Abstract
The transition from vegetative growth to reproductive development is a complex process that requires an integrated response to multiple environmental cues and endogenous signals. In Arabidopsis thaliana, which has a facultative requirement for vernalization and long days, the genes of the autonomous pathway function as floral promoters by repressing the central repressor and vernalization-regulatory gene FLC. Environmental regulation by seasonal changes in daylength is under control of the photoperiod pathway and its key gene CO. The root and leaf crop species Beta vulgaris in the caryophyllid clade of core eudicots, which is only very distantly related to Arabidopsis, is an obligate long-day plant and includes forms with or without vernalization requirement. FLC and CO homologues with related functions in beet have been identified, but the presence of autonomous pathway genes which function in parallel to the vernalization and photoperiod pathways has not yet been reported. Here, this begins to be addressed by the identification and genetic mapping of full-length homologues of the RNA-regulatory gene FLK and the chromatin-regulatory genes FVE, LD, and LDL1. When overexpressed in A. thaliana, BvFLK accelerates bolting in the Col-0 background and fully complements the late-bolting phenotype of an flk mutant through repression of FLC. In contrast, complementation analysis of BvFVE1 and the presence of a putative paralogue in beet suggest evolutionary divergence of FVE homologues. It is further shown that BvFVE1, unlike FVE in Arabidopsis, is under circadian clock control. Together, the data provide first evidence for evolutionary conservation of components of the autonomous pathway in B. vulgaris, while also suggesting divergence or subfunctionalization of one gene. The results are likely to be of broader relevance because B. vulgaris expands the spectrum of evolutionarily diverse species which are subject to differential developmental and/or environmental regulation of floral transition.
Keywords: Autonomous pathway, circadian clock, FLK, floral transition, FVE, sugar beet
Introduction
Floral transition is a major developmental switch which is tightly controlled by a network of proteins that perceive and integrate environmental and developmental signals to promote or inhibit the transition to reproductive growth. In the model species Arabidopsis thaliana, several regulatory pathways which differ in their response to distinct cues have been defined, including the vernalization, photoperiod, and autonomous pathway (for reviews, see He and Amasino, 2005; Bäurle and Dean, 2006; Jung and Müller, 2009; Michaels, 2009). The central regulator of the vernalization response is FLOWERING LOCUS C (FLC), which acts as a repressor of flowering and is down-regulated in response to prolonged exposure to cold over winter. The promotion of floral transition by long days is mediated by CONSTANS (CO), a key protein of the photoperiod pathway which activates the floral integrator gene FLOWERING LOCUS T (FT). Plant genome and expressed sequence tag (EST) sequencing projects in species other than Arabidopsis, together with functional studies have begun to unveil the presence and evolutionary conservation of floral regulatory genes across taxa (e.g. Hecht et al., 2005; Albert et al., 2005; Remay et al., 2009; Mouhu et al., 2009). While components of the photoperiod pathway are widely conserved, the regulation of the vernalization requirement and response appears to have diverged considerably during evolution, as exemplified by distinct mechanisms in Arabidopsis and temperate cereals (Turck et al., 2008; Colasanti and Coneva, 2009; Distelfeld et al., 2009; Greenup et al., 2009; Jung and Müller, 2009). The phylogenetic lineage leading to Beta vulgaris, which includes the biennial crop subspecies sugar beet (Beta vulgaris L. ssp. vulgaris) as well as annual and perennial wild beets, diverged from that leading to Arabidopsis ∼120 million years ago—that is, relatively soon after the monocot–dicot divergence (Chaw et al., 2004; Davies et al., 2004). In beet, the vernalization requirement is under the control of the bolting gene B (Munerati, 1931; Abegg, 1936; Boudry et al., 1994; El-Mezawy et al., 2002), which is not related to FLC (Reeves et al., 2007; A. E. Müller et al., unpublished), and a second, unlinked locus B2 which may act epistatically to B (Büttner et al., 2010). In the absence of the dominant early bolting allele at the B locus, beets possess an obligate requirement for both vernalization and long photoperiods, and under high temperature and short-day conditions are prone to reversion to a vegetative state by devernalization. Despite apparent differences in the regulation of floral transition between A. thaliana and B. vulgaris, the recent identification of beet homologues of FLC and CO suggest at least partial conservation of the genetic basis of the plants’ responses to the environment (Reeves et al., 2007; Chia et al., 2008). The FLC-like gene BvFL1 in beet is regulated by vernalization, and delays flowering in transgenic Arabidopsis plants, suggesting that BvFL1 may also be a floral repressor (Reeves et al., 2007). Similarly, evolutionary conservation of CO homologues was suggested by overexpression of the CO-like gene BvCOL1 in Arabidopsis, which complements the late-flowering phenotype of a loss-of-function co mutation and activates FT expression (Chia et al., 2008).
Floral transition in Arabidopsis is also regulated by the autonomous pathway of flowering time control whose genes are thought to function largely in parallel to the vernalization pathway upstream of FLC and the photoperiod pathway (for reviews, see Boss et al., 2004; Simpson, 2004; Quesada et al., 2005). Autonomous pathway genes repress FLC and thus act as promoters of floral transition, and include FLOWERING LOCUS CA (FCA), FLOWERING LOCUS D (FLD), FLOWERING LOCUS KH DOMAIN (FLK), FLOWERING LOCUS PA (FPA), FLOWERING LOCUS VE (FVE), FLOWERING LOCUS Y (FY), and LUMINIDEPENDENS (LD) (Simpson, 2004). They have in common that mutations in these genes are generally recessive and delay flowering under both long-day and short-day conditions, while the inhibitory effect of the mutations can be overcome by vernalization. Mutations at FLC eliminate the late-flowering phenotype caused by mutations in autonomous pathway genes (Koornneef et al., 1994; Lee et al., 1994; Sanda and Amasino, 1996; Michaels and Amasino, 2001).
Although some genes of the autonomous pathway interact genetically and all share a common target, they do not form a single linear pathway with a hierarchical order of activities, but rather constitute different regulatory subgroups (or ‘subpathways’; Marquardt et al., 2006). Autonomous pathway genes regulate FLC expression through RNA-based control mechanisms and/or by chromatin modification (Boss et al., 2004; Simpson, 2004; Quesada et al., 2005; Bäurle et al., 2007; Bäurle and Dean, 2008). Four genes mediate RNA regulatory processes, FCA, FPA, FY, and FLK. FCA and FPA encode plant-specific RNA-binding proteins which both carry multiple RNA recognition motifs (RRMs; Macknight et al., 1997; Schomburg et al., 2001). FCA physically and genetically interacts with the RNA 3' end processing factor FY, and this interaction is required both for correct processing of transcripts derived from FCA itself and (directly or indirectly) for down-regulation of FLC expression (Quesada et al., 2003; Simpson et al., 2003). FCA and FPA interact genetically with FLD, which encodes a chromatin regulatory protein of the autonomous pathway (see below), and at least part of the effect of FCA and FPA on FLC expression and flowering time depends on FLD (Liu et al., 2007; Bäurle and Dean, 2008). Thus, FCA and FPA appear to link RNA and chromatin level control of gene expression.
An analysis of flowering time in various autonomous pathway double mutants indicated that the fourth protein predicted to function in RNA regulation, FLK, acts independently of both FCA and FPA (Bäurle and Dean, 2008; Ripoll et al., 2009). FLK expression was not detectably affected in any of the other six autonomous pathway mutants analysed (fca, fpa, fy, fld, fve, and ld), and, vice versa, the expression of all autonomous pathway genes tested (FCA, FPA, FVE, and LD) was unaltered in an flk mutant (Lim et al., 2004). FLK encodes a plant-specific putative RNA-binding protein which contains three K-homology (KH)-type RNA-binding domains (Lim et al., 2004; Mockler et al., 2004). The mode of action of the FLK product is not known, but other KH domain proteins in Arabidopsis, including HUA ENHANCER 4 (HEN4) (harbouring five KH domains) and RS2-INTERACTING KH PROTEIN (RIK), were shown to be part of protein complexes which mediate pre-mRNA processing or have been implicated in RNA-directed chromatin regulation of gene expression, respectively (Cheng et al., 2003; Phelps-Durr et al., 2005). Also, both correctly spliced FLC transcripts and intron-retaining variants accumulated to higher levels in an flk mutant than in wild-type plants (Ripoll et al., 2009), and repression of AtSN1, a retroelement which is subject to RNA-directed chromatin silencing, was at least partially released in mutant plants (Bäurle and Dean, 2008; Veley and Michaels, 2008). Together, these findings have been interpreted to indicate that FLK may suppress FLC at least partially at the transcriptional level, perhaps through RNA-directed chromatin silencing (Veley and Michaels, 2008; Ripoll et al., 2009). Ripoll et al. (2009) further found that PEPPER (PEP), a paralogue of FLK in Arabidopsis, acts as a positive regulator of FLC. The authors showed that pep mutations can at least partially rescue the flowering time phenotype of flk mutants, and that overexpression of PEP resulted in a similar effect to mutation of FLK on flowering time. Overexpression of PEP in an flk mutant background neither further delayed flowering nor led to an increase of FLC expression when compared with flk mutant plants not carrying the PEP transgene, suggesting that FLK and PEP may interact in the same genetic pathway (Ripoll et al., 2009).
