10.1016/j.soc.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Low PS, Antony AC. Folate receptor-targeted drugs for cancer and inflammatory diseases. Adv Drug Deliv Rev. 2004;56:1055–1058. doi: 10.1016/j.addr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Salazar M, Ratnam M. The folate receptor: what does it promise in tissue-targeted therapeutics? Cancer Metastasis Rev. 2007;26:141–152. doi: 10.1007/s10555-007-9048-0. [DOI] [PubMed] [Google Scholar]
- 12.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Simnick AJ, Valencia CA, Liu R, Chilkoti A. Morphing low-affinity ligands into high-avidity nanoparticles by thermally triggered self-assembly of a genetically encoded polymer. ACS Nano. 2010;4:2217–2227. doi: 10.1021/nn901732h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng Y, Liu J, Hong R. Translocation of liposomes into cancer cells by cell-penetrating peptides penetratin and TAT: a kinetic and efficacy study. Mol Pharmacol. 2002;62:864–872. doi: 10.1124/mol.62.4.864. [DOI] [PubMed] [Google Scholar]
- 15.Console S, Marty C, Garcia-Echeverria C, Schwendener R, Ballmer-Hofer K. Antennapedia and HIV transactivator of transcription (TAT) “Protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J Biol Chem. 2003;278:35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 16.Sugita T, Yoshikawa T, Mukai Y, Yamanada N, Imai S, Nagano K, et al. Improved cytosolic translocation and tumor-killing activity of TAT-shepherdin conjugates mediated by co-treatment with TAT-fused endosome-disruptive HA2 peptide. Biochem Biophys Res Commun. 2007;363:1027–1032. doi: 10.1016/j.bbrc.2007.09.077. [DOI] [PubMed] [Google Scholar]
- 17.Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, et al. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res. 2007;67:2938–43. doi: 10.1158/0008-5472.CAN-06-4535. [DOI] [PubMed] [Google Scholar]
- 18.Zhao P, Wang H, Yu M, Cao S, Zhang F, Chang J, et al. Paclitaxel-loaded, folic-acid-targeted and TAT-peptide-conjugated polymeric liposomes: in vitro and in vivo evaluation. Pharm Res. 2010;27:1914–1926. doi: 10.1007/s11095-010-0196-5. [DOI] [PubMed] [Google Scholar]
- 19.Thorén PE, Persson D, Karlsson M, Nordén B. The antennapedia peptide penetratin translocates across lipid bilayers - the first direct observation. FEBS Lett. 2000;482:265–268. doi: 10.1016/s0014-5793(00)02072-x. [DOI] [PubMed] [Google Scholar]
- 20.Ziegler A, Seelig J. Binding and clustering of glycosaminoglycans: a common property of mono- and multivalent cell-penetrating compounds. Biophys J. 2008;94:2142–2149. doi: 10.1529/biophysj.107.113472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–96. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Mark Saltzman W. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vauthier C, Schmidt C, Couvreur P. Measurement of the density of polymeric nanoparticulate drug carriers by isopycnic centrifugation. J Nanopart Res. 1999;1:411–418. [Google Scholar]
- 24.Goodson HV, Dzurisin JS, Wadsworth P. Generation of stable cell lines expressing GFP-tubulin and photoactivatable-GFP-tubulin and characterization of clones. Cold Spring Harb Protoc. 2010:2010. doi: 10.1101/pdb.prot5480. pdb.prot5480. [DOI] [PubMed] [Google Scholar]
- 25.Uster PS, Allen TM, Daniel BE, Mendez CJ, Newman MS, Zhu GZ. Insertion of poly(ethylene glycol) derivatized phospholipid into pre-formed liposomes results in prolonged in vivo circulation time. FEBS Lett. 1996;386:243–246. doi: 10.1016/0014-5793(96)00452-8. [DOI] [PubMed] [Google Scholar]
- 26.Silvander M, Johnsson M, Edwards K. Effects of PEG-lipids on permeability of phosphatidylcholine/cholesterol liposomes in buffer and in human serum. Chem Phys Lipids. 1998;97:15–26. doi: 10.1016/s0009-3084(98)00088-7. [DOI] [PubMed] [Google Scholar]
