Abstract
A recombinant adenovirus (rAd) expressing Cre recombinase derived from bacteriophage P1 has already been extensively used for the conditional gene activation and inactivation strategies in mammalian systems. In this study, we generated AxCAFLP, a rAd expressing FLP recombinase derived from Saccharomyces cerevisiae and carried out quantitative comparisons with Cre-expressing rAd in both in vitro and in cultured cells to provide another efficient gene regulation system in mammalian cells. In the in vitro experiments, the relative recombination efficiency of FLP expressed in 293 cells infected with FLP-expressing rAd was approximately one-thirtieth that of Cre even at 30°C, the optimum temperature for FLP activity, and was approximately one-ninetieth at 37°C. Co-infection experiments in HeLa cells using a target rAd conditionally expressing LacZ under the control of FLP showed that an FLP-expressing rAd, infected at a multiplicity of infection (MOI) of 5, was able to activate the transgene in almost 100% of HeLa cells whereas the Cre-expressing rAd was sufficient at an MOI of 0.2. Since an MOI of 5 is ordinarily used in rAd experiments, these results showed that the FLP-expressing rAd is useful for gene activation strategies and is probably applicable to a sequential gene regulation system in combination with Cre-expressing rAd in mammalian cells.
INTRODUCTION
Site-specific recombinases have become a powerful tool for the regulation of the expression of transgenes (for reviews see 1–3). The site-specific recombinase Cre, derived from bacteriophage P1, has been widely applied to the gene activation and inactivation strategies in mammalian systems, in cell lines (e.g. 4,5) and in transgenic mice (e.g. 6–8). Moreover, a Cre-expressing recombinant adenovirus (rAd) has provided a ‘molecular switch’ to turn on or off the transgene expression in cell lines (9–18) or in mice (19–33). Despite their widespread application in many fields, these strategies of transgene regulation can only be carried out once, i.e. ‘off to on’ or ‘on to off’, when manipulated by Cre alone. However, ‘off-on-off’ or ‘on-off-on’ regulation is more attractive when verifying the function of a given gene, and would be valuable for stage-specific serial gene regulation in developmental studies. However, to regulate the expression of transgenes in serial steps, we need novel regulation systems mediated by recombinases other than Cre.
Another well-characterized site-specific recombinase FLP (34–38), derived from Saccharomyces cerevisiae, has also been applied to gene regulation in plants (39,40), Drosophila (41–43) and mammals (44–48). The use of FLP in combination with Cre may facilitate sequential gene regulation; however, only a few reports have been reported to date (49), probably due to the relatively low activity of FLP at 37°C, the optimum temperature of FLP being 30°C (50). In fact, a mutant FLP, FLPe, has been developed recently (51), which has overcome its low activity at 37°C.
In this report, we generated a rAd expressing wild-type FLP with the aim of using it as a ‘second molecular switch’ in cultured mammalian cells. We quantified the recombination efficiency of FLP expressed in 293 cells at 37°C and found that the activity of FLP was several-tens of times lower than that of Cre. However, we showed that such low activity can be readily overcome as long as an efficient rAd expression system under the control of a potent CAG promoter (52) was used. The FLP-expressing rAd was able to activate a transgene in virtually 100% of cultured mammalian cells, suggesting the usefulness of FLP-expressing rAd as a secondary regulation system in combination with the Cre-expressing rAd.
MATERIALS AND METHODS
Cell lines
The human embryonic kidney cell line, 293 (53), and the human cervical cancer-derived cell line, HeLa, were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS). The 293 cells constitutively express adenoviral E1 genes and support replication of E1-substituted rAds. After the infection of rAds, cells were maintained in DMEM supplemented with 5% FCS.
Construction of the target unit CAFNF and the FLP expression unit
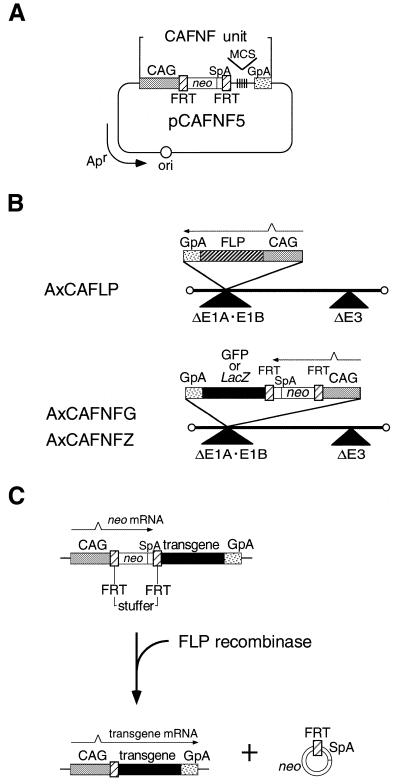
The FLP recognition target (FRT) sequence used in this work was a synthesized nucleotide of 54 nt (5′-AAATtccgga[BspEI]GAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCgacgtc[AatII]ATTT-3′), where the 34 nt functional FRT site (underlined) was flanked by BspEI and AatII sites (lowercase letters) in that order. A head-to-tail dimer was cloned at the SmaI site of pUC18 (termed pUFwF) and used as a donor of a pair of FRT sequences. pCAFNF5 (Fig. 1A) was constructed as a cassette plasmid containing a target unit, CAFNF, consisting of the CAG promoter (52), the first FRT sequence, a neo-resistance gene (1.0 kb BglII–SmaI fragment derived from pSV2neo), the second FRT sequence and a rabbit β-globin polyadenylation [poly(A)] signal in that order followed by multi-cloning sites (MCS) (SwaI–EcoRI–SacI–KpnI–SmaI). pCAFNFG and pCAFNFZ, the target plasmids, were constructed by inserting green fluorescent protein (GFP) and LacZ genes, respectively, as the reporter gene at the MCS of pCAFNF5. The source of the GFP gene was pxCAEGFP, originally derived from pEGFP-C1 (EGFP vector; Clontech), introducing a termination codon at the BglII site. The coding sequence of LacZ was derived from pSRLacZ (54). pCAFNFG was also used as substrate DNA for the in vitro recombination assay of the FLP/FRT system.