Chromatin level control of FLC expression is mediated by FLD, FVE, and LD. FLD is a homologue of the human histone H3K4 demethylase LSD1 (LYSINE-SPECIFIC HISTONE DEMETHYLASE1) and, in A. thaliana, represses FLC by H3K4 demethylation and H4 deacetylation of FLC chromatin, possibly as part of a co-repressor complex (He et al., 2003; Jiang et al., 2007), and is dependent on the sumoylation state of FLD (Jin et al., 2008). The Arabidopsis genome contains three additional homologues of FLD, LSD1-LIKE1 (LDL1), LDL2, and LDL3, two of which (LDL1 and LDL2) have been shown to act in partial redundancy with FLD to repress FLC (Jiang et al., 2007). Furthermore, LDL1 (also termed SWP1, SWIRM DOMAIN PAO PROTEIN 1) interacts with the histone methyltransferase CZS (C2H2 ZINC FINGER-SET DOMAIN PROTEIN) and is part of a co-repressor complex which represses FLC by H4 deacetylation and H3K9 and H3K27 methylation at the FLC locus (Krichevsky et al., 2007). LD, a unique nuclear-localized protein in Arabidopsis which contains a homeodomain-like domain (Lee et al., 1994; Aukerman et al., 1999), also appears to regulate FLC expression by histone modification, including H3K4 demethylation and H3 deacetylation (Domagalska et al., 2007), but may also repress FLC by a negative regulatory interaction with a transcriptional activator of FLC, SUF4 (SUPPRESSOR OF FRIGIDA 4; Kim et al., 2006). FVE [also termed MSI4 (MULTICOPY SUPPRESSOR OF IRA1 4) and ACG1 (ALTERED COLD-RESPONSIVE GENE EXPRESSION 1)] is homologous to MSI1 (MULTICOPY SUPPRESSOR OF IRA1) in yeast and retinoblastoma-associated proteins in animals, which are components of chromatin assembly complexes. FVE is part of a small family of MSI1-like WD40 repeat proteins in Arabidopsis (Kenzior and Folk, 1998; Ausin et al., 2004; Kim et al., 2004; Hennig et al., 2005). In fve mutants, histones H3 and H4 are hyperacetylated in FLC chromatin, suggesting that FVE, perhaps together with FLD, is part of a complex which represses FLC expression by chromatin modification (He et al., 2003; Amasino, 2004; Ausin et al., 2004). FVE and other MSI1-like proteins are generally thought of as structural proteins without catalytic function and may provide a scaffold for assembly of larger complexes. FVE has also been implicated in temperature-dependent regulation of flowering time, and appears to promote flowering in response to elevated ambient temperatures through an FLC-independent, thermosensory pathway which also includes FCA (Blázquez et al., 2003). In addition, FVE mediates the plant's response to intermittent cold stress and may provide a link between cold stress response and flowering time control (Kim et al., 2004; Franklin and Whitelam, 2007). FVE is expressed in all major plant organs, but appears to be preferentially expressed in actively dividing cells, and has been assigned a more general role in the regulation of cellular differentiation and developmental transitions (Morel et al., 2009). Recent grafting experiments in A. thaliana suggest that FVE mRNA is phloem mobile and may contribute to long-distance signalling in plant development (Yang and Yu, 2010).
There is increasing evidence that several, if not all, autonomous pathway genes also regulate developmental processes other than floral transition. In particular, double mutant analyses showed that fpa fld, fpa fve, and fpa ld mutants have pleiotropic, FLC-independent effects on growth rate, chlorophyll content, leaf morphology, flower development, and fertility, and that the corresponding genes have partially redundant functions (Veley and Michaels, 2008). Furthermore, mutations in FCA, FVE, and LD result in an increase in the period length of the circadian clock, thus implicating autonomous pathway genes in the regulation of the clock (Salathia et al., 2006). Finally, transposon and transgene silencing assays in mutants indicated that FCA, FPA, FLK, and FVE have a more widespread role in RNA-directed chromatin silencing of a range of target genes (Bäurle et al., 2007; Bäurle and Dean, 2008; Veley and Michaels, 2008). For FCA, FVE, FY, and LD at least partial conservation of floral regulatory functions was shown in monocots (van Nocker et al., 2000; Lee et al., 2005; Lu et al., 2006; Baek et al., 2008; Jang et al., 2009).
Here, the components of the autonomous pathway in B. vulgaris have begun to be dissected through a survey of ESTs with homology to autonomous pathway genes and isolation of the corresponding genes. For beet homologues of four autonomous pathway genes, termed BvFLK, BvFVE1, BvLD, and BvLDL1, the full-length genomic and coding sequences were identified and the genes were mapped on a reference map of the sugar beet genome. Exon–intron structure and domain organization were found to be conserved between beet and Arabidopsis in all four genes. One homologue each of autonomous pathway genes implicated in RNA or chromatin regulatory mechanisms, BvFLK and BvFVE1, respectively, was further characterized by overexpression and complementation analysis in A. thaliana wild type and mutants. BvFLK was able to accelerate bolting time in A. thaliana wild type and complement the late-bolting phenotype of an flk mutant. In contrast, BvFVE1 was unable to complement an fve mutant, and was found to be under circadian clock regulation in beet, which has not been reported for FVE in Arabidopsis. Together, the data suggest conservation of autonomous pathway components in B. vulgaris, while also providing first evidence for divergence or subfunctionalization of at least one autonomous pathway gene homologue.
Materials and methods
Bioinformatic analyses
The B. vulgaris EST database BvGI (Beta vulgaris Gene Index, versions 2.0 and 3.0; http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=beet) and the B. vulgaris subsets of the NCBI EST and nt/nr databases (http://www.ncbi.nlm.nih.gov) were used to identify beet homologues of flowering time genes in A. thaliana. Database searches were performed using the tblastn algorithm (Altschul et al., 1990) and A. thaliana protein sequences from the Arabidopsis Genome Initiative (AGI; http://www.arabidopsis.org/tools/bulk/sequences/index.jsp) as queries. To help infer orthology by bidirectional best hit (BBH) analysis (Overbeek et al., 1999), the beet sequences retrieved through this analysis were used as queries for blastx searches against A. thaliana protein sequences at NCBI (http://www.ncbi.nlm.nih.gov) and TAIR (http://www.arabidopsis.org). Beta vulgaris sequences were annotated using pairwise sequence alignments (BLAST2; http://www.ncbi.nlm.nih.gov) against putative A. thaliana orthologues and the FGENESH+ and FGENESH_C gene prediction programs (http://linux1.softberry.com/berry.phtml) for annotation of exon–intron structures, TSSP (http://linux1.softberry.com/berry.phtml) and PLACE for annotation of promoter regions (http://www.dna.affrc.go.jp/PLACE; Higo et al., 1999), and PFAM (http://pfam.sanger.ac.uk) for identification of conserved protein domains. Multiple sequence alignments were made using CLUSTAL W (http://www.ebi.ac.uk/Tools/clustalw/index.html). Amino acid identity was calculated as the percentage of identical residues in two homologues divided by the total number of residues in the reference gene. For phylogenetic analysis, putative FLK and FVE orthologues in other plant species were identified by blastp searches of the NCBI Reference Sequence (RefSeq) protein database (http://www.ncbi.nlm.nih.gov/refseq) and BBH analysis essentially as described above, except that the blastp algorithm was used. The sequences were aligned using CLUSTAL W, and unrooted phylogenetic trees were constructed using the Neighbor–Joining algorithm and the Dayhoff PAM matrix as implemented in the MEGA4 software (Tamura et al., 2007).
