Abstract
Bardet-Biedl Syndrome (BBS) is a rare human hereditary disorder associated with several features including obesity, retinopathy, renal defects, polydactyly, learning disabilities and hypogenitalism. This article discusses the abnormalities accounting for energy imbalance leading to obesity in BBS with emphasis on the recent evidence pointing to aberrations in hypothalamic action of leptin. Indeed, BBS proteins have emerged as important mediators of leptin receptor trafficking, and loss of BBS genes results in leptin resistance that may be due to abnormal leptin receptor handling in a subset of leptin responsive neurons. These recent discoveries hold promise for improved clinical management of BBS patients. The relevance of these findings to non-syndromic common obesity is also discussed.
Genetic aspects of Bardet-Biedl syndrome (BBS)
BBS is a highly pleiotropic autosomal recessive disorder associated with wide array of phenotypes, but there are six clinical features that are considered the cardinal manifestations: obesity, retinitis pigmentosa, renal anomalies, postaxial polydactyly, learning disabilities and defects in the urogenital tract [1;2]. BBS is also associated with increased susceptibility to several other disorders including hearing loss, neurological impairment, speech deficit, hepatic fibrosis, diabetes mellitus, hypertension and congenital heart disease [2–5].
In the last two decades, significant progress has been made in understanding the genetic basis of BBS. To date, twelve BBS genes (BBS1–12) have been identified. Hypomorphic mutants in two additional genes (meckel syndrome 1 [MKS1] and centrosomal protein 290 [CEP290]) were found to be associated with BBS phenotypes and were classified as BBS13 and BBS14, respectively [6;7]. Both genes encode centrosomal proteins [8;9]. More recently, the human gene encoding Fritz protein, which controls planar cell polarity, was identified as a BBS gene (BBS15) [10]. These 15 genes account for about 80% of the known cases of BBS, indicating that additional BBS genes are yet to be identified [6].
Usually, BBS is transmitted in autosomal recessive manner, but emerging evidence indicates that BBS can be inherited in a complex fashion. Indeed, some families were found to exhibit “triallelic” mode of inheritance [11–14] where the clinical manifestation of the syndrome require two mutations at one BBS gene plus one mutation at a second BBS gene. Such multilocus inheritance is not observed in large pedigrees and the vast majority of cases display autosomal recessive inheritance [15]. However, inter and intrafamilial variability in the expressivity of BBS phenotypes raises the possibility that other genes modify the BBS phenotype. These genetic modifiers may be the BBS genes themselves, or other genes that do not independently cause BBS when mutated [11;12;16].
BBS is rare in the general population, with a prevalence of 1 in 125,000–160,000 in people of European descent. However, due to high consanguinity rate some isolated populations particularly in Middle East (Bedouin tribes) and Canada (Newfoundland Island) exhibit much higher incidence of BBS (with prevalence up to 1 in 13,500) [17;18]. Identifying the BBS genes and underlying molecular mechanisms of the disorders associated with this syndrome have garnered great interest due to the fact that components of the phenotype such as obesity, diabetes and cardiovascular diseases are common in the general population. Of note, individuals carrying BBS heterozygous mutations are found to be more susceptible to develop obesity than non carriers, without displaying other BBS phenotypes [19]. Moreover, mutation analysis of the coding and conserved regions of several BBS genes performed in lean and obese French-Caucasian population revealed an association between four single nucleotide polymorphisms (SNPs) in three genes (BBS2, BBS4 and BBS6) and common obesity [20]. In BBS2 gene, one SNP, rs4784675, exhibited significant differences in allele frequency between lean and obese adults. In BBS4 gene, an association was detected between allele frequency in one SNP, rs7178130, and early-onset childhood obesity as well as morbid adult obesity. Two SNPs in BBS6 gene rs6108572 and rs221667, were found to be associated with childhood obesity while SNP rs221667 was also found to be significantly different in allele frequency in the severely obese adults [20]. These observations linking variants of BBS genes with obesity in non-BBS individuals further support the relevance of BBS genes to common human obesity.
In this review, we discuss the recent developments in this emerging field, with emphasis on the function and defects that are relevant for obesity in BBS.
Link between BBS and cilia
Cilia are hairlike cellular projections found in virtually all organs and cell types of the mammalian body. These evolutionary conserved organelles play a fundamental role in the regulation of several biological processes [21]. The best known function of cilia is their action as sensors of extracellular cues and the transduction of those cues into cellular signaling ultimately affecting gene transcription, cell division and differentiation [22].
Typically, a cilium projects from the apical surface of a cell and is composed of two structures known as the axoneme and basal body (Figure 1). The axoneme is formed by a microtubule-based protrusion that extends into the extracellular space and is covered by a specialized plasma membrane. The basal body represents the anchorage point of the axoneme and is derived from the centriole. The various components of the cilium are put together and maintained by a multiprotein machinery termed intraflagellar transport (IFT) that moves cargo vesicles (such as structural components, receptors and signaling molecules) into (anterograde) and from (retrograde) the cilium [23]. BBS is considered a prototype for the illustration of the role of cilia in human disease [6]. In addition to BBS, there are several other human diseases which are thought to be related to ciliary dysfunction (Box 1).