Figure 1.
Transgene activation strategy mediated by FLP recombinase. (A) Structure of the cassette plasmid, pCAFNF5, possessing the CAFNF target unit. CAG, CAG promoter; SpA, SV40 early poly(A) signal; MCS, multi-cloning site; GpA, rabbit β-globin poly(A) signal. The stuffer region is composed of the neo gene and the SpA flanked by a pair of FRT. (B) Structure of the FLP-expressing rAd, AxCAFLP, and the target rAds, AxCAFNFG and AxCAFNFZ. A triangle under the adenovirus genome represents the deletion of an adenovirus sequence. (C) Activation of the transgene by CAFNF target unit mediated by FLP recombinase. The horizontal arrows indicate the orientation of the transcription. FLP-mediated excisional deletion removes the stuffer region, resulting in the expression of the reporter genes, GFP or LacZ, under the control of the CAG promoter.
The initiation codon of the coding region of FLP (1.5 kb SphI–XbaI fragment from pUC19-Flp, a generous gift from Dr M. Jayaram) was altered to fit Kozak’s rule (55). The SphI digests at the initiation site including the initiation codon, so that the synthesized nucleotide (5′-agctt[HindIII end]ctgcag[PstI]CAGACCGTGCATcatg[SphI-compatible end]-3′) fitting Kozak’s rule was ligated to the SphI–XbaI fragment derived from pUC19-Flp. The resulting fragment was then cloned at the HindIII–XbaI site of pUC19 (termed pUkFLP). The pUkFLP was used as a donor of the FLP sequence.
Since our aim is to compare the recombination efficiency between FLP and Cre, we used Cre-expression unit as described (9) whose initiation codon has also been altered to fit Kozak’s rule by tagging SV40 T antigen-derived nuclear localization signal (NLS, underlined) (5′-ctgcag[PstI]CAGACCGTGCATCATGAGCGGCCCTCCAAAAAAGAAGAGAAAGGTAGAAGACCCGGgcggccgc[NotI]-3′).
Generation of rAds
The rAds were generated by the COS-TPC method as described (54). Briefly, both FLP target units, CAFNFG and CAFNFZ, excised from pCAFNFG and pCAFNFZ, respectively, as a SalI–HindIII fragment, were cloned into a cassette cosmid pAxcw (54). The FLP gene was excised from pUkFLP with PstI and FspI and cloned into an expression cassette cosmid, pAxCAwt (termed pAxCAFLP), containing CAG promoter and poly(A) signal. Each cosmid was mixed with adenovirus DNA-terminal protein complex and transfected into 293 cells, resulting in the generation of rAds through homologous recombination. Target rAds containing the CAFNFG unit (AxCAFNFG), CAFNFZ unit (AxCAFNFZ) and FLP-expressing rAd (AxCAFLP) were generated (Fig. 1B). A target rAd, AxCALNLG, which expresses the GFP gene described above under the regulation of Cre, was also generated by inserting a target unit, CALNLG, into pAxcw. A purified and concentrated virus stock was prepared as described (56).
In vitro recombination assay
The in vitro recombination assay of the FLP/FRT system in this paper was essentially as described (57). FLP-containing lysate was prepared as follows. In total, 2 × 107 of 293 cells were infected with AxCAFLP at an MOI of 5. Twenty-two hours later, cells were harvested by scraping and collected by centrifugation at 1000 r.p.m. for 5 min at 4°C using Avanti HP-25I (Beckman-Coulter). The cell pellet was washed twice with PBS(–) and resuspended with 1 ml of FLP storage buffer (10% glycerol, 20 mM Tris–HCl pH 7.5, 300 mM NaCl, 1 mM EDTA and 0.02 mM phenylmethylsulfonyl fluoride). The cell suspension was then sonicated for 3 min (six cycles, 30 s each) using Bioruptor II (CosmoBio; Tokyo, Japan) at maximum power (200 W) and immediately cetrifuged at 10 000 r.p.m. for 20 min at 4°C. The supernatant was stored at –80°C.
The reaction was started by mixing HindIII-linearized pCAFNFG with FLP lysate (2.5–25 µl), and incubated for 30 min at 37 or 30°C in a 50 µl volume of FLP reaction buffer (25 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 5 mM DTT). The reaction was terminated by phenol/chloroform extraction followed by two cycles of chloroform extraction and ethanol precipitation. The recovered DNA was then digested with FspI and DNA digests were detected by ethidium bromide staining after agarose gel electrophoresis. The recombination efficiency was calculated by quantifying the density of the DNA bands on the photograph based on densities of the quantification-standard DNA digests on the same gel as described (58). To compare the recombination efficiency of FLP and Cre, an in vitro recombination assay of Cre/loxP system was also carried out by using EcoRI-linearized pCALNLZ (9) and Cre lysate (0.1–5.0 µl) as described (58).