Plant material and growth conditions
For expression analysis in different tissues, the biennial B. vulgaris accession A906001 (El-Mezawy et al., 2002) was grown in the greenhouse under long-day conditions supplemented with artificial light [Son-T Agro 400W (Koninklijke Philips Electronics NV, Eindhoven, The Netherlands) for 16 h]. Six-week-old plants were vernalized under short-day (8 h light) conditions at 5 °C for 3 months. Plants were returned to the greenhouse and continued to be grown under the same conditions as before vernalization until the first flowers opened (BBCH scale 60; Meier, 2001). For diurnal and circadian expression, the commercial biennial cultivar Roberta (KWS Saatzucht GmbH, Einbeck, Germany) was grown under defined light regimes (long days of 16 h and short days of 8 h) in Sanyo Gallenkamp MLR 350 growth chambers at 22 °C. Lighting was supplied by 36 W fluorescent Daystar lamps (CEC Technology) providing 300 μmol m−2 s−1 of photosynthetically active radiation.
A. thaliana flowering time gene mutants SALK_112850 (flk-1; Alonso et al., 2003; Lim et al., 2004) and SALK_013789 (Alonso et al., 2003), which will be referred to as fve-7 (following on from the fve mutant number in Morel et al., 2009), were received from the Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info/). The fve-7 mutant carries a T-DNA insertion in intron 1 at nucleotide position 2835 of the genomic sequence entry for FVE in GenBank (accession number AF498101). Mutants homozygous for the T-DNA inserts in FLK or FVE, respectively, were identified by PCR using a T-DNA-specific primer (A479 for flk-1, B478 for fve-7; for primer sequences see Supplementary Table S1 available at JXB online) in combination with a gene-specific primer (A477 for flk-1, B476 for fve-7). The absence of the wild-type alleles was confirmed by PCR with gene-specific primers which flank the insert on either side (A477 and A478 for flk-1, and B476 and B477 for fve-7). A. thaliana Col-0 (wild-type) plants, mutants, and the T1 and T2 generations of transgenic plants were phenotyped for bolting time under long-day conditions (16 h light, Osram L58 W77 Fluora and Osram L58 W840 Lumilux Cool White Hg) at 22 °C in a growth chamber (BBC Brown Boveri York, Mannheim, Germany).
Bacterial artificial chromosome (BAC) library screening
EST sequence information was used to generate genomic fragments for use as probes to screen the B. vulgaris BAC library described by Schulte et al. (2006). Probe fragments were generated by PCR amplification using primer combinations A039–A064 for BvFLK (717 bp), A066–A042 for BvFVE1 (1089 bp), A043–A067 for BvLD (589 bp), and A033–A065 for BvLDL1 (678 bp), and genomic DNA of A906001 as template, and purified using the Montage PCR96 Cleanup Kit (Millipore Corporation, Bedford, CA, USA). The probes were labelled and hybridized to high-density BAC filters essentially as described by Hohmann et al. (2003). Positive clones were verified by PCR analysis using the same primer combinations as for PCR amplification of probe fragments. BAC DNA was isolated using the NucleoBond BAC 100 kit according to the manufacturer's protocol (Macherey-Nagel, Dürren, Germany). The genes of interest were sequenced by primer walking (BvFLK) or whole BAC sequencing (BvFVE1, BvLD, and BvLDL1) on a GS20 sequencing machine (MWG, Ebersberg, Germany).
RT-PCR and RACE
Total RNA was extracted from roots, stems, leaves, and flowers of adult plants of accession A906001 using the Plant RNAeasyKit™ (Qiagen, Hilden, Germany) and DNase treated (Ambion, Austin, TX, USA). A 1.5 μg aliquot of RNA was reverse transcribed using the First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany), and the cDNA was diluted 10 times for RT-PCR. The complete coding sequences of BvFLK, BvFVE1, and BvLDL1 were amplified using leaf cDNA as template and primer combinations A396–A397, A836–A837, and A823–A825, respectively. The coding sequence of BvLD was amplified by RT-PCR of several overlapping fragments, using primer combinations A067–A108, B391–A046, A043–B392, A068–A069, and A045–B396. Primers for RT-qPCR were designed and optimized to 94.6% amplification efficiency for BvFLK (primers B042-B043), and 92.9% for BvFVE1 (primers A066–A042). Fluorescence of the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen Corporation, Carlsbad, CA, USA) was measured in an CFX96 real-time PCR machine (Bio-Rad, Munich, Germany) over 40 cycles at annealing temperatures of 65 °C for BvFLK and BvFVE1, and 60 °C for glyceraldehyde 3-phosphate dehydrogenase (BvGAPDH; BvGI 2.0 accession number TC1351; RT-qPCR amplification efficiency 96.3%). Expression levels were measured in triplicate and normalized against the reference gene BvGAPDH.
For diurnal and circadian expression analysis, first-strand cDNA was prepared and analysed by RT-qPCR as described (Chia et al., 2008) except that an in-solution DNase treatment (Ambion) was used and that expression data were normalized against three B. vulgaris housekeeping genes, BvGAPDH (see above), elongation factor 1-α (BvEF1α; BvGI 2.0 accession number TC5), and elongation factor 2 (BvEF2; BvGI 2.0 accession number TC64). A normalization factor (NF) was generated for each sample using the geNorm Software v3.5 (http://medgen.ugent.be/∼jvdesomp/genorm/; Vandesompele et al., 2002). The NF was used to normalize and calculate the relative expression values for the genes of interest. All primers for normalizer genes and BvFVE1 were optimized to 98–110% amplification efficiency using serial dilutions of sugar beet leaf cDNA. Amplified fragments were sequenced to confirm specificity. Fluorescence of the bound SYBR-GREEN (Stratagene, La Jolla, CA, USA) was detected in an MX3000 real-time PCR machine (Stratagene) over 40 cycles at 60 °C annealing temperature.
The 5' end of BvFVE1 was identified by 5′-rapid amplification of cDNA ends (RACE) (Frohman et al., 1988) using the GeneRacer™ kit according to the manufacturer's protocol (Invitrogen Corporation). 5′-RACE was performed on double-stranded adaptor-ligated cDNA synthesized from 5 μg of total RNA from leaves of 4-week-old sugar beet plants using exon-specific primers (5′-RACE primer A830 and 5′-RACE nested primer A831). 5′-RACE fragments were cloned into the pGEM-T vector (Promega Corporation, Madison, WI, USA) and sequenced using standard Sp6 and T7 primers.
For expression analysis of FLC and FLK in A. thaliana, total RNA was isolated from rosette leaves of 30-day-old plants using the Plant RNAeasyKit™ (Qiagen) and DNase treated (Fermentas). A 2 μg aliquot of RNA was reverse transcribed using the First Strand cDNA Synthesis Kit (Fermentas), and the cDNA was diluted 10 times for RT-qPCR. Amplification efficiency of FLC (GenBank accession number NM_121052, primers B336-B337), FLK (GenBank accession number AC011437, B281-B282), and GAPDH (GenBank accession number NM_111283; B349-B350) was 100, 97.4, and 94.2%, respectively.