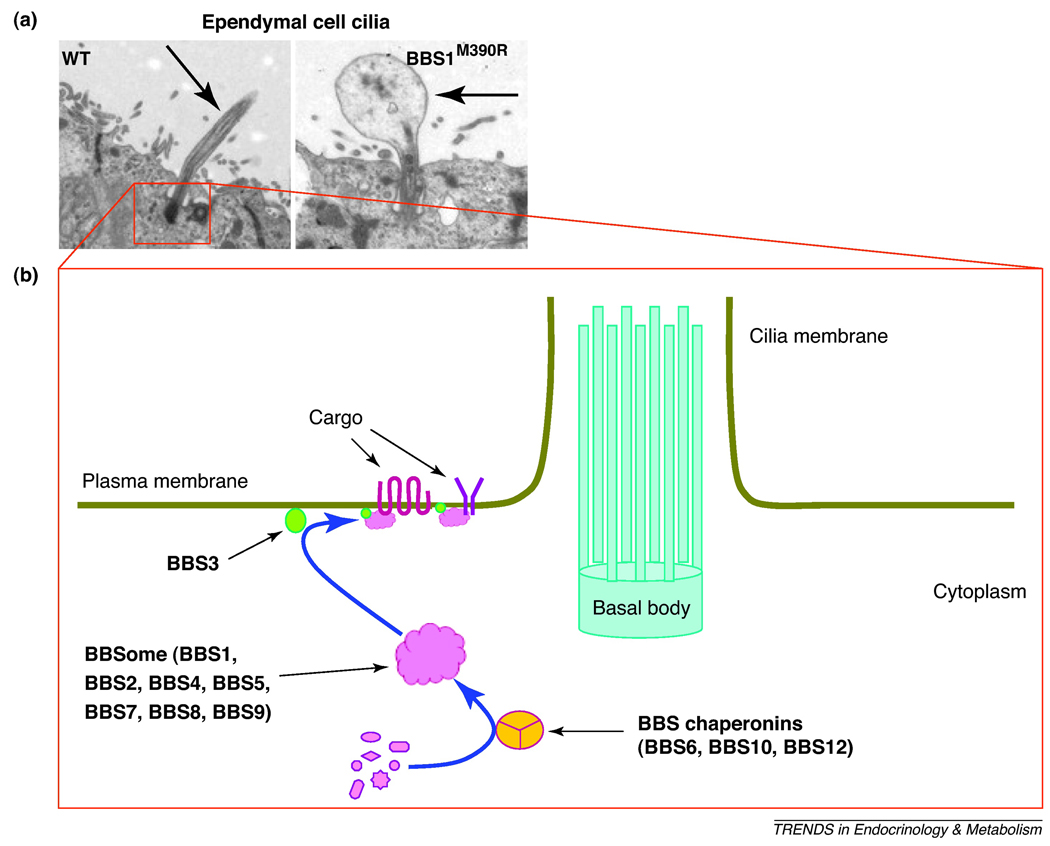
Figure 1.
BBS proteins and ciliary function. A) Abnormal ependymal cell cilia in BBS mice. Analysis by transmission electron microscopy of ependymal cell cilia (indicated by the arrow) lining the ventral portion of the third ventricle (adjacent to the arcuate nucleus of the hypothalamus) in BBS1 M290R mutant and wild type (WT) mice demonstrates enlargement of the distal region of cilia in BBS1 mutant animals. These results are representative of data published elsewhere (ref. 32). B) Schematic illustration depicting the cilium structure and the function of various BBS proteins. The BBS chaperonin complex mediate the assembly of the BBSome core which then interact with BBS3 at the base of the cilium to sort and mediate the trafficking of cargos including receptors to cilia and/or cell membrane.
Box 1. Cilia-related human diseases
The role of cilia in human disease, termed ciliopathies, has expanded significantly as a wide spectrum of human disorders have been attributed to defects that impact the function of cilia [73]. Human ailments that are long known to be cilia-related include polycystic kidney disease and primary cilia dyskinesia [74;75]. Polycystic kidney disease, one the most common genetic disorders, is commonly associated with altered differentiation and proliferation of renal and other epithelial cells resulting in cystic kidneys [21]. Interestingly, the protein products encoded by the genes responsible for inherited forms of cystic kidney diseases (e.g. PKD1 and PKD2 genes) are expressed in primary cilia, basal bodies or the centrosome [76;77]. Primary cilia dyskinesia is characterized by situs inversus totalis, the complete mirror image reversal of the internal organ positioning, retinal degeneration, infertility, recurring respiratory infections and some instances of hydrocephalus [78;79]. Two of the genes commonly mutated in patients with primary cilia dyskinesia, DNAH5 and DNAI1, encode dynein motor proteins, components that are critical for cilia movement [79;80].
Besides BBS, a number of rare pleiotropic human syndromes such as Alström syndrome, Jeune syndrome, Joubert syndrome, McKusick-Kaufman syndrome, Meckel-Gruber syndrome, orofacial digital syndrome 1 and Senior-Loken syndrome are now considered as cilia-related diseases [73;81]. Although each of these syndromes has a unique clinical feature there are several overlapping phenotypes such as retinal degeneration and cystic lesions in the kidneys, pancreas and liver [73]. For instance, in addition to displaying a number of phenotypes that are reminiscent of BBS such as obesity and retinitis pigmentosa, patients with Alström syndrome exhibit sensory deafness, but do not develop polydactyly or mental retardation [82;83]. Many of the genes responsible for these various human syndromes have been identified [81]. Consistent with the shared phenotypes, there is evidence for genetic overlap between the celiopathies. Such genetic overlap is exemplified by the finding that several mutations in McKusick-Kaufman syndrome gene cause BBS [84]. Meckel-Gruber syndrome causing genes such as MKS1 and MKS3 were also found to be involved in BBS, both as causative and modifying alleles [7].
The evidence implicating ciliary defects as the main cause of the phenotypes associated with BBS is extensive. This includes the localization of all the analyzed BBS proteins to the basal body and the ciliary axoneme, the restricted evolution of BBS genes to ciliated species, and their expression in ciliated cells [22;24–26]. Additional evidence derives from loss of function studies in several animal models. For instance, cilia of BBS mutant C. elegans are found to be relatively short and display functional defects and compromised IFT [27]. In zebrafish, suppression of BBS genes results in failure of development of an early ciliated structure known as Kupffer’s vesicle [28]. Additionally, zebrafish embryos lacking BBS genes have delayed retrograde melanosome transport. In line with the prevalence of renal defects in BBS, zebrafish models of BBS were found to develop kidney cysts [29]. These findings support the requirement of BBS proteins for normal cilia formation and function, and normal vesicle trafficking suggesting a role for these proteins in IFT.