GFP or LacZ gene activation by recombinase-expressing rAds
To compare the recombination efficiency of FLP and Cre recombinase in cultured mammalian cells, target rAds and the recombinase-expressing rAds were co-infected into HeLa cells. Either AxCAFLP or AxCANCre (NLS-tagged Cre-expressing rAd) (9) was infected to express each recombinase. Either AxCAFNFG or AxCAFNFZ was infected to introduce the reporter genes, GFP or LacZ, respectively, to assay the regulation of FLP. Similarly, either AxCALNLG or AxCALNLZ (9) was infected to assay the regulation of Cre. When each recombinase recognizes their target units, it excises the stuffer region, composed of the neo gene and the poly(A) signal flanked by a pair of recombinase target sequences, resulting in the expression of the transgene (Fig. 1C). The recombination efficiency of FLP and Cre recombinase was compared by measuring the fluorescence intensity of expressed GFP or by staining LacZ-expressed cells with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). Three days after co-infection, the fluorescence intensity of GFP was quantified by Fluoroskan Ascent FL (Labsystems). For cell staining, 3 days after co-infection, cells were washed with PBS(–) twice, fixed with 0.25% glutaraldehyde and stained with 0.1% X-gal.
RESULTS
Construction of the target unit CAFNF
We first constructed a cassette plasmid pCAFNF5 (Fig. 1A), which can activate the expression of a transgene inserted into its MCS mediated by FLP, based on the strategy mediated by Cre established previously (9,12). The stuffer region, composed of the neo gene and the SV40 early poly(A) signal (SpA) flanked by a pair of target sequences, hampers the expression of the foreign gene so that the target unit CAFNF initially expresses the neo gene. When functional FLP is supplied, the FLP-mediated excisional deletion removes the stuffer region, resulting in the expression of the transgene under the control of the CAG promoter (Fig. 1C). We confirmed whether the CAFNF unit works in cultured mammalian cells as described above by transfection experiments (see below).
Establishment of FLP/FRT-mediated recombination assay in vitro
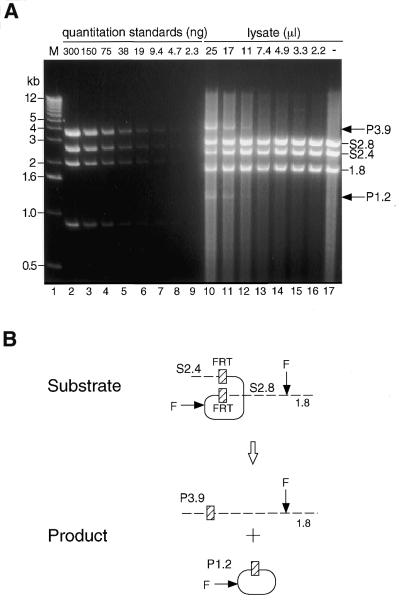
To compare the recombination efficiency of FLP and Cre produced in mammalian cells, an in vitro assay to detect and to quantify the excisional deletion mediated by FLP was established. We adopted the same strategy as the in vitro assay of Cre (58), using FLP expressed in mammalian cultured cells, since our aim was to assess whether FLP works efficiently in mammalian systems. When various doses of the FLP lysate were mixed with 1 µg of linearized substrate DNA, pCAFNFG, and incubated for 30 min at 30°C, which is the optimum temperature for FLP activity, recombined products were detected in a dose-dependent manner (Fig. 2A, lanes 10–16). As a result of the recombination, when the DNA recovered from the reaction mixture was digested with FspI, the recombined products were detected as 3.9 and 1.2 kb fragments derived from 2.8 and 2.4 kb substrates (Fig. 2B). The maximum rate of the recombination efficiency reached ∼30–40% when the maximum amount of FLP lysate (25 µl) in our experimental conditions of 50 µl total reaction volume was used. We did not detect any bands of the ‘intermediate’ product, which was reported in the Cre/loxP system (58).
Figure 2.
In vitro recombination of the FLP/FRT system at 30°C. (A) Quantitation of the recombination products. M (lane 1), 1 kb DNA ladder marker (Gibco BRL); P and S, product and substrate, respectively. Quantitation standards (lanes 2–9) represent the NcoI fragment of the EcoRI-digested pCALNLZ. HindIII-linearized pCAFNFG and each denoted amount of the FLP lysate was incubated for 30 min at 30°C. The recovered DNA was then digested with FspI and the density of the recombined 3.9 kb products (P3.9) were quantified based on densities of the quantitation standards. One representative result out of three is shown. (B) Recombination process of the FLP/FRT system. F, FspI site.
Recombination efficiency of FLP and Cre in vitro
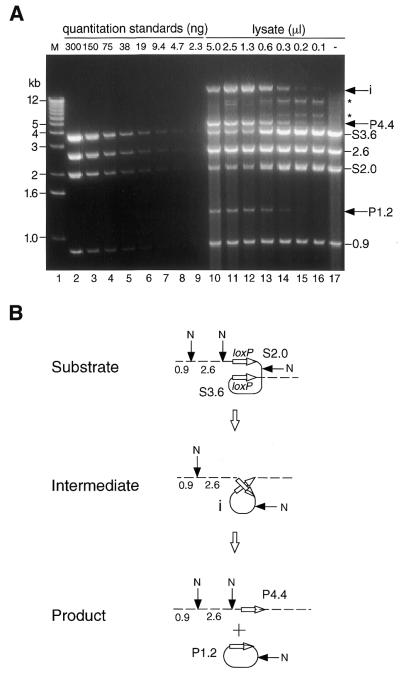
A comparative in vitro recombination experiment mediated by Cre/loxP was also carried out as described (58) using pCALNLZ as a substrate DNA. Cre lysate was prepared by same procedure used to prepare the FLP lysate. Cre showed lysate-dependent recombination at 37°C using 1 µg of linearized pCALNLZ (Fig. 3A, lanes 10–16), reaching a plateau level of ∼40% with >1.3 µl of the lysate as described (58). As shown in Figure 3B, after NcoI digestion, the recombined products were detected as 4.4 and 1.2 kb fragments derived from 3.6 and 2.0 kb substrates, along with the ‘intermediate’ product as described previously (58).