Vector construction and transformation of A. thaliana
The coding sequences of BvFLK and BvFVE1 were amplified as described above. The BvFLK coding sequence was cloned into pGEM-T (Promega Corporation) to yield plasmid pFT002, and re-amplified from pFT002 with primers A680-XhoI and A652-SpeI. The PCR product was restricted with XhoI and SpeI (Fermentas) and cloned into the corresponding restriction enzyme sites of the binary vector pSR752Ω (kindly provided by Chonglie Ma and Richard Jorgensen, University of Arizona, Tucson, AZ, USA) which carries the cauliflower mosaic virus (CaMV) 35S promoter. The resulting construct was designated pFT013. A 1813 bp fragment carrying the endogenous promoter region of BvFLK and 363 bp of the 5′ region of the coding sequence was amplified from BAC DS 794 using primers A896-EcoRI and A821-HpaI. The fragment was restricted with EcoRI and HpaI and inserted into the corresponding restriction sites of pFT013, thus effectively replacing the CaMV 35S promoter by the 1435 bp endogenous beet sequence upstream of the BvFLK start codon in the resultant plasmid (pFT033). A plasmid carrying the coding region of FLK (GenBank accession number BX823281) was kindly provided by the French Genomic Resource Center (INRA-CNRGV, Castanet Tolosan cedex, France). The coding sequence of AtFLK was amplified using primers A901-XhoI and A938-SmaI, and inserted into the corresponding restriction sites of pSR752Ω. The resulting vector carries the FLK coding sequence under the control of the CaMV 35S promoter and was designated pFT016. The coding sequence of BvFVE1 was cloned into pDONOR221 using the Gateway Cloning System (Invitrogen Corporation) to yield plasmid pBS355 The coding sequence was subsequently transferred into the pEarleyGate 100 vector (Earley et al., 2006) to yield the binary vector pBS356, in which the BvFVE1 coding sequence is under the control of the CaMV 35S promoter.
The intactness of the binary vectors and the sequence of all inserts were confirmed by restriction enzyme digests, PCR amplification, and sequencing (Institute of Clinical Molecular Biology, Kiel, Germany). The constructs were transferred by electroporation into Agrobacterium tumefaciens LBA 4404 using Electromax (Invitrogen Corporation) competent cells (pFT013, pFT016, and pFT033) or A. tumefaciens GV2260 competent cells prepared according to the protocol of Mersereau et al. (1990) (pBS356), and transformed into A. thaliana by the floral dip method (Clough and Bent, 1998). Primary transformants (T1 plants) were selected by spraying BASTA (Bayer CropScience, Wolfenbüttel, Germany) at a concentration of 1.7 g l−1 at the two-leaf stage. The presence of the transgene in BASTA-resistant plants was confirmed by PCR analysis using primer combinations B042–B043 for pFT013 and pFT033, B281–B282 for pFT016, and A875–A876 for pBS356. The transformants were propagated by selfing to produce T2 seed. Genomic DNA was extracted using the NucleoSpin 96 Plant DNA isolation kit (Macherey and Nagel, Düren, Germany).
Genetic mapping and statistical analysis
Flowering time genes were mapped genetically in the D2 (100 F2 individuals) and K1 (97 F2 individuals) reference populations described by Schneider et al. (2007). Polymorphisms were identified by PCR amplification and sequencing of genomic fragments using DNA from the parent and F1 plants as template. Map positions were calculated using Join Map 3.0 (Van Ooijen and Voorrips, 2001) and the Kosambi mapping function (Kosambi, 1944) at a LOD score of 4.0.
Analysis of variance (ANOVA) and t-tests were performed using SAS 9.1 TS level 1M3 (SAS Institute, Cary, NC, USA). Sample groups with significantly different means were further analysed using Fisher's least significant difference (LSD) test at a 5% probability level (SAS 9.1 TS level 1M3).
Results
Beta vulgaris homologues of autonomous pathway genes
To identify autonomous pathway gene candidates in beet, the B. vulgaris EST database BvGI (versions 2.0 and 3.0) and the B. vulgaris subsets of the NCBI EST and nt/nr databases were searched for homologues to autonomous pathway genes in A. thaliana (see Materials and methods). Among the seven classical genes assigned to the autonomous pathway in Arabidopsis (FCA, FLD, FLK, FPA, FVE, FY, and LD), four were found to have putative orthologues in beet [FLK (BQ586739), FPA (BQ489608), FVE (BQ592158, EG550040), and LD (BQ589018, BQ594506); Table 1]. Besides FLK, one EST (BQ590839) was identified whose closest homologue in A. thaliana is PEP, an FLK paralogue (Ripoll et al., 2009; see Introduction). In addition, one EST (CV301493) with homology to FLD appears to be orthologous to LDL1/SWP1, an FLD-like gene in Arabidopsis which recently was also assigned to the autonomous pathway (Jiang et al., 2007; Krichesky et al., 2007; see Introduction). Orthologous ESTs were not identified for FLD, FCA and FY.
Table 1.
Beta vulgaris ESTs with homology to Arabidopsis thaliana autonomous pathway genes
| At locus number | Gene | Best hit(s) in B. vulgaris (GenBank accession no)a | E-value | Best hit in A. thaliana (At locus no or gene name)b |
| At4g16280 | FCA | TC13484 | 6.1e-17 | At1g44910 |
| At3g10390 | FLD | CV301493 | 6.6e-71 | LDL1/SWP1 |
| At3g04610 | FLK | BQ586739 | 8.4e-83 | FLK |
| BQ590839 | 8.8e-44 | PEP | ||
| At2g43410 | FPA | BQ489608 | 7.4e-17 | FPA |
| At2g19520 | FVE | EG550040 | 4.4e-85 | FVE |
| BQ592158 | 2.4e-62 | FVE | ||
| At5g13480 | FY | BQ588779 | 1.3e-15 | At4g02730 |
| At4g02560 | LD | BQ589018 | 4.0e-27 | LD |
| BQ594506 | 1.6e-18 | LD |
Beta vulgaris ESTs or TCs (tentative consensus sequences) were identified by tblastn sequence similarity searches in BvGI 3.0 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=beet).
Best hits in A. thaliana were identified by blastx in the TAIR9 protein database (http://www.arabidopsis.org).
For four putative autonomous pathway genes, named BvFLK, BvFVE1 (corresponding to one of the two homologous ESTs), BvLD, and BvLDL1, the complete genomic sequence was identified by BAC library screening and BAC sequencing or primer walking. The two ESTs with homology to LD were both found to derive from the same gene. The exon–intron structure of BvFLK, BvFVE1, BvLD, and BvLDL1 was determined by RT-PCR and sequencing. The number of exons and the sites of introns were found to be conserved between the corresponding A. thaliana and B. vulgaris genes (Fig. 1A; Supplementary Fig. S1 at JXB online). However, several of the B. vulgaris introns are substantially larger than the respective introns in A. thaliana and contain repetitive elements such as minisatellites and various short low complexity and/or simple repeat regions [e.g. (AT)n], but longer transposons or retroelements were not identified. BvLDL1, like LDL1 in A. thaliana (but unlike FLD), contains only a single exon. The coding sequences of the four B. vulgaris genes are somewhat shorter than those of their Arabidopsis counterparts (BvFLK 1674 bp versus FLK 1731 bp; BvFVE1 1413 bp versus FVE 1524 bp; BvLD 2829 bp versus LD 2859 bp; BvLDL1 2487 bp versus LDL1 2532 bp).
Fig. 1.
Sequence and structure of the autonomous pathway gene homologues BvFLK and BvFVE1. (A) Exon–intron structure of BvFLK, BvFVE1, and the respective A. thaliana genes (FLK, accession number AAX51268; FVE, accession number AF498101). Exons are indicated as black rectangles, and the position of start and stop codons is indicated by arrows and vertical bars, respectively. (B) Pairwise sequence alignments and domain organization. The alignments were generated using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Identical and similar residues are highlighted by black or grey boxes, respectively. The position of protein domains according to Pfam 22.0 (http://pfam.sanger.ac.uk/) is marked by horizontal lines above the alignment. In FLK, the positions of three perfect eight-residue repeats and the core residues of K-homology RNA-binding (KH) domains (Mockler et al., 2004) are indicated by arrows and asterisks, respectively. The first and sixth WD40 repeat domains (WD1 and WD6) were not identified by Pfam and were annotated according to Ausin et al. (2004). A putative nuclear localization signal (NLS; black line) in FVE according to Ausin et al. (2004) and a potential zinc-binding site (unfilled box) in WD6 (Kenzior and Folk, 1998) are also indicated. WD, WD40 repeat domain; CAF1c, CAF1 subunit C/histone-binding protein RBBP4 domain.