- 27.Fahmy TM, Samstein RM, Harness CC, Mark Saltzman W. Surface modification of biodegradable polyesters with fatty acid conjugates for improved drug targeting. Biomaterials. 2005;26:5727–36. doi: 10.1016/j.biomaterials.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 28.Duncanson WJ, Figa MA, Hallock K, Zalipsky S, Hamilton JA, Wong JY. Targeted binding of PLA microparticles with lipid-PEG-tethered ligands. Biomaterials. 2007;28:4991–4999. doi: 10.1016/j.biomaterials.2007.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moghimi S. The effect of methoxy-PEG chain length and molecular architecture on lymph node targeting of immuno-PEG liposomes. Biomaterials. 2006;27:136–144. doi: 10.1016/j.biomaterials.2005.05.082. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Huang L. Nanoparticles evading the reticuloendothelial system: Role of the supported bilayer. BBA Biomembranes. 2009;1788:2259–2266. doi: 10.1016/j.bbamem.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tirosh O, Barenholz Y, Katzhendler J, Priev A. Hydration of polyethylene glycol-grafted liposomes. Biophys J. 1998;74:1371–1379. doi: 10.1016/S0006-3495(98)77849-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avgoustakis K. Pegylated poly(lactide) and poly(lactide-co-glycolide) nanoparticles: preparation, properties and possible applications in drug delivery. Curr Drug Deliv. 2004;1:321–333. doi: 10.2174/1567201043334605. [DOI] [PubMed] [Google Scholar]
- 33.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 34.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gindy ME, Ji S, Hoye TR, Panagiotopoulos AZ, Prud'homme RK. Preparation of poly(ethylene glycol) protected nanoparticles with variable bioconjugate ligand density. Biomacromolecules. 2008;9:2705–2711. doi: 10.1021/bm8002013. [DOI] [PubMed] [Google Scholar]
- 36.Rux, Gref R, Couvreur P, Barratt G, Mysiakine E. Surface-engineered nanoparticles for multiple ligand coupling. Biomaterials. 2003;24:4529–4537. doi: 10.1016/s0142-9612(03)00348-x. [DOI] [PubMed] [Google Scholar]
- 37.Garbuzenko O, Barenholz Y, Priev A. Effect of grafted PEG on liposome size and on compressibility and packing of lipid bilayer. Chem Phys Lipids. 2005;135:117–129. doi: 10.1016/j.chemphyslip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Chan JM, Gu FX, Rhee J, Wang AZ, Radovic-Moreno AF, et al. Self-assembled lipid−polymer hybrid nanoparticles: a robust drug delivery platform. ACS Nano. 2008;2:1696–1702. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Li K, Pan J, Liu B, Feng S. Folic acid conjugated nanoparticles of mixed lipid monolayer shell and biodegradable polymer core for targeted delivery of Docetaxel. Biomaterials. 2010;31:330–338. doi: 10.1016/j.biomaterials.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 40.Torchilin VP, Rammohan R, Weissig V, Levchenko TS. TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors. Proc Natl Acad Sci U S A. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashok B, Arleth L, Hjelm RP, Rubinstein I, Onyüksel H. In vitro characterization of PEGylated phospholipid micelles for improved drug solubilization: effects of PEG chain length and PC incorporation. J Pharm Sci. 2004;93:2476–2487. doi: 10.1002/jps.20150. [DOI] [PubMed] [Google Scholar]
- 42.Anderson MT, Martin JE, Odinek JG, Newcomer PP. Effect of methanol concentration on CTAB micellization and on the formation of surfactant-templated silica (STS). Chem Mater. 1998;10:1490–1500. [Google Scholar]
- 43.Akhter MS, Alawi SM. The effect of organic additives on critical micelle concentration of non-aqueous micellar solutions. Colloids Surf A Physicochem Eng Asp. 2000;175:311–320. [Google Scholar]
- 44.MacKay JA, Li W, Huang Z, Dy EE, Huynh G, Tihan T, et al. HIV TAT peptide modifies the distribution of DNA nanolipoparticles following convection-enhanced delivery. Mol Ther. 2008;16:893–900. doi: 10.1038/mt.2008.36. [DOI] [PubMed] [Google Scholar]
- 45.Lorenz C, Hadwiger P, John M, Vornlocher H, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Sabharanjak S, Mayor S. Folate receptor endocytosis and trafficking. Adv Drug Deliv Rev. 2004;56:1099–1109. doi: 10.1016/j.addr.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, et al. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 48.Lee RJ, Low PS. Folate-mediated tumor cell targeting of liposome-entrapped doxorubicin in vitro. BBA Biomembranes. 1995;1233:134–144. doi: 10.1016/0005-2736(94)00235-h. [DOI] [PubMed] [Google Scholar]
- 49.Wan CP, Park CS, Lau BH. A rapid and simple microfluorometric phagocytosis assay. J Immunol Methods. 1993;162:1–7. doi: 10.1016/0022-1759(93)90400-2. [DOI] [PubMed] [Google Scholar]
- 50.Haigler HT, Maxfield FR, Willingham MC, Pastan I. Dansylcadaverine inhibits internalization of 125I-epidermal growth factor in BALB 3T3 cells. J Biol Chem. 1980;255:1239–1241. [PubMed] [Google Scholar]
- 51.Zolnik BS, Burgess DJ. Effect of acidic pH on PLGA microsphere degradation and release. J Control Release. 2007;122:338–344. doi: 10.1016/j.jconrel.2007.05.034. [DOI] [PubMed] [Google Scholar]