Figure 3.
In vitro recombination of the Cre/loxP system at 37°C. (A) Quantitation of the recombination products. M (lane 1), 1 kb DNA ladder marker (Gibco BRL); P and S, product and substrate, respectively; i, recombination intermediate; *, unexpected bands (may be due to the site preference of NcoI). The same quantitation standards (lanes 2–9) as used in the FLP/FRT system were applied. EcoRI-linearized pCALNLZ and each denoted amount of the Cre lysate was incubated for 30 min at 37°C. The recovered DNA was then digested with NcoI and the densities of the 4.4 kb products (P4.4) were quantified based on those of the quantitation standards. One representative result out of three is shown. (B) Recombination process of the Cre/loxP system. N, NcoI site.
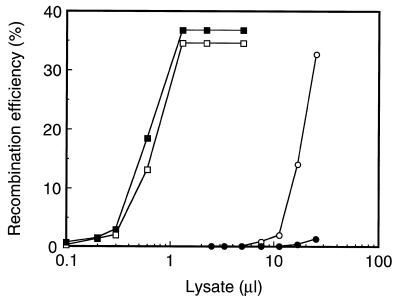
Since the optimum temperature of FLP activity is 30°C (50), it has been considered that FLP does not work efficiently in mammalian expression systems (51). Therefore, we carried out the in vitro recombination assay at both 37 and 30°C, using FLP and Cre lysate expressed in 293 cells and the amount of the recombined products were quantified based on the band intensity of the quantitation standards (Figs 2A and 3A, for example). As shown in Figure 4, Cre showed almost the same recombination efficiency at 37 and 30°C. In contrast, FLP showed significantly higher recombination efficiency at its optimum temperature, 30°C. The kinetics of recombination efficiency of Cre appeared to be a three-phase or sigmoid curve; a gradual increase up to ∼2%, linear dose-dependency from 2 to 35%, and saturation at ∼35%. The kinetics of FLP at 30°C appears similar at least up to the second phase. Since the maximum recombination efficiency of FLP at 37°C in our experiment was ∼2%, we compared the relative recombination efficiency as the reciprocal number of the lysate amount yielding 2% recombination. The relative recombination efficiency of FLP at 30°C was approximately one-thirtieth of Cre at 30°C, and was approximately one-ninetieth of Cre at 37°C. Therefore, the recombination efficiency of FLP at 37°C was only 3-fold greater than that at 30°C. The large difference in the relative recombination efficiency between FLP and Cre seems to be due to the enzymatic activity, but we cannot rule out the possibility that it is due to the expression level of these proteins in cultured mammalian cells.
Figure 4.
Recombination efficiency of FLP and Cre in vitro. The recombination efficiency per lysate of FLP at 37°C (filled circle) or 30°C (open circle) and that of Cre at 37°C (filled square) or 30°C (open square) were quantified as described. The maximum amount of the reacted lysate was 25 µl in our experimental condition of 50 µl total reaction volume. The quantitative results were normally reproducible within ±5%. One representative result out of three is shown.
Quantitative gene activation by FLP-expressing rAd
To assess whether the FLP/FRT system works efficiently in cultured mammalian cells, we generated three rAds (Fig. 1B); one expressing FLP (AxCAFLP), and the others bear the target unit with reporter genes, GFP and LacZ (AxCAFNFG and AxCAFNFZ, respectively). Corresponding rAds using the Cre/loxP system instead of the FLP/FRT system were also prepared. AxCAFLP was infected into HeLa cells together with the target rAd. The infection titer of the target rAd was kept constant at MOI of 5 and AxCAFLP was co-infected at various MOIs.
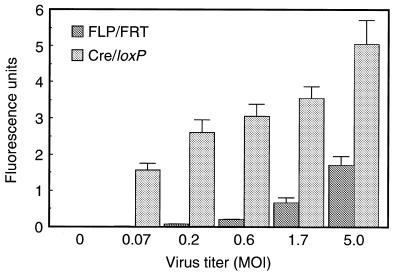
To measure the expression level of the activated transgene, we first compared the fluorescence intensity of GFP infected with the recombinase-expressing rAds, AxCAFLP or AxCANCre, and the target rAds, AxCAFNFG or AxCALNLG, respectively (Fig. 5). Each recombinase-expressing rAd showed MOI-dependent expression of GFP, while no fluorescence was detected after infection of the target rAd alone. The fluorescence intensity of GFP of AxCAFLP infection at MOI of 5 almost corresponded to that of AxCANCre infection at MOI of 0.07, suggesting that the recombination efficiency of FLP in mammalian cultured cells was approximately one-seventieth of that of Cre. This result agreed well with that of the in vitro recombination assay compared at 37°C described above.
Figure 5.
GFP gene activation mediated by recombinase-expressing rAds. Various MOIs of recombinase-expressing rAds, AxCAFLP or AxCANCre, and target rAds, AxCAFNFG or AxCALNLG kept constantly at an MOI of 5, were co-infected into HeLa cells. Three days later, the fluorescence intensity of GFP was quantified by Fluoroskan Ascent FL (Labsystems). All data represent means ± SD of several regions of the wells measured.
Frequency of FLP-mediated gene activation in mammalian cultured cells
While confirming whether CAFNF works as a target unit of FLP by co-transfecting plasmids, pCAFNFZ and pxCAFLP, into 293 cells, we also compared the number of LacZ-expressing cells by co-transfecting pCALNLZ and pxCANCre (9). The number of LacZ-expressed cells of FLP/FRT was no less than 25% of that obtained with the Cre/loxP system (data not shown), apparently different from when GFP-expressing rAd was infected into HeLa cells.