Mouse models lacking BBS genes also exhibit ciliary defects as indicated by the lack of flagella formation during spermatogenesis [24;30;31]. Although these animal models develop other motile and primary cilia, structural and functional ciliary alterations have been documented [31]. For instance, in BBS mice, cilia of ependymal cells which line the brain ventricles displayed abnormal length and enlarged distal regions (Figure 1A). Given the importance of ependymal cell cilia for cerebrospinal fluid circulation, such defects may account for the hydrocephalus and enlarged brain ventricles observed in BBS mice [32]. Certain G protein-coupled receptors including somatostatin receptor 3 (sst3) and serotonin receptor 6 (5-HTR6) localize to cilia [33;34] and loss of BBS genes disrupt cilia localization of these receptors in certain neurons indicating that BBS proteins are important for the trafficking receptors to the neuronal cilium.
Evidence implicating cilia in the regulation of energy homeostasis derives from the observation that systemic suppression of IFT-related elements such as Ift88 or Kif3a genes which disrupt cilia results in hyperphagia and obesity in mice [35]. Notably, these phenotypes can largely be recapitulated by the disruption of neuronal cilia throughout the central nervous system or in a specific hypothalamic neuronal populations (proopiomelanocortin (POMC)-expressing neurons) that are critical for the control of energy balance [35]. In C. elegans, perturbation of ciliated neurons promotes fat accumulation and obesity [36] supporting further the importance of neuronal cilia in the regulation of energy homeostasis.
Function of BBS proteins
Major progress in understanding the biochemical tasks of BBS proteins and their role in ciliary function has been made in recent years (Figure 1B). Seven of the most conserved BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8 and BBS9) were found to form a core complex, known as the BBSome that is critical for ciliary function [37]. This complex associates with the ciliary membrane and function as a unit that sorts and directs protein/vesicle trafficking to the ciliary membrane through a mechanism involving Rabin8, a GDP/GTP exchange factor for the small GTPase RAB8. BBS3 gene encodes a protein, ARL6, which is a member of the ADP-ribosylation factor-like family of proteins that belong to the Ras superfamily of small GTP-binding proteins. Consistent with its localization in close proximity to the site of vesicle docking at the base of cilia, BBS3 protein appears critical for the recruitment of the BBSome complex onto the ciliary membrane [38].
The three chaperonin-like BBS proteins (BBS6, BBS10 and BBS12) were also found to interact together and form another complex in combination with other chaperonin proteins of the CCT (also known as TRiC) family [39]. One component of the BBSome, BBS7, appears to directly associate with this chaperonin-like complex. In addition, ablation of BBS6, BBS10 or BBS12 abolishes the interaction of the proteins that form the BBSome indicating the importance of this chaperonin complex in mediating the assembly of the BBSome [39].
The function of the remaining BBS proteins is less well characterized. We know that BBS11 is an E3 ubiquitin ligase involved in protein ubiquitination [40;41], BBS13 is located at the centrosomes in dividing cells and is required for cell branching morphogenesis [8], BBS14 is implicated in microtubule-associated protein transport [9] and that BBS15 is a critical determinant of planar cell polarity [10]. Also, the exact mechanism involved in assembling the BBS complexes and the molecular activities these complexes may mediate beyond those relevant to ciliary functions remain to be identified. However, the existence of such functional connections and interdependency between the different BBS proteins provide a compelling explanation for the similarity in the phenotypes between patients carrying mutations in various BBS genes. Finally, the possibility that besides their involvement in the BBS complexes, individual BBS proteins may be engaged in ciliary function and/or other cellular tasks requires further investigation.
Metabolic alterations in BBS
Despite the wide spectrum of disorders that BBS patients may carry, according to the BBS society obesity appears to be one the most disturbing symptoms of the syndrome [42]. Obesity is highly prevalent in BBS individuals and present in almost all patients carrying homozygous mutations [17;43]. Of note, relative to non-BBS subjects with comparable body mass index (BMI), BBS subjects tends to have higher adiposity [42;44]. Additionally, patients with BBS were found to have significantly more abdominal visceral fat compared to BMI-matched control subjects [44]. Conversely, lean mass tends to be lower in BBS patients relative to non-BBS subjects [41]. Predominance of morbid obesity (BMI over 40 kg/m2) is also elevated in BBS patients [45].
The observation that obesity is frequent in BBS heterozygotes individuals raises the possibility that carrying one mutant allele of BBS genes predispose to obesity [19]. In addition, variants of several BBS genes seem to increase susceptibility to obesity in non-BBS patients [20;46]. The mechanisms by which variation in BBS genes may influence adiposity is not clear, but this may occur in conjunction with other known genes (e.g. MC4R [melanocortin 4 receptor] and FTO [fat mass and obesity-associated gene]) [47] or unidentified loci that influence fat mass. Alternatively, the dominant negative mode of action of those variants of the BBS genes could explain their effects on adiposity [48].
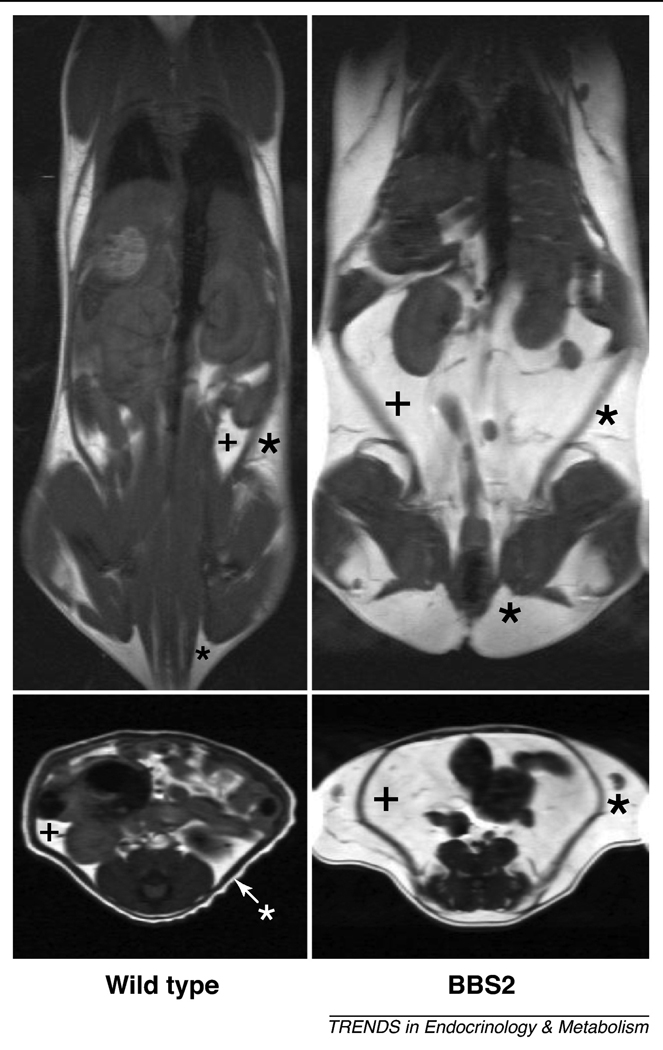
Importantly, inactivation of any BBS gene gives rise to the obesity phenotype across animal phylogeny. For instance, mice with deletion of Bbs2, Bbs4 or Bbs6 genes or carrying an M390R mutation in Bbs1 gene develop obesity [32;49;50] attributable to increased subcutaneous and visceral fat mass (Figure 2). Bbs4−/−mice also exhibited fatty livers and abnormal lipid and cholesterol profiles [49]. Usually, obesity results from energy imbalance between ingested and expended calories, and BBS mice are hyperphagic with 20–30% elevation in daily food intake [49;50]. However, despite pair-feeding, BBS knockout mice continued to have increased adiposity indicating that BBS mice also have decreased energy expenditure [50].