The predicted protein sequences were aligned against the corresponding A. thaliana genes (Fig. 1B; Supplementary Fig. S1 at JXB online). For FLK and FVE, homologues had also been identified in rice (Lim et al., 2004; Baek et al., 2008) and were included in the alignment. The overall amino acid sequence identity between the proteins in A. thaliana and B. vulgaris was highest for FVE (72%), intermediate for FLK (57%) and LDL1 (58%), and relatively low for LD (43%). The domain organization of all four proteins was largely conserved, and the degree of sequence conservation between homologues was highest within domains. In particular, all domains in BvFLK (three KH-type RNA-binding domains, with 77–88% amino acid identity to the corresponding domains in the A. thaliana homologue) and in BvFVE1 were highly conserved [a chromatin assembly factor 1 subunit C (CAF1c) domain with 96% amino acid identity, and six WD40 repeat domains with 83–95% amino acid identity]. In contrast, in BvFLK, BvFVE1, and BvLDL1, the N-terminal protein regions are only slightly conserved. Furthermore, 50 amino acids at the N-terminus of FVE including a putative nuclear localization signal (amino acids 20–30; Ausin et al., 2004) are absent in BvFVE1.
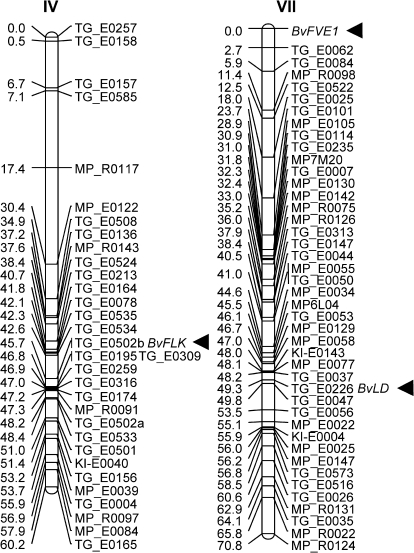
Genetic map positions
The genes were mapped on a single nucleotide polymorphism (SNP)-based genetic reference map of expressed genes in sugar beet (Schneider et al., 2007). BvFLK carries a SNP in the 3′ untranslated region (UTR) and was mapped to chromosome IV, where it colocalizes with marker TG_0502b at a cumulative genetic distance of 45.7 cM (Fig. 2). BvFVE1 carries three SNPs within a 1457 bp fragment of intron 4 and was mapped to a position at the very distal end of chromosome VII. This map position is consistent with the presence of several sequences on the BAC clone that carries BvFVE1, which show homology to ApaI and RsaI satellite sequences known to be located in subtelomeric regions in sugar beet (Dechyeva and Schmidt, 2006), including several subtelomeric repeats located just upstream of BvFVE1. The two ESTs corresponding to BvLD had been mapped previously to chromosome VII by Schneider et al. (2007) and are represented by markers TG_E0240 (BQ589018) and TG_E0226 (BQ594506; Fig. 2). BvLDL1 was mapped to position 10.90 cM on chromosome IX (data not shown).
Fig. 2.
Genetic map positions. BvFLK and BvFVE1 (arrowheads) were mapped to position 45.7 cM on chromosome IV and the top end of chromosome VII, respectively, on a reference map of the sugar beet genome (Schneider et al., 2007). The map position of an EST which had been mapped previously (Schneider et al., 2007) and corresponds to BvLD is also indicated. Genetic distances in centiMorgans (cM) are given on the left, and marker names on the right.
BvFLK accelerates the time to bolting in Arabidopsis and complements the flk-1 mutation
Overexpression and complementation analyses in A. thaliana wild-type and mutant plants were employed to assess whether the function of autonomous pathway gene homologues is conserved between beet and Arabidopsis. BvFLK and BvFVE1 were chosen as homologues of autonomous pathway genes which, respectively, are putative RNA regulatory genes or exert their function at the level of chromatin.
The coding sequence of BvFLK driven by the CaMV 35S promoter (‘35S::BvFLK’), the BvFLK coding sequence driven by the 1435 bp genomic region upstream of the BvFLK coding sequence in beet, which will be referred to as the endogenous BvFLK promoter (‘endo::BvFLK’), and the coding sequence of FLK under the control of the CaMV 35S promoter (‘35S::AtFLK’) were transformed into A. thaliana Col-0 and the flk mutant SALK_112850 [Alonso et al., 2003; corresponding to flk-1 in Lim et al. (2004) and to flk-4 in Mockler et al. (2004)]. Under long-day conditions, bolting in the flk-1 mutant was found to be delayed by 31–33 d when compared with Col-0, the genetic background of the mutant (Table 2).
Table 2.
Number of days to bolting (DTB) and total number of leaves at bolting (TNL) of primary transformants (T1 generation) and transgenic plants in segregating T2 populations derived from transformation of BvFLK and FLK into A. thaliana Col-0 and the flk mutant SALK_112850 (flk-1)
| Overexpression in Col-0 |
Complementation in flk-1 |
|||||
| Genotype | No. of plants | DTB (mean± SD) | TNL (mean ±SD) | No. of plants | DTB (mean ±SD) | TNL (mean ±SD) |
| T1 generation | ||||||
| 35S::BvFLK | 32 | 29.59±2.43 a | 10.59±1.83 a | 25 | 32.00±2.10 a | 13.08±1.55 a |
| endo::BvFLK | 15 | 30.00±2.56 a | 11.20±1.52 a | 9 | 34.89±3.79 a | 13.33±2.29 a |
| 35S::AtFLK | 10 | 30.90±2.08 a | 12.00±1.94 b | 4 | 31.50±4.20 a | 10.57±1.26 a |
| Col-0 | 16 | 35.69±1.70 b | 13.31±1.14 b | 16 | 35.69±1.70 a | 13.31±1.14 a |
| flk-1 mutant | – | – | – | 22 | 67.71±7.15 b | 66.35±1.32 b |
| F-value (probability) | 27.45 (0.00) | 10.03 (0.00) | 383.00 (0.00) | 3397.00 (0.00) | ||
| LSD0.05a | 2.14 | 1.46 | 4.42 | 2.05 | ||
| T2 generation | ||||||
| 35S::BvFLK | 13 | 27.77±2.59 a | 15.23±1.79 a | 14 | 34.14±3.06 a | 15.43±1.16 a |
| endo::BvFLK | 13 | 28.46±1.66 a | 15.31±1.71 a | 12 | 33.92±3.62 a | 15.51±1.29 a |
| 35S::AtFLK | 14 | 27.23±2.05 a | 15.00±2.00 a | 15 | 34.13±5.60 a | 15.80±1.08 a |
| Col-0 | 17 | 37.88±1.50 b | 17.53±1.01 b | 17 | 37.88±1.50 a | 17.53 ±1.01 a |
| flk-1mutant | – | – | – | 17 | 69.82±4.63 b | 67.82±2.01 b |
| F-value (probability) | 95.38 (0.00) | 8.34 (0.00) | 222.44 (0.00) | 2307.00 (0.00) | ||
| LSD0.05a | 1.85 | 1.41 | 4.06 | 1.86 | ||
Fisher's least significant difference at α=0.05. The letters a and b indicate significant differences between the mean values given in a table column (i.e. mean values in table cells including the letter ‘b’ are significantly different from the mean values in table cells including the letter ‘a’)
Selection with BASTA and PCR analysis of transgene integration identified 9–32 primary (T1) transformants derived from transformation of the BvFLK transgene cassettes into Col-0 or the flk-1 mutant, and 10 and 4 transformants, respectively, derived from transformation of the 35S::AtFLK transgene cassette. Bolting time in Col-0 plants transformed with either of the transgene cassettes was ∼5–6 d earlier than in Col-0 control plants, and this effect did not differ significantly between the three sets of transformants (Table 2). The total number of leaves at bolting was only slightly reduced in the transformants, but differed significantly from the Col-0 control plants in the 35S::BvFLK and endo::BvFLK transgenic plants. Expression of the transgene cassettes in the flk-1 mutant background fully rescued the phenotype in regard to both bolting time and numbers of leaves at bolting. All three sets of transformants bolted ∼33–36 d earlier than the flk-1 mutant.