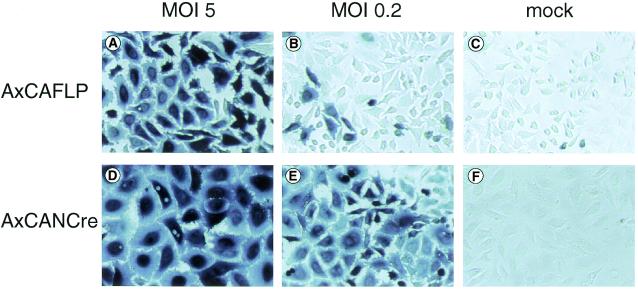
Since gene transfer mediated by rAd is rather uniform and more quantitative than transfection methods, we then co-infected AxCAFLP or AxCANCre with LacZ-expressing target rAds, AxCAFNFZ or AxCALNLZ, into HeLa cells and compared the LacZ-expression by X-gal staining (Fig. 6). When only each target Ad was infected, very few cells, if any, were stained blue (Fig. 6C and F). To obtain staining of 100% of HeLa cells, the FLP-expressing rAd, AxCAFLP, had to be infected at least at MOI of 5 (Fig. 6A) while an MOI of 0.2 of Cre-expressing rAd, AxCANCre was sufficient (Fig. 6E). Since AxCAFLP can be prepared as a purified high titer stock at >1010 plaque forming units (p.f.u.)/ml, infection at MOI of 5 is readily obtained in cultured cells. Therefore, the FLP-expressing rAd could activate the transgene in nearly 100% of cultured mammalian cells.
Figure 6.
LacZ gene activation mediated by recombinase-expressing rAds. Denoted MOI of AxCAFLP (A–C) or AxCANCre (D–F) and target rAds, AxCAFNFZ or AxCALNLZ kept constant at an MOI of 5, were co-infected into HeLa cells. Three days later, cells were fixed and stained with X-gal. The results of MOI 5 (A and D), MOI 0.2 (B and E) and non-infected (C and F) of recombinase-expressing rAds are shown.
DISCUSSION
Here we demonstrated that the rAd expressing FLP worked efficiently in the gene-activation strategies in cultured mammalian cells. This conclusion is based on the fact that the activation of the LacZ gene was observed in nearly 100% of cultured cells by co-infection of the FLP-expressing rAd, AxCAFLP, and the target rAd, AxCAFNFZ.
We showed that the infection of the Cre-expressing rAd, AxCANCre, at an MOI of 0.2 gave transgene activation in almost 100% of cultured cells (Fig. 6E) (9). Since one p.f.u. generally corresponds to ∼10 infectious viral particles (9), the results showed that the Cre-expressing rAd is so effective that only a few Cre-expressing DNA copies per cell are sufficient for successful gene activation. On the other hand, the FLP-expressing rAd required an MOI of 5 for gene activation in nearly 100% of cultured cells, suggesting that ∼50 copies of FLP-expressing DNA per cell are required. Transduction of >50 copies per cell in 100% of cultured cells is easily achieved by adenovirus vector, but not necessarily by other methods of gene transfer. Therefore, the FLP-expressing rAd described here may be a useful tool for the gene regulation strategies by FLP. In this work, the target CAFNF unit was present extrachromosomally on the adenovirus genome. We also observed that the FLP-expressing rAd was able to activate the CAFNF unit located on the cell choromosome after isolating neo-resistant cell clones in nearly 100% of cultured cells (Y.K., M.N. and I.S., unpublished data).
Although the efficiency of gene activation by FLP is much lower than by Cre, a number of groups successfully applied FLP-mediated gene regulation in cultured mammalian cells as long as the transfection method was used (44,45,59). We also observed that, in transient co-transfection experiments using target and recombinase-expressing plasmids, the number of conditionally LacZ-expressing cells mediated by FLP was as much as 25% of that mediated by Cre, and not more than 10-fold lower. This may be because, in transient transfection, most of the cells having received plasmids tend to accept many copies, >50 copies for instance, of plasmid DNA. However, this has meant that FLP-mediated gene activation has been successfully used in transfection experiments as long as gene activation is sufficient for only a fraction of cultured cells. The approach using FLP-expressing rAd, therefore, will be especially valuable when gene activation is desirable in almost all cultured cells.
Buchholz et al. (50) demonstrated lower recombination efficiency by FLP above 35°C in mammalian cultured cells. Recently, a thermostabilized FLP, FLPe, which retains its activity even at 37°C, was developed (51) and was reported to work efficiently in mice (60). Our in vitro results, however, suggested that the temperature sensitivity of FLP may not be the major reason for the poor recombination efficiency, since the recombination efficiency of FLP at 37°C was only 3-fold different from that at 30°C, while the relative recombination efficiency of FLP expressed by infecting AxCAFLP to 293 cells was approximately one-thirtieth of Cre even at 30°C, the optimum for FLP. Other reasons, such as lower activity of the FLP enzyme itself or a lower expression rate of FLP in mammalian cells than Cre, may be major reason(s) for such low recombination efficiency of FLP. Ringrose et al. reported (61), based on their in vitro recombination assay, that ∼10-fold more FLP was required than Cre to recombine a given quantity of substrate. Therefore, although the use of a thermostabilized FLP is undoubtedly important for achieving efficient recombination by FLP, a strategy of high-level expression of FLP in mammalian cells, for instance using an adenovirus vector with a potent CAG promoter, may be more effective, enabling the use of FLP for sequential gene regulation in combination with Cre.