Figure 2.
Obesity phenotype in BBS mice. Representative MRI images demonstrating the increased fat mass in a BBS2 mutant mouse relative to a wild-type littermate control. Coronal (top panels) and axial abdominal (bottom panels) sections of are shown. Fat appears white whereas lean mass appears gray or black; (+) indicate subcutaneous fat and (*) visceral fat pads. These results are representative of data published elsewhere (ref. 50).
We also noticed a marked decrease in locomotor activity in the obese BBS null mice relative to non-BBS diet-induced obese mice that have similar body weights [50]. This finding is in line with the reported lower level of physical activity in BBS subjects compared to non-BBS control subjects with comparable degree of obesity [42]. Therefore, perturbation of both energy intake and energy expenditure appears to contribute to fat accumulation and obesity in BBS.
Molecular abnormalities in BBS
The search for the molecular defects in BBS unraveled aberrations in hypothalamic action of leptin as a major abnormality accounting for energy imbalance in this syndrome. Leptin is a 167-amino acid protein expressed mainly by adipocytes and released in the blood in proportion to fat mass [51]. Leptin action in the central nervous system promotes weight loss by reducing food intake and increasing energy expenditure [52;53]. The severe obesity and the hyperphagia caused by the absence of leptin in rodents and humans demonstrate the importance of this hormone for the control of body weight and food intake in mammals [54]. BBS patients and mice are hyperleptinemic [44;49;50;55]. Moreover, BBS mutant mice displayed increased circulating levels of leptin even at an early age [50]. These findings exclude leptin deficiency as a mechanism of obesity in BBS. In addition, administration of leptin failed to alter body weight and food intake in BBS mice indicating that the hyperleptinemia in these animals is associated with leptin resistance [50]. Remarkably, eliminating obesity and hyperleptinemia by calorie-restriction does not restore leptin sensitivity in BBS mice indicating that leptin resistance is intrinsic rather than secondary to hyperleptinemia and obesity [56]. Consistent with these findings in mouse models, patients with BBS were found to exhibit greater leptin resistance than would be predicted from their degree of adiposity. This is based on the significantly higher serum leptin in BBS patients compared with BMI-matched non-BBS subjects [44].
To determine the specific defect leading to leptin resistance in BBS mice, several possibilities were examined. First, a potential defect in leptin transport across the blood brain barrier was considered. Plasma leptin is transported to the brain by a saturable, unidirectional system [57], involving binding of leptin to the short form of the leptin receptor located at the endothelium of the vasculature and the epithelium of the choroid plexus [53]. The high leptin levels in the cerebrospinal fluid and the inability of cerebroventricular administration of leptin to affect feeding and body weight in BBS mice argue against a meaningful role for transport defects in the resistance to leptin [50]. Absence of a normal leptin receptor such as in db/db mice or in Zucker obese rats is a known cause of severe obesity and leptin resistance [58–63]. However, hypothalamic expression of the long form of leptin receptor, known as ObRb, that mediates most of the biological actions of leptin was unaltered in BBS mice [56].
Alterations in the signaling capacity of the ObRb in BBS mice was evaluated by examining the ability of leptin to activate hypothalamic signal transducer and activator of transcription-3 (STAT3), a major signaling pathway of the ObRb [64]. Importantly, body weight and leptin levels in BBS mice were normalized by calorie-restriction to eliminate leptin resistance and obesity as confounding factors. In control mice, leptin caused a robust increase in STAT3 phosphorylation, but this response was greatly reduced in BBS mice despite normal serum leptin levels. Given that specific disruption of leptin-induced activation of the STAT3 pathway leads to severe obesity in mice [65], these data demonstrate that leptin resistance in BBS mice is caused by the inability of leptin to activate the downstream intracellular machinery associated with its receptor.
An increase in suppressor of cytokine signaling-3 (SOCS-3), one of the targets induced by STAT3 and a negative feedback inhibitor of the ObRb signaling, is considered an important molecular mechanism of leptin resistance in diet-induced obese mice [53;64]. However, SOCS-3, both at the mRNA and protein levels, were decreased in BBS animals, indicating that leptin resistance in these mice was not due to increased expression of SOCS-3 in contrast to what is observed in typical obesity.
The alteration in leptin action was associated with changes in the downstream neuropeptides involved in the control of energy homeostasis. Several brain regions ranging from cortex to brainstem are known to be involved in leptin regulation of energy homeostasis, with the hypothalamus playing a key role [66]. Within the hypothalamus, the arcuate nucleus is considered a major site for the control of energy balance and regulation of physiological processes by leptin [52]. Although the arcuate nucleus contains several neuronal populations, two classes of neurons have been well characterized with regard to leptin action (Figure 3): POMC neurons (which also express cocaine- and amphetamine-related transcripts (CART) that are activated by leptin; and neuropeptide Y (NPY) neurons (which also express agouti-related protein (AgRP) that are inhibited by leptin [52]. In POMC neurons, leptin is known to increase the gene expression of POMC and CART genes. In contrast, in NPY neurons, leptin inhibited the expression of NPY and AgRP genes.