Nine to 10 T1 plants of each of the sets of transformants carrying the 35S::BvFLK or endo::BvFLK transgene cassettes in either the Col-0 or flk-1 mutant background, and all 35S::AtFLK transformants were selfed to produce T2 seed. In the T2 families derived from transformation into Col-0, plants which bolted ≥5 d earlier than the mean of the Col-0 control plants (37.88±1.50 d to bolting) were initially considered as candidates for transgenic segregants. According to this criterion, 3–9 of the T2 families from each of the three sets of transformants exhibited segregation of the number of early-bolting plants (24–33 d to bolting) to the number of plants which bolted within the time range observed for the control plants (35–40 d to bolting) of 13:4 to 16:1, which did not deviate significantly from the 3:1 or 15:1 ratios expected for one to two transgene loci (as tested by χ2 analysis; Supplementary Table S2 at JXB online). In the remaining families, all plants bolted ≥5 d earlier than the control plants. The T2 families derived from transformation into the flk-1 mutant consisted of plants which bolted either as late as the flk-1 mutant controls (63–77 d to bolting, with a mean and SD of 69.82±4.63), or much earlier (25–44 d to bolting). In 1–3 of the T2 families from each of the three sets of transformants, early- and late-bolting plants segregated at ratios as above, while the remaining families only contained early-bolting plants.
From all six sets of transformants, one T2 family each which segregated for early and late bolting (at a ratio which did not deviate significantly from 3:1) was selected for co-segregation analysis of the early-bolting phenotype and the transgene. As expected, all plants which bolted early were found to be transgenic, whereas none of the remaining plants tested positive for the transgene (Supplementary Table S2 at JXB online). ANOVA of days to bolting and the number of leaves at bolting revealed highly significant differences for both traits between the transgenic subfamilies in T2 and the respective untransformed control plants (Table 2). Similarly, t-tests between the transgenic and non-transgenic individuals within a T2 family showed highly significant differences for both traits in all six families (Supplementary Table S3 at JXB online). For transgenic plants in the flk-1 mutant background, the values for neither of the traits differed significantly from those for Col-0, thus confirming that the transgenes fully rescue the phenotype (Table 2; Fig. 3A).
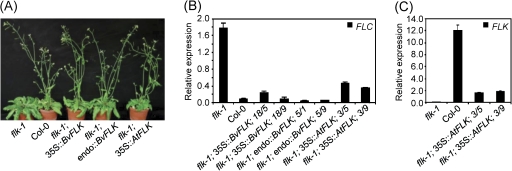
Fig. 3.
BvFLK complements the A. thaliana flk-1 mutant. (A) Phenotypes at 51 d after sowing of the A. thaliana flk-1 mutant, the ecotype Col-0, and the flk-1 mutant transformed with BvFLK driven by the CaMV 35S promoter (35S::BvFLK), BvFLK driven by the endogenous promoter of BvFLK in sugar beet (endo::BvFLK), or A. thaliana FLK driven by the CaMV 35S promoter (35S::AtFLK) (T1 plants). Plants were grown under long-day conditions. (B) RT-qPCR expression analysis of FLC in flk-1, Col-0, and transgenic T3 plants carrying the 35S::BvFLK, endo::BvFLK, or 35S::AtFLK transgene in the flk-1 mutant background. For each of the transgenic lines, two T3 plants were tested that were derived from different transgenic individuals of a T2 family. (C) RT-qPCR expression analysis of FLK in flk-1, Col-0, and transgenic 35S::AtFLK T3 plants. Expression levels in B and C were normalized against GAPDH and measured in triplicate. Error bars indicate the standard deviations of the mean. (This figure is available in colour at JXB online.)
BvFLK represses FLC expression in Arabidopsis
To investigate further the functional conservation of BvFLK and FLK, it was tested whether the effect of BvFLK on bolting time in transgenic A. thaliana plants is mediated through FLC. Consistent with previous results obtained with different flk mutants (Lim et al., 2004; Mockler et al., 2004), FLC expression was significantly higher in the flk-1 mutant than in Col-0 (Fig. 3B). In contrast, FLC expression levels in flk-1 plants carrying the BvFLK transgene driven by either the 35S promoter or its endogenous promoter were strongly reduced compared with the untransformed mutant, and resembled the low expression of FLC in Col-0. FLC expression was also down-regulated in flk-1 plants expressing the 35S::AtFLK transgene, although expression was somewhat less reduced than in BvFLK transgenic plants or the Col-0 wild type. RT-qPCR of FLK confirmed the previous finding that FLK is not detectably expressed in flk-1 (Lim et al., 2004), and showed further that expression of FLK in the 35S::AtFLK transgenic plants which were assayed for FLC expression was lower than in Col-0 wild type (Fig. 3C).
BvFVE1 does not complement an fve mutation in Arabidopsis
To assess the possible role of BvFVE1 in regulating floral transition, the coding sequence of BvFVE1 driven by the CaMV 35S promoter (‘35S::BvFVE1’) was transformed into A. thaliana Col-0 and the fve mutant SALK_013789 (Alonso et al., 2003; fve-7). Bolting under long-day conditions in fve-7 was found to be delayed by 29–34 d when compared with Col-0 (Supplementary Table S4 at JXB online, and data not shown). Selection with BASTA and PCR analysis identified 16 and 27 transgenic events in the Col-0 and fve-7 mutant background, respectively. All primary transformants were phenotyped for initiation of bolting, but showed a similar phenotype to the respective untransformed control plants (data not shown). Ten T1 plants from each set of transformants were selfed for further analysis in the T2 generation. Because the phenotypic data for the primary transformants suggested that the transgene may not or may only weakly affect bolting time, BASTA selection was used to identify T2 families in which the transgene is segregating. For each of the two sets of transformants, one family was identified in which the presence and absence of the transgene segregated at a ratio which did not deviate significantly from 3:1, and one family with a segregation ratio of ≥15:1. For each of these four families, an additional 25 plants were grown without selection, phenotyped for bolting time, and tested for the presence of the transgene by PCR. However, the time to bolting was generally very similar in the transgenic and the non-transgenic plants, and neither the means of numbers of days to bolting nor the means of numbers of leaves at bolting differed significantly between the two groups within a given family (Supplementary Table S4). In one exception, a T2 family which carried the 35Spro::BvFVE1 transgene in the fve-7 mutant background and only contained transgenic plants among 21 plants tested (T2 family #32) bolted ∼6 d earlier (59.95±7.26 d to bolting) than control plants (65.75±4.33 d to bolting; Supplementary Table S4). This difference was statistically significant by conventional criteria, but only marginally so at a P-value of 0.04. There was no significant difference between the means of total numbers of leaves at bolting between the two groups. The intactness of the 35S::BvFVE1 transgene cassette and transgene expression was confirmed by PCR amplification and sequencing of the complete promoter and coding sequence of the transgene, and by RT-PCR, respectively (data not shown).
Transcript accumulation of BvFVE1, but not BvFLK, is under circadian clock control
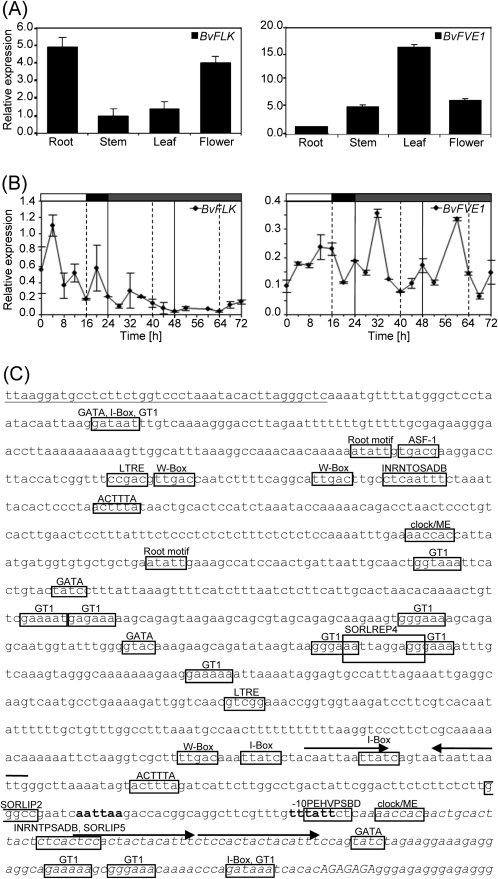
Expression of BvFLK and BvFVE1 in sugar beet was analysed in four major plant organs, root, stem, leaf, and flower, of adult plants. Both genes were found to be expressed in all samples analysed. BvFVE1 is relatively abundant in leaves and only slightly expressed in roots, whereas BvFLK is relatively highly expressed in roots and flowers (Fig. 4A). Because various autonomous pathway genes in Arabidopsis have been implicated in the regulation of the circadian clock (Salathia et al., 2006; see Introduction), and many regulators are subject to feedback regulation by the clock (Pruneda-Paz and Kay, 2010), diurnal and circadian regulation of BvFLK and BvFVE1 expression was investigated by RT-qPCR of transcripts in leaves from 8- to 10-week-old plants (Fig. 4B). Under long-day (16 h light) conditions, BvFLK transcript levels fluctuated during the course of the day and appeared to be highest at 4 h of the light phase. To investigate circadian clock regulation, long day-entrained plants were moved to continuous light and transcript levels were monitored over the subsequent 48 h. However, BvFLK expression did not oscillate and remained low throughout this period. BvFVE1 expression, under long-day conditions, rises to a peak at 12 h, decreases during the first 4 h of darkness, and rises again at the end of the night. In sharp contrast to BvFLK, BvFVE1 expression continued to oscillate robustly throughout each 24 h period under continuous light, although peak amplitude was higher than during the entrainment phase. A promoter sequence analysis for both genes identified a region ∼0.5–1 kb upstream of the transcription start site of BvFLK in which multiple root motifs (Elmayan and Tepfer, 1995) and phytohormone-response elements, including four cytokinin-response elements (Fusada et al., 2005), are clustered, whereas consensus sequences for light-regulated elements appear to be relatively scarce (Supplementary Fig. S2 at JXB online). Sequence elements involved in circadian regulation were not identified. In contrast, the BvFVE1 promoter contains a number of elements which have been implicated in light or circadian regulation, including two SORLIPs (sequences over-represented in light-induced promoters), one SORLREP (sequence over-represented in light-repressed promoters), 11 GT1 consensus sequence motifs (Gilmartin et al., 1990; Zhou, 1999; Hudson and Quail, 2003), and a –10 promoter element (–10PEHVPSPD) from a circadian clock-regulated gene in barley (Thum et al., 2001) (Fig. 4C).