Since an appropriate FLP antibody to detect the FLP protein has not been available, we could not quantify the amount of each recombinase contained in the lysate. However, the difference of the recombination efficiency between FLP and Cre observed in this study does not seem to be due to the difference of expression level of each recombinase-encoding mRNA, since we normally observed similar mRNA expression levels among several genes as long as expressed by the identical rAd under the control of CAG promoter (data not shown). Moreover, although the NLS sequence tagged to Cre improved the efficiency by ∼3-fold (9), it does not seem to be the major reason for the difference in apparent activity between FLP and Cre.
Recent advances in conditional gene activation or inactivation strategies mediated by Cre-expressing rAd have identified the functions of a number of key molecules in several signal transduction cascades (13–16,24,29,33). The strategy using Cre-expressing rAd has advantages such as stable regulated effects, very low background and a wide selection of promoters, including tissue-specific promoters, over other strategies (tetracycline-mediated regulation for example). However, it can only activate or inactivate a given gene once, i.e. ‘off to on’ or ‘on to off’. To better understanding the contribution of such molecules to signal cascades, sequential gene regulation such as a novel ‘off-on-off’ or ‘on-off-on’ strategy mediated by FLP-expressing rAd in combination with Cre-expressing rAd may be a valuable tool, especially in time-course experiments, to obtain further functional information.
Cre-expressing rAd has facilitated extensive manipulations in transgenic mice (19–26,28,29,31). Since the recombination efficiency of FLP at 37°C was markedly lower than that of Cre, FLP is probably not appropriate for gene regulation strategies in mice. FLP was reported, however, to be useful for gene activation or for removal of the selectable marker in murine ES cells in transfection experiments (59,62). FLP- and Cre-expressing rAds may therefore have complementary functions through the generation of transgenic mice; FLP for the removal of the selectable marker in ES cells and Cre for the conditional gene regulation in mature transgenic mice. Especially, FLP-expressing rAd probably serves as an attractive tool for removing selectable markers.
Regulation of the expression of foreign genes introduced by a viral vector is necessary for both safety and efficacy of clinical gene therapy. For example, we have established a specific- and a high-level expression strategy using a tissue-specific promoter for controlled Cre production (27). Sequential gene regulation mediated by FLP- and Cre-expressing rAds will provide further sophisticated strategies for gene therapy. Whenever the therapeutic gene may cause side effects harmful to the patient, we could immediately remove the expression unit, and when transgene expression was insufficient, further therapy could be continued with an another therapeutic gene using a secondary regulation system.
FLP-expressing rAd, AxCAFLP, and FRT-containing plasmids, pUFwF and pCAFNF5, will be available from the Riken DNA Bank (http://www.rtc.riken.go.jp).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr M.Jayaram for pUC19-Flp, Dr J.Miyazaki for the CAG promoter and Ms T.Shino and Ms K.Yasueda for their excellent secretarial support. This work was supported in part by grants-in-aid from Ministry of Science, Education, Sports and Culture to Y.K. and to I.S.
References
- 1.Barinaga M. (1994) Knockout mice: round two [published erratum appears in Science (1994) 265, 855]. Science, 265, 26–28. [DOI] [PubMed] [Google Scholar]
- 2.Rossant J. and Nagy,A. (1995) Genome engineering: the new mouse genetics. Nat. Med., 1, 592–594. [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky K., Gu,H., Kuhn,R., Betz,U.A., Muller,W., Roes,J. and Schwenk,F. (1996) Conditional gene targeting. J. Clin. Invest., 98, 600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer B. and Henderson,N. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA, 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukushige S. and Sauer,B. (1992) Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 89, 7905–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakso M., Sauer,B., Mosinger,B.,Jr, Lee,E.J., Manning,R.W., Yu,S.H., Mulder,K.L. and Westphal,H. (1992) Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu H., Zou,Y.R. and Rajewsky,K. (1993) Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre–loxP-mediated gene targeting. Cell, 73, 1155–1164. [DOI] [PubMed] [Google Scholar]
- 8.Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 9.Kanegae Y., Lee,G., Sato,Y., Tanaka,M., Nakai,M., Sakaki,T., Sugano,S. and Saito,I. (1995) Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res., 23, 3816–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anton M. and Graham,F.L. (1995) Site-specific recombination mediated by an adenovirus vector expressing the Cre recombinase protein: a molecular switch for control of gene expression. J. Virol., 69, 4600–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai K., Mitani,K. and Miyazaki,J. (1995) Efficient regulation of gene expression by adenovirus vector-mediated delivery of the CRE recombinase. Biochem. Biophys. Res. Commun., 217, 393–401. [DOI] [PubMed] [Google Scholar]