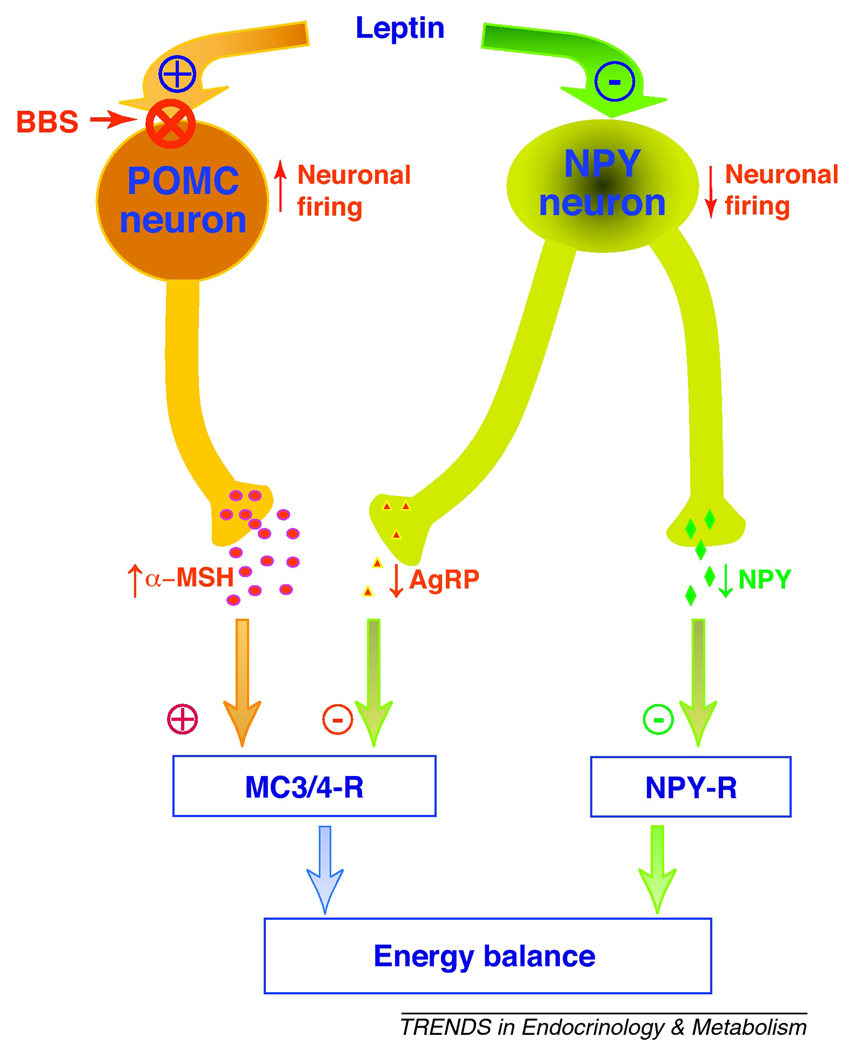
Figure 3.
Schematic illustration of the leptin-sensitive neuronal populations in the hypothalamic arcuate nucleus; those activated (catabolic pathway represented by proopiomelanocortin (POMC) neurons) and those inhibited (anabolic pathway represented by the neuropeptide Y (NPY) neurons which also express agouti related protein, AgRP). In POMC neurons, leptin increases neuronal firing and POMC gene expression promoting the secretion of alpha-melanocyte stimulating hormone (α-MSH), an agonist of the melanocortin 3 and 4 receptors (MC3/4-R) located in the second order neurons. Conversely, in NPY neurons, leptin inhibits neuronal firing rate and decreases the expression and secretion of NPY and AgRP (antagonist of the MC3/4-R), promoting activation of MC3/4-R and inhibition of NPY-R, respectively. Leptin suppression of the NPY anabolic pathway and stimulation the POMC catabolic pathway reduce food intake and promote thermogenesis (resulting in decrease in body weight). BBS appears to be associated with a specific defect in leptin action in POMC neurons, but not NPY neurons.
In BBS mice, the orexigenic Agrp and Npy genes were normal, whereas the expression level of the anorexigenic Pomc gene, a gene activated by leptin via STAT3, was significantly lower [50]. These findings suggest that BBS is associated with defective leptin action in POMC, but not NPY, neurons (Figure 3). Moreover, in BBS mice, changes in Agrp and Npy mRNA levels in response to fasting were similar to that of controls. In contrast, fasting-induced decreases in Pomc mRNA levels was significantly less in BBS mice relative to control animals [56]. These data indicate that POMC neurons of BBS mice do not respond properly to changes in peripheral energy balance. It is interesting to note that abrogating cilia in these POMC neurons increases food intake and causes obesity in mice [35]. Together, these findings indicate that POMC neurons are selectively affected in BBS mice downstream of ObRb signaling. This is further supported by the fact that treatment with MTII, a melanocortin receptor agonist, causes anorexia and weight loss in BBS mice suggesting that the hypothalamic abnormality is upstream of melanocortin receptors in the leptin-melanocortin pathway [56].
In addition to the hypothalamic defects, abnormalities in peripheral tissues, particularly adipogenesis, may contribute to the pathogenesis of obesity in BBS. The expression levels of several BBS genes were found to vary significantly during adipogenesis [67]. In adipocytes, suppression of BBS10 and BBS12 genes enhance glycogen synthase kinase 3 and peroxisome proliferator-activated receptor nuclear pathways, promoting adipogenesis and fat accumulation [68]. However, the contribution of these alterations in adipogenesis to obesity in BBS remains to be determined. Also, knowing that BBS genes are expressed in almost every cell type in the body, the potential contribution of other peripheral tissues such as the liver and skeletal muscle to energy imbalance in BBS needs further investigation.