Fig. 4.
Expression of BvFLK and BvFVE1 in B. vulgaris. (A) Expression across major plant organs in the biennial genotype A906001. Plants were vernalized and grown under long-day conditions. RT-qPCR expression levels were normalized against BvGAPDH and measured in triplicate. (B) Diurnal and circadian RT-qPCR expression profiles. Relative expression levels in leaves are shown for a 24 h period under long-day conditions, followed by 48 h under continuous low light at a constant temperature of 22 °C. Expression was measured every 4 h. Expression was normalized using BvGAPDH, BvEF2, and BvTUB. Error bars indicate the standard deviations of the mean. (C) BvFVE1 promoter and 5′ UTR. A total of 1116 bp of the genomic sequence upstream of the start codon are shown. The transcription start site was determined by RACE. The 5′ UTR sequence is printed in italics. Bold letters at positions –9 and –36 (relative to the transcription start site) indicate a TATA-box-like sequence (de Pater et al., 1990) and a TATA-box according to the transcription start site prediction program TSSP (http://www.softberry.ru/berry.phtml), respectively. A GA repeat motif (Santi et al., 2003) in the 5′ UTR just upstream of the ATG start codon is shown in upper case letters. Putative light- and circadian clock-regulated promoter elements are boxed [SORLIP and SORLREP (Hudson and Quail, 2003), GT1 consensus sequence (Terzaghi and Cashmore, 1995), IBOX core motif (Terzaghi and Cashmore, 1995), GATA box (Gilmartin et al., 1990), INRNTPSADB (Nakamura et al., 2002), –10PEHVPSBD (Thum et al., 2001), and a six nucleotide motif (clock/ME) which is common to a promoter element over-represented in circadian clock-regulated genes and a morning element (Harmer and Kay, 2005)]. Arrows above the sequence denote inverted and tandem repeat units. The 3′ end of a sequence tract with homology to the subtelomeric satellite AM076746 of B. vulgaris [clone pAv34-32 (Dechyeva and Schmidt, 2006)] is underlined.
Discussion
The key regulatory genes of the vernalization and photoperiod pathways in Arabidopsis, FLC and CO, had been reported previously to have functionally related homologues in beet (Reeves et al., 2007; Chia et al., 2008). In the present study the question was addressed of whether the genes of the third major regulatory pathway of flowering time control in Arabidopsis are conserved between the two species. Putative B. vulgaris orthologues of four autonomous pathway genes in Arabidopsis, FLK, FVE, LD, and LDL1, were identified and genetically mapped. Three of these genes (BvFVE1, BvLD, and BvLDL1) are homologous to autonomous pathway genes which are thought to regulate FLC expression by chromatin modification, whereas the fourth gene (BvFLK) encodes a putative RNA-binding protein. The beet genes are highly similar to their Arabidopsis counterparts in terms of exon–intron structure and domain organization. With the exception of BvLD, the degree of overall sequence conservation with Arabidopsis is similar to (BvFLK and BvLDL1) or higher than (BvFVE1) that of the only two previously characterized flowering time genes in beet, BvFL1 and BvCOL1 (Reeves et al., 2007; Chia et al., 2008). For another gene of the autonomous pathway, FPA, a sugar beet EST with only moderate homology was identified (Table 1). However, ∼3 kb of the genomic sequence of the corresponding gene (BvFPA) was sequenced and found to include the coding region for three RRM-type RNA-binding domains also present in FPA, which substantiated that BvFPA and FPA are likely orthologues (data not shown). For the remaining three classical autonomous pathway genes, FCA, FY, and FLD, orthologous ESTs were not identified. The currently available EST and transcript sequence collection for B. vulgaris, however, only represents 17 184 genes (BvGI 3.0, release date June 16, 2010; http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=beet), which is approximately one-half to two-thirds of the total number of genes in beet (Herwig et al., 2002). Orthologues of FCA and FY with partially conserved functions (Lee et al., 2005; Lu et al., 2006; Jang et al., 2009) and a gene with high homology to FLD (Lu et al., 2006) have been identified in rice, suggesting that homologues may also be present in beet. Together, these sequence data and observations indicate that at least some autonomous pathway genes are conserved in beet.
Two genes were chosen, BvFLK and BvFVE1, whose homologues in Arabidopsis are thought to regulate FLC expression either through an RNA-based control mechanism (FLK) or by chromatin modification (FVE), to test the hypothesis that at least some of the autonomous pathway gene homologues are also functionally conserved. Among the genes identified here, these two genes also showed the highest degree of sequence conservation within conserved domains. Transgenic expression of BvFLK from both the constitutive CaMV 35S promoter and the endogenous promoter of BvFLK was found to accelerate bolting in A. thaliana and, in an flk mutant background, to fully rescue the phenotype to the bolting time of wild-type plants. FLC expression in transgenic plants carrying the BvFLK transgene was strongly reduced compared with untransformed controls, suggesting that the effect of BvFLK on bolting time is mediated through repression of FLC. In both 35S::BvFLK and endo::BvFLK transgenic plants, FLC expression is similarly low as in Col-0 wild-type plants, which indicates that all regulatory protein domains required for regulation of FLC expression are functionally conserved between FLK and BvFLK. At the sequence level, this is consistent with the high degree of homology within the KH-type RNA-binding domains and the strict conservation between Arabidopsis and beet of the core consensus sequence of KH domains (Mockler et al., 2004; see asterisks in Fig. 1B). The low sequence conservation outside the KH domains, in particular in the N-terminal region of the proteins [which in FLK contains three perfect eight-residue repeats of unknown function (Mockler et al., 2004) which are not conserved in BvFLK], further suggests that this region is less critical for FLK function. The B. vulgaris homologue of FLC, BvFL1, complements FLC function in A. thaliana flc mutants (Reeves et al., 2007), suggesting that BvFLK may also regulate BvFL1. However, the regulation of vernalization requirement and response appears to have diverged considerably during evolution (Colasanti and Coneva, 2009; Distelfeld et al., 2009; Greenup et al., 2009; Jung and Müller, 2009), and possible interactions between BvFLK and BvFL1 in B. vulgaris have yet to be experimentally verified. Nevertheless, the complementation data for both BvFLK in this study and BvFL1 in the study of Reeves et al. (2007) suggest that regulatory interactions between these genes contribute to flowering time control in beet.