- 12.Kanegae Y., Takamori,K., Sato,Y., Lee,G., Nakai,M. and Saito,I. (1996) Efficient gene activation system on mammalian cell chromosomes using recombinant adenovirus producing Cre recombinase. Gene, 181, 207–212. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M., Nagata,S., Kita,Y., Nakatsu,N., Ihara,S., Kaibuchi,K., Kuroda,S., Ui,M., Iba,H., Konishi,H., Kikkawa,U., Saitoh,I. and Fukui,Y. (1997) Expression of a constitutively active phosphatidylinositol 3-kinase induces process formation in rat PC12 cells. Use of Cre/loxP recombination system. J. Biol. Chem., 272, 16089–16092. [DOI] [PubMed] [Google Scholar]
- 14.Sato N., Milligan,C.E., Uchiyama,Y. and Oppenheim,R.W. (1997) Cloning and expression of the cDNA encoding rat caspase-2. Gene, 202, 127–132. [DOI] [PubMed] [Google Scholar]
- 15.Piston D.W., Knobel,S.M., Postic,C., Shelton,K.D. and Magnuson,M.A. (1999) Adenovirus-mediated knockout of a conditional glucokinase gene in isolated pancreatic islets reveals an essential role for proximal metabolic coupling events in glucose-stimulated insulin secretion. J. Biol. Chem., 274, 1000–1004. [DOI] [PubMed] [Google Scholar]
- 16.Shintani Y., Yotsuyanagi,H., Moriya,K., Fujie,H., Tsutsumi,T., Kanegae,Y., Kimura,S., Saito,I. and Koike,K. (1999) Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J. Gen. Virol., 80, 3257–3265. [DOI] [PubMed] [Google Scholar]
- 17.Kurihara H., Zama,A., Tamura,M., Takeda,J., Sasaki,T. and Takeuchi,T. (2000) Glioma/glioblastoma-specific adenoviral gene expression using the nestin gene regulator. Gene Ther., 7, 686–693. [DOI] [PubMed] [Google Scholar]
- 18.Ueno T., Matsumura,H., Tanaka,K., Iwasaki,T., Ueno,M., Fujinaga,K., Asada,K. and Kato,I. (2000) Site-specific integration of a transgene mediated by a hybrid adenovirus/adeno-associated virus vector using the Cre/loxP-expression-switching system. Biochem. Biophys. Res. Commun., 273, 473–478. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Krushel,L.A. and Edelman,G.M. (1996) Targeted DNA recombination in vivo using an adenovirus carrying the Cre recombinase gene. Proc. Natl Acad. Sci. USA, 93, 3932–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakso M., Pichel,J.G., Gorman,J.R., Sauer,B., Okamoto,Y., Lee,E., Alt,F.W. and Westphal,H. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA, 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohlmann A., Gotthardt,M., Willnow,T.E., Hammer,R.E. and Herz,J. (1996) Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat. Biotechnol., 14, 1562–1565. [DOI] [PubMed] [Google Scholar]
- 22.Akagi K., Sandig,V., Vooijs,M., Van der Valk,M., Giovannini,M., Strauss,M. and Berns,A. (1997) Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res., 25, 1766–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata H., Toyama,K., Shioya,H., Ito,M., Hirota,M., Hasegawa,S., Matsumoto,H., Takano,H., Akiyama,T., Toyoshima,K., Kanamaru,R., Kanegae,Y., Saito,I., Nakamura,Y., Shiba,K. and Noda,T. (1997) Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science, 278, 120–123. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y.H., Sauer,B., Johnson,P.F. and Gonzalez,F.J. (1997) Disruption of the c/ebp α gene in adult mouse liver. Mol. Cell. Biol., 17, 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner K.U., Wall,R.J., St-Onge,L., Gruss,P., Wynshaw-Boris,A., Garrett,L., Li,M., Furth,P.A. and Hennighausen,L. (1997) Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res., 25, 4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H.G., Bilbao,G., Zhou,T., Contreras,J.L., Gomez-Navarro,J., Feng,M., Saito,I., Mountz,J.D. and Curiel,D.T. (1998) Application of a Fas ligand encoding a recombinant adenovirus vector for prolongation of transgene expression. J. Virol., 72, 2483–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato Y., Tanaka,K., Lee,G., Kanegae,Y., Sakai,Y., Kaneko,S., Nakabayashi,H., Tamaoki,T. and Saito,I. (1998) Enhanced and specific gene expression via tissue-specific production of Cre recombinase using adenovirus vector. Biochem. Biophys. Res. Commun., 244, 455–462. [DOI] [PubMed] [Google Scholar]
- 28.Wakita T., Taya,C., Katsume,A., Kato,J., Yonekawa,H., Kanegae,Y., Saito,I., Hayashi,Y., Koike,M. and Kohara,M. (1998) Efficient conditional transgene expression in hepatitis C virus cDNA transgenic mice mediated by the Cre/loxP system. J. Biol. Chem., 273, 9001–9006. [DOI] [PubMed] [Google Scholar]
- 29.Chang B.H., Liao,W., Li,L., Nakamuta,M., Mack,D. and Chan,L. (1999) Liver-specific inactivation of the abetalipoproteinemia gene completely abrogates very low density lipoprotein/low density lipoprotein production in a viable conditional knockout mouse. J. Biol. Chem., 274, 6051–6055. [DOI] [PubMed] [Google Scholar]
- 30.Okuyama T., Fujino,M., Li,X.K., Funeshima,N., Kosuga,M., Saito,I., Suzuki,S. and Yamada,M. (1998) Efficient Fas-ligand gene expression in rodent liver after intravenous injection of a recombinant adenovirus by the use of a Cre-mediated switching system. Gene Ther., 5, 1047–1053. [DOI] [PubMed] [Google Scholar]
- 31.Stec D.E., Davisson,R.L., Haskell,R.E., Davidson,B.L. and Sigmund,C.D. (1999) Efficient liver-specific deletion of a floxed human angiotensinogen transgene by adenoviral delivery of Cre recombinase in vivo. J. Biol. Chem., 274, 21285–21290. [DOI] [PubMed] [Google Scholar]
- 32.Kijima T., Osaki,T., Nishino,K., Kumagai,T., Funakoshi,T., Goto,H., Tachibana,I., Tanio,Y. and Kishimoto,T. (1999) Application of the Cre recombinase/loxP system further enhances antitumor effects in cell type-specific gene therapy against carcinoembryonic antigen-producing cancer. Cancer Res., 59, 4906–4911. [PubMed] [Google Scholar]