A novel mechanism of leptin resistance accounting for obesity in BBS
The discoveries discussed above firmly established defective leptin receptor signaling as an important contributor to the obesity phenotype of BBS. The exclusion of classic mechanisms of leptin resistance described in common obesity prompted the search for alternative explanations for the loss of leptin receptor signaling in BBS. Of note, the leptin receptor was found to localize in cilia of the rat olfactory neurons, mostly in their proximal part, but also in their very distal part [69]. Inactivation of BBS genes in the retina leads to mislocalization of rhodopsin which precedes apoptotic death of the photoreceptors and retinal degeneration leading to blindness [30]. These data in conjunction with other evidence implicating BBS proteins in intracellular trafficking of receptors to cilia [28;37] raised the possibility that BBS proteins may be involved in transporting the leptin receptor to cilia and/or cell membrane and that loss of BBS genes cause abnormal trafficking and localization of the leptin receptor.
Consistent with such a hypothesis, BBS1 protein, but none of the other tested BBSome proteins, was found to interact directly with the leptin receptor in transient transfection and co-immunoprecipitation assays [56]. Interestingly, the interaction between BBS1 and leptin receptor appears specific to the signaling isoform of the receptor, ObRb, as the non-signaling isoform, ObRa, did not interact with BBS1. This was further confirmed by demonstrating that the C-terminal cytoplasmic domain that is unique to ObRb was sufficient to interact with BBS1.
The key question then becomes whether ObRb trafficking or localization was affected in the absence of BBS proteins. The first evidence for this derives from the finding that the M390R mutation in BBS1 disrupts its interaction with the leptin receptor. Additionally, when BBS1 or BBS2 expression is suppressed, ObRb displayed altered cellular localization and its absence in the plasma membrane of cells. The fact that depletion of BBS2 protein recapitulated the mistrafficking of ObRb suggests that the BBSome complex may be involved in ObRb trafficking. The importance of BBS proteins for microtubule stability [38] indicate that microtubule dysfunction may account for the mislocalization of ObRb when BBS genes are suppressed. Regardless of the mechanism, these data demonstrate that a fundamental mechanism underlying the pathophysiology of obesity in BBS is the requirement of BBS proteins for the proper trafficking and cellular localization of the leptin receptor.
Perturbation in the neuronal system involved in energy balance in BBS is not unique to the leptin receptor, as neurons of BBS null mice also exhibited improper localization of the melanin-concentrating hormone 1 receptor (MCHR1) [33]. However, the relevance of neuronal mistrafficking of MCHR1 for the energy imbalance and obesity in BBS is not clear, as the MCH pathway is orexigenic. Indeed, chronic infusion of MCH results in hyperphagia and obesity, whereas mice deficient of MCH are hypophagic and lean [70;71]. Also, pharmacological blockade or genetic ablation of MCHR1 reduces feeding behavior and promotes leanness [71]. Given the ciliary localization of 5-HTR6, its mistrafficking in neurons lacking BBS proteins has been suggested as a possible mechanism of obesity in BBS [5]. However, mice deficient in 5-HTR6 eat less and are lean and resistant to diet-induced obesity [72]. These data indicate that a potential perturbation in 5-HTR6 is unlikely to contribute to BBS-associated obesity.
Conclusions and perspectives
Recent years have witnessed dramatic advances in deciphering the function of BBS proteins and the molecular biology of BBS-related phenotypes including obesity. The identification of BBS protein complexes (BBSome and the chaperon BBS complex) and their involvement in ciliary function explains the identical or overlapping phenotypes arising from mutations in different BBS genes. We also learned that a common and primary function of BBS proteins is the mediation and regulation of intracellular transport of several membrane receptors including the leptin receptor. Consistent with this, BBS was found to be associated with leptin resistance perhaps due to an abnormal leptin receptor handling in a subset of leptin receptor expressing neurons.
The progress in defining the biological basis for energy imbalance and obesity in BBS may help the patients with this disorder to dispel any perception of obesity as a lack of discipline and personal behavioral failure. Such progress will also be helpful for clinical management of BBS patients and for rationale mechanism-based therapies. For instance, the finding that, in animal models of BBS, activation of brain melanocortin receptors can suppress appetite and causes weight loss is an indication that if safe drugs targeting these receptors are developed, they may be useful for the management of obesity in BBS patients. Despite the tremendous progress achieved in understanding the biology of BBS much research is needed to address important unanswered questions with potential implications in health and disease beyond BBS (Box 2).
Box 2. Important questions for future consideration
The diversity of clinical manifestations and complex genetic etiology of BBS as well as other cilia-related diseases is remarkable [73]. However, this also raises questions about how disruption of the same cellular organelle leads to overlapping, but distinct phenotypes and whether all clinical manifestations of ciliopathies are due to ciliary defects. The idea that BBS proteins and other cilia-related proteins might be involved in cellular mechanisms other that IFT and ciliary functions must be considered. Such possibility is supported by the finding that in zebrafish knockdown of BBS genes disrupt retrograde melanosome transport along microtubules, indicating a functional role in vesicle transport [28].
Given that obesity is a common feature of several cilia-related disorders, it remains to be determined whether the defects found in BBS such as the loss of leptin action in POMC neurons underlie the obesity associated with other ciliopathies. The availability of animal models that recapitulate the humans conditions of these ciliopathies [35;85;86] should facilitate the investigation of these aspects. Similarly, exploration of the relevance of BBS proteins for common obesity with regard to leptin receptor trafficking is a worthy effort given the reported association between polymorphisms in BBS genes and adiposity in a general population [20]. Interestingly, rats that are genetically predisposed to develop diet-induced obesity have a significantly reduced number of leptin binding sites in the hypothalamic nuclei including the arcuate nucleus [87]. A potential involvement of BBS genes in dietary obesity-associated reduction in hypothalamic leptin receptors remains to be determined.
The exact mechanism that underlies leptin receptor trafficking by BBS proteins is unclear. In part, this is due to the fact that we know very little about the mechanisms involved in leptin receptor transport to the cell membrane. It is also puzzling why and how absence of BBS proteins affects leptin receptor signaling selectively in POMC neurons versus NPY neurons – two neuronal populations located in the same hypothalamic nucleus. One possibility is that BBS proteins are expressed only in POMC neurons. Alternatively, BBS proteins may be expressed in POMC as well as NPY neurons, but leptin receptor trafficking in NPY neurons is mediated by other systems independent from BBS proteins. Finally, whether the decreased POMC gene expression in BBS can be explained entirely by loss of leptin action in these neurons remain to be investigated. For instance, receptors for insulin and other adipokines are present in POMC neurons and participate in the modulation of the function of these neurons including POMC gene expression [52–54]. Examining the action in POMC neurons of insulin or other adipokines will help address this issue.
Acknowledgements
The authors work is supported by the National Institute of Health grant HL084207 and American Diabetes Association grant 1–11-BS-127.
Footnotes
Conflict of interest
The authors declare that they have no financial conflict of interest.
References
- 1.Bardet G. Sur un syndrome d'obesite infantile avec polydactylie et retinite pigmentaire (contribution a l'etude des formes cliniques de l'obesite hypophysaire) 1920 [Google Scholar]
- 2.Green JS, et al. The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N. Engl. J. Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- 3.Harnett JD, et al. The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N. Engl. J. Med. 1988;319:615–618. doi: 10.1056/NEJM198809083191005. [DOI] [PubMed] [Google Scholar]
- 4.Elbedour K, et al. Cardiac abnormalities in the Bardet-Biedl syndrome: echocardiographic studies of 22 patients. Am. J. Med. Genet. 1994;52:164–169. doi: 10.1002/ajmg.1320520208. [DOI] [PubMed] [Google Scholar]
- 5.Blacque OE, Leroux MR. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell. Mol. Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 8.Dawe HR, et al. The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum. Mol. Genet. 2007;16:173–186. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- 9.Chang B, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol, Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SK, et al. Planar Cell Polarity Acts Through Septins to Control Collective Cell Movement and Ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badano JL, et al. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Hum. Mol. Genet. 2003;12:1651–1659. doi: 10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- 12.Beales PL, et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am. J. Hum. Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsanis N, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 14.Katsanis N. The oligogenic properties of Bardet-Biedl syndrome. Hum. Mol. Genet. 2004;1:65–71. doi: 10.1093/hmg/ddh092. [DOI] [PubMed] [Google Scholar]
- 15.Mykytyn K, et al. Evaluation of complex inheritance involving the most common Bardet-Biedl syndrome locus (BBS1) Am. J. Hum. Genet. 2003;72:429–437. doi: 10.1086/346172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan Y, et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 17.Moore SJ, et al. Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: A 22-year prospective, population-based, cohort study. Am. J. Med. Genet. Part A. 2005;132A:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheffield VC. Use of isolated Populations in the study of a human obesity syndrome, the Bardet-Biedl syndrome. Pediatr. Res. 2004;55:908–911. doi: 10.1203/01.pdr.0000127013.14444.9c. [DOI] [PubMed] [Google Scholar]
- 19.Croft JB, et al. Obesity in Heterozygous Carriers of the Gene for the Bardet-Biedl Syndrome. Am. J. Med. Genet. 1995;55:12–15. doi: 10.1002/ajmg.1320550105. [DOI] [PubMed] [Google Scholar]
- 20.Benzinou M, et al. Bardet-Biedl syndrome gene variants are associated with both childhood and adult common obesity in French Caucasians. Diabetes. 2006;55:2876–2882. doi: 10.2337/db06-0337. [DOI] [PubMed] [Google Scholar]
- 21.Davenport JR, Yoder BK. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am. J. Physiol.-Renal Physiol. 2005;289:F1159–F1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 22.Singla V, Reiter JF. The primary cilium as the cell's antenna: Signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 23.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 24.Mykytyn K, et al. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Nat. Acad. Sci. U. S.A. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JB, et al. Comparative and basal genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 26.Kim JC, et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 27.Blacque OE, et al. Loss of C-elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yen HJ, et al. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 29.Tobin JL, Beales PL. Restoration of renal function in zebrafish models of ciliopathies. Pediatr. Nephrol. 2008;23 doi: 10.1007/s00467-008-0898-7. 2095-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura DY, et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Nat. Acad. Sci. U. S.A. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mykytyn K, et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat. Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 32.Davis RE, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc. Nat. Acad. Sci. U. S.A. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berbari NF, et al. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Nat. Acad. Sci. U. S.A. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brailov I, et al. Localization of 5-HT6 receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000;872:271–275. doi: 10.1016/s0006-8993(00)02519-1. [DOI] [PubMed] [Google Scholar]
- 35.Davenport JR, et al. Disruption of intraflagellar in adult mice leads to transport obesity and slow-onset cystic kidney disease. Curr. Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mak HY, et al. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 37.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 38.Loktev AV, et al. A BBSome Subunit Links Ciliogenesis, Microtubule Stability, and Acetylation. Dev. Cell. 2008;15:854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Seo SJ, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. U. S.A. 2010;107:2054–2059. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang AP, et al. Homozygosity mapping with SNP arrays identifies TRIM32 an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc. Natl. Acad. Sci. U. S.A. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Locke M, et al. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009;18:2344–2358. doi: 10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grace C, et al. Energy metabolism in Bardet-Biedl syndrome. Int. J. Obes. 2003;27:1319–1324. doi: 10.1038/sj.ijo.0802420. [DOI] [PubMed] [Google Scholar]
- 43.Green JS, et al. The Cardinal Manifestations of Bardet-Biedl Syndrome, A Form of Laurence-Moon-Biedl Syndrome. N. Engl. J. Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- 44.Feuillan PP, et al. Patients with Bardet-Biedl Syndrome Have Hyperleptinemia Suggestive of Leptin Resistance. J. Clin. Endocrinol. Metab. 2011 doi: 10.1210/jc.2010-2290. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore SJ, et al. Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am. J. Med. Genet. A. 2005;132:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beales PL. Lifting the lid on Pandora's box: the Bardet-Biedl syndrome. Curr. Opin. Genet. Dev. 2005;15:315–323. doi: 10.1016/j.gde.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaghloul NA, et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc. Nat. Acad. Sci. U. S.A. 2010;107:10602–10607. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eichers ER, et al. Phenotypic characterization of Bbs4 null mice reveals age-dependent penetrance and variable expressivity. Hum. Genet. 2006;120:211–226. doi: 10.1007/s00439-006-0197-y. [DOI] [PubMed] [Google Scholar]