- 33.Li X.K., Fujino,M., Guo,L., Okuyama,T., Funeshima,N., Hashimoto,M., Okabe,K., Yaginuma,H., Mikoshiba,K., Enosawa,S., Amemiya,H. and Suzuki,S. (2000) Inhibition of Fas-mediated fulminant hepatitis in CrmA gene-transfected mice. Biochem. Biophys. Res. Commun., 273, 101–109. [DOI] [PubMed] [Google Scholar]
- 34.Broach J.R., Guarascio,V.R. and Jayaram,M. (1982) Recombination within the yeast plasmid 2µ circle is site-specific. Cell, 29, 227–234. [DOI] [PubMed] [Google Scholar]
- 35.Andrews B.J., Proteau,G.A., Beatty,L.G. and Sadowski,P.D. (1985) The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell, 40, 795–803. [DOI] [PubMed] [Google Scholar]
- 36.Senecoff J.F., Bruckner,R.C. and Cox,M.M. (1985) The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proc. Natl Acad. Sci. USA, 82, 7270–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gates C.A. and Cox,M.M. (1988) FLP recombinase is an enzyme. Proc. Natl Acad. Sci. USA, 85, 4628–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayaram M. (1997) The cis–trans paradox of integrase. Science, 276, 49–51. [DOI] [PubMed] [Google Scholar]
- 39.Riou-Khamlichi C., Huntley,R., Jacqmard,A. and Murray,J.A. (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science, 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- 40.Luo H., Lyznik,L.A., Gidoni,D. and Hodges,T.K. (2000) Technical Advance: FLP-mediated recombination for use in hybrid plant production. Plant J., 23, 423–430. [DOI] [PubMed] [Google Scholar]
- 41.Golic K.G. and Lindquist,S. (1989) The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell, 59, 499–509. [DOI] [PubMed] [Google Scholar]
- 42.Golic M.M., Rong,Y.S., Petersen,R.B., Lindquist,S.L. and Golic,K.G. (1997) FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res., 25, 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rong Y.S. and Golic,K.G. (2000) Gene targeting by homologous recombination in Drosophila. Science, 288, 2013–2018. [DOI] [PubMed] [Google Scholar]
- 44.O’Gorman S., Fox,D.T. and Wahl,G.M. (1991) Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science, 251, 1351–1355. [DOI] [PubMed] [Google Scholar]
- 45.Aladjem M.I., Brody,L.L., O’Gorman,S. and Wahl,G.M. (1997) Positive selection of FLP-mediated unequal sister chromatid exchange products in mammalian cells. Mol. Cell. Biol., 17, 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vooijs M., van der Valk,M., te Riele,H. and Berns,A. (1998) Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene, 17, 1–12. [DOI] [PubMed] [Google Scholar]
- 47.Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. [DOI] [PubMed] [Google Scholar]
- 48.Koch K.S., Aoki,T., Wang,Y., Atkinson,A.E., Gleiberman,A.S., Glebov,O.K. and Leffert,H.L. (2000) Site-specific integration of targeted DNA into animal cell genomes. Gene, 249, 135–144. [DOI] [PubMed] [Google Scholar]
- 49.Meyers E.N., Lewandoski,M. and Martin,G.R. (1998) An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet., 18, 136–141. [DOI] [PubMed] [Google Scholar]
- 50.Buchholz F., Ringrose,L., Angrand,P.O., Rossi,F. and Stewart,A.F. (1996) Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res., 24, 4256–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchholz F., Angrand,P.O. and Stewart,A.F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol., 16, 657–662. [DOI] [PubMed] [Google Scholar]
- 52.Niwa H., Yamamura,K. and Miyazaki,J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–199. [DOI] [PubMed] [Google Scholar]
- 53.Graham F.L., Smiley,J., Russell,W.C. and Nairn,R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol., 36, 59–74. [DOI] [PubMed] [Google Scholar]
- 54.Miyake S., Makimura,M., Kanegae,Y., Harada,S., Sato,Y., Takamori,K., Tokuda,C. and Saito,I. (1996) Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc. Natl Acad. Sci. USA, 93, 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozak M. (1981) Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res., 9, 5233–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanegae Y., Makimura,M. and Saito,I. (1994) A simple and efficient method for purification of infectious recombinant adenovirus. Jpn J. Med. Sci. Biol., 47, 157–166. [DOI] [PubMed] [Google Scholar]
- 57.Meyer-Leon L., Senecoff,J.F., Bruckner,R.C. and Cox,M.M. (1984) Site-specific genetic recombination promoted by the FLP protein of the yeast 2-micron plasmid in vitro. Cold Spring Harb. Symp. Quant. Biol., 49, 797–804. [DOI] [PubMed] [Google Scholar]
- 58.Lee G. and Saito,I. (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene, 216, 55–65. [DOI] [PubMed] [Google Scholar]
- 59.Dymecki S.M. and Tomasiewicz,H. (1998) Using Flp-recombinase to characterize expansion of Wnt1-expressing neural progenitors in the mouse. Dev. Biol., 201, 57–65. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez C.I., Buchholz,F., Galloway,J., Sequerra,R., Kasper,J., Ayala,R., Stewart,A.F. and Dymecki,S.M. (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet., 25, 139–140. [DOI] [PubMed] [Google Scholar]
- 61.Ringrose L., Lounnas,V., Ehrlich,L., Buchholz,F., Wade,R. and Stewart,A.F. (1998) Comparative kinetic analysis of FLP and cre recombinases: mathematical models for DNA binding and recombination. J. Mol. Biol., 284, 363–384. [DOI] [PubMed] [Google Scholar]
- 62.Seibler J., Schubeler,D., Fiering,S., Groudine,M. and Bode,J. (1998) DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry, 37, 6229–6234. [DOI] [PubMed] [Google Scholar]