- 50.Rahmouni K, et al. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J. Clin. Invest. 2008;118:1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Barone M, Leopold L, Friedman JM, et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 52.Morton GJ, et al. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 53.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- 54.Coll AP, et al. The hormonal control of food intake. Cell. 2007;129:251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fath MA, et al. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum. Mol. Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 56.Seo SJ, et al. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum. Mol. Genet. 2009;18:1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banks WA, et al. Leptin enters the brain by a saturable system independent of insulin. Peptides. 1996;17:305–311. doi: 10.1016/0196-9781(96)00025-3. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 59.Iida M, et al. Substitution at codon 269 (glutamine --> proline) of the leptin receptor (OB-R) cDNA is the only mutation found in the Zucker fatty (fa/fa) rat. Biochem.Biophys.Res.Commun. 1996;224:597–604. doi: 10.1006/bbrc.1996.1070. [DOI] [PubMed] [Google Scholar]
- 60.Iida M, et al. Phenotype-linked amino acid alteration in leptin receptor cDNA from Zucker fatty (fa/fa) rat. Biochem.Biophys.Res.Commun. 1996;222:19–26. doi: 10.1006/bbrc.1996.0691. [DOI] [PubMed] [Google Scholar]
- 61.Lee GH, et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 62.Phillips MS, et al. Leptin receptor missense mutation in the fatty Zucker rat. Nat.Genet. 1996;13:18–19. doi: 10.1038/ng0596-18. [DOI] [PubMed] [Google Scholar]
- 63.Takaya K, et al. Molecular cloning of rat leptin receptor isoform complementary DNAs--identification of a missense mutation in Zucker fatty (fa/fa) rats. Biochem.Biophys.Res.Commun. 1996;225:75–83. doi: 10.1006/bbrc.1996.1133. [DOI] [PubMed] [Google Scholar]
- 64.Myers MG, Cowley MA, Munzberg H, et al. Mechanisms of leptin action and leptin resistance. Annu. Rev. Physiol. 2008;70:537–556. doi: 10.1146/annurev.physiol.70.113006.100707. [DOI] [PubMed] [Google Scholar]
- 65.Bates SH, et al. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003;421:856–859. doi: 10.1038/nature01388. [DOI] [PubMed] [Google Scholar]
- 66.Myers MG, et al. The Geometry of Leptin Action in the Brain: More Complicated Than a Simple ARC. Cell Metab. 2009;9:117–123. doi: 10.1016/j.cmet.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forti E, et al. Temporal expression pattern of Bardet-Biedl syndrome genes in adipogenesis. Int. J. Biochem. Cell Biol. 2007;39:1055–1062. doi: 10.1016/j.biocel.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 68.Marion V, et al. Transient ciliogenesis involving Bardet-Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc. Nat. Acad. Sci. U.S.A. 2009;106:1820–1825. doi: 10.1073/pnas.0812518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baly C, et al. Leptin and its receptors are present in the rat olfactory mucosa and modulated by the nutritional status. Brain Res. 2007;1129:130–141. doi: 10.1016/j.brainres.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 70.Antal-Zimanyi I, Khawaja X. The Role of Melanin-Concentrating Hormone in Energy Homeostasis and Mood Disorders. J. Mol. Neurosci. 2009;39:86–98. doi: 10.1007/s12031-009-9207-6. [DOI] [PubMed] [Google Scholar]
- 71.Pissios P. Animals models of MCH function and what they can tell us about its role in energy balance. Peptides. 2009;30:2040–2044. doi: 10.1016/j.peptides.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frassetto A, et al. Reduced sensitivity to diet-induced obesity in mice carrying a mutant 5-HT6 receptor. Brain Res. 2008;1236:140–144. doi: 10.1016/j.brainres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 73.Badano JL, et al. The ciliopathies: An emerging class of human genetic disorders. Annu. Rev. Genom. Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 74.Saeki H, et al. Immotile Cilia Syndrome Associated with Polycystic Kidney. J. Urol. 1984;132:1165–1166. doi: 10.1016/s0022-5347(17)50080-4. [DOI] [PubMed] [Google Scholar]
- 75.Afzelius BA. Human Syndrome Caused by Immotile Cilia. Science. 193:317–319. doi: 10.1126/science.1084576. (1076) [DOI] [PubMed] [Google Scholar]
- 76.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 77.Yoder BK, et al. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
- 78.Afzelius BA. Cilia-related diseases. J. Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan SY, et al. Heterotaxy and complex structural heart defects in a mutant mouse model of primary ciliary dyskinesia. J. Clin. Invest. 2007;117:3742–3752. doi: 10.1172/JCI33284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hornef N, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respiratory and Critical Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cardenas-Rodriguez M, Badano JL. Ciliary Biology: Understanding the Cellular and Genetic Basis of Human Cillopathies. Am. J. Med. Genet. Part C-Seminars in Med. Genet. 2009;151C:263–280. doi: 10.1002/ajmg.c.30227. [DOI] [PubMed] [Google Scholar]
- 82.Collin GB, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat. Genet. 2002;31:74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- 83.Hearn T, et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat. Genet. 2002;31:79–83. doi: 10.1038/ng874. [DOI] [PubMed] [Google Scholar]
- 84.Slavotinek AM, et al. Mutations in MKKS cause Bardet-Biedl syndrome. Nat. Genet. 26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- 85.Collin GB, et al. Alms1-disrupted mice recapitulate human Alstrom syndrome. Hum. Mol. Genet. 2005;14:2323–2333. doi: 10.1093/hmg/ddi235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arsov T, et al. Fat Aussie - A new Alstrom syndrome mouse showing a critical role for ALMS1 in obesity, diabetes, and spermatogenesis. Mol. Endocrinol. 2006;20:1610–1622. doi: 10.1210/me.2005-0494. [DOI] [PubMed] [Google Scholar]
- 87.Irani BG, et al. Altered hypothalamic leptin, insulin, and melanocortin binding associated with moderate-fat diet and predisposition to obesity. Endocrinology. 2007;148:310–316. doi: 10.1210/en.2006-1126. [DOI] [PubMed] [Google Scholar]