The present data also show for the first time, to our knowledge, that transgenic expression of the endogenous A. thaliana gene complements an flk mutant. The fact that full phenotypic complementation was achieved by expression from the CaMV 35S promoter suggests that developmental or environmental regulation of FLK transcription is not a precondition for the gene's function in flowering time control. Interestingly, FLC expression in 35S::AtFLK transgenic plants was strongly reduced but was not as low as in BvFLK transformants or wild-type controls. Incomplete repression of FLC correlated to some extent with the relatively low expression of FLK in 35S::AtFLK plants compared with the wild type. Because the early bolting phenotype of the wild type was fully restored in the transformants, the plant appears to tolerate moderately elevated expression levels of FLC without a significant delay in bolting.
Expression of BvFLK across major plant organs of sugar beet was strongest in roots and flowers, but was also clearly detectable in leaves and stems, and resembled the relative expression levels of FLK in A. thaliana (Lim et al., 2004). While expression in aerial parts of the plant may well be associated with a functional role in flowering time control, the high expression level in roots, which was also observed for FLK in Arabidopsis, may reflect an additional, unknown function of BvFLK (as has also been suggested for FLK; Lim et al., 2004). FLK was implicated in RNA-directed chromatin silencing of retroelements (Bäurle and Dean, 2008; Veley and Michaels, 2008). Although speculative, it is conceivable that BvFLK is involved in repression of other targets such as, for example, developmental genes which may not be required at certain developmental stages (e.g. after floral transition, and/or in certain organs such as roots). The clustering of putative phytohormone response elements in the promoter of BvFLK may further suggest hormonal regulation of BvFLK activity. Finally, BvFLK does not appear to be under circadian clock control. Microarray data for A. thaliana indicate that expression of FLK is not circadian regulated either (Edwards et al., 2006; http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl, accession number NASCARRAYS-108). Together, the data suggest that FLK and BvFLK are functionally related regulators of floral transition, and provide the first evidence for evolutionary conservation of FLK function outside the model species A. thaliana. In the apparent absence of an FLC orthologue in rice, sequence conservation in rice of the same regions which are conserved between A. thaliana and B. vulgaris may support the notion that FLK also has additional functions. Consistent with this possibility, putative orthologues of FLK are present in non-angiosperm species including the gymnosperm Piceasitchensis and the lycophyte Selaginella moellendorffii (Supplementary Fig. S3A at JXB online). For the rice orthologue of another RNA regulatory autonomous pathway gene, OsFCA, the possibility of other functions has been raised by detection of various protein–protein interactions (Lee et al., 2005; Lu et al., 2006; Jang et al., 2009).
In contrast to BvFLK, transgenic expression of BvFVE1 did not complement the bolting time phenotype of an A. thaliana fve mutant. This result differs from data reported by Baek et al. (2008) for CaMV 35S promoter-driven expression of a rice homologue, OsFVE (Fig. 1B), which was shown to at least partially rescue the flowering time phenotype of an fve mutant in Arabidopsis in ∼29% of independent primary transformants. So what are the reasons for the apparent absence of functional conservation between Arabidopsis and beet? The degree of sequence conservation between FVE and BvFVE1 (72% amino acid identity) is very similar to that between FVE and OsFVE (Baek et al., 2008), and BvFVE1 and OsFVE are also highly similar to each other (71% amino acid identity). The degree of sequence conservation with Arabidopsis is also higher than that for BvFLK, BvFL1, and BvCOL1, and much higher than for other autonomous pathway genes in rice (OsFCA and OsFY) with at least partially conserved functions (Lee et al., 2005; Lu et al., 2006). Thus, the degree of overall sequence conservation alone does not appear to be a good indicator of functional conservation, and may simply be a consequence of the large proportion of amino acid residues within functional or structural domains, as the CAF1c domain and the WD40 repeat domains thought to be required for the formation of a β-propeller structure (Kenzior and Folk, 1998; Murzina et al., 2008) make up most of the protein. The regions outside the conserved domains vary considerably between FVE homologues. In particular, an N-terminal region which is present in FVE is missing in both BvFVE1 and OsFVE. The finding that OsFVE at least partially complements FVE function suggests that this region is not absolutely required and, by extrapolation, its absence in BvFVE1 cannot be the reason for the lack of functional complementation. Other regions with low sequence conservation are located between the WD40 repeat domains 2 and 3, and between the WD40 repeat domains 5 and 6. In the human FVE homologue RbAp46, the latter region carries a negatively charged loop which contributes to recognition of histone H4 and is not present in other WD40 β-propeller structures (Murzina et al., 2008). It seems possible that this region determines binding specificity and/or the type of interaction with other proteins. For structural proteins such as FVE which are vitally involved in protein complex formation, or formation of various protein complexes with different functions, as suggested by Amasino (2004), selection of binding partners is likely to be a crucial determinant of protein function. In addition, specific protein–protein interactions depend on co-evolution of binding sites. It is thus conceivable that the binding specificity determinants of FVE and BvFVE1 have diverged sufficiently to prevent wild-type-like interactions between BvFVE1 and interacting proteins in A. thaliana. If the absence of functional complementation of FVE by BvFVE1 is indeed a result of divergent evolution of protein-binding sites, as suggested above, the possibility cannot formally be excluded that BvFVE1 still exerts a similar function in beet to that of FVE in Arabidopsis, for example as part of a co-evolved protein complex.
The B. vulgaris genome appears to carry a second gene, represented by EST EG550040 (Table 1), which may be a paralogue of BvFVE1 and will be referred to as BvFVE2. In the region represented by the EST, BvFVE2 shares a degree of homology to FVE similar to that of BvFVE1 (with the exception of the C-terminal region of the putative partial translation product; see Supplementary Fig. S4 at JXB online). Interestingly, the valine and lysine residues located in the variable region between WD40 repeat domains 5 and 6 and conserved between FVE and OsFVE are also conserved in BvFVE2 (Supplementary Fig. S4 at JXB Online). Although Baek et al. (2008) alluded to the presence of a second rice sequence with homology to FVE (OsJ_003110), this sequence was removed from GenBank, and no other close rice homologue of FVE in GenBank was identified. However, several other species were found to carry putative paralogous pairs (or groups) of FVE homologues, notably including two species in the Malpighiales order of dicotyledonous angiosperms (Populus trichocarpa, PtFVE1–PtFVE3, and Ricinus communis, RcFVE1 and RcFVE2; Supplementary Fig. S3B, C at JXB Online), which (like B. vulgaris) are also only distantly related to the Brassicales. Thus, it is conceivable that a gene duplication event occurred relatively early in the course of dicot evolution, possibly followed by gene loss in some lineages (including the lineage to Arabidopsis), and that the two paralogues underwent subfunctionalization.
The present expression data may provide further indications for subfunctionalization of BvFVE1. First, BvFVE1 appears to be most strongly expressed in leaves of adult plants, whereas the data for FVE indicate highest expression in flowers (Ausin et al., 2004). Secondly, BvFVE1 is diurnally regulated and under circadian control, a finding which appears consistent with the presence of multiple putative light-regulatable promoter elements. It is worth noting that BvFVE1 also has an unusual map position close to the telomere and immediately adjacent to subtelomeric repeats, which may have contributed to the evolution of cis regulation of BvFVE1 expression. Although FVE was shown to affect circadian period length (Salathia et al., 2006), the gene itself (or any of the other classical autonomous pathway genes) has not been reported to be under control of the circadian clock. Furthermore, the microarray data of Edwards et al. (2006) indicate that FVE is not circadian regulated, as determined after entrainment under intermediate daylength conditions (12 h light/dark cycles). Interestingly, the small glycine-rich RNA-binding protein GRP7 in Arabidopsis which has long been known to be under circadian clock control was recently assigned to the autonomous pathway (Streitner et al., 2008). Like FVE, this protein has also been implicated in various other biological processes including cold stress response (Carpenter et al., 1994; Heintzen et al., 1994). Circadian regulation of autonomous pathway genes or homologues may indicate a certain level of cross-talk between different floral regulatory pathways upstream of the floral integrator FT, as has been observed for FLC and the circadian clock (Edwards et al., 2006; Salathia et al., 2006; Spensley et al., 2009). A comparison with the circadian-regulated, putative photoperiod pathway gene BvCOL1, which was analysed for circadian oscillations in the same plant samples as BvFVE1 (Chia et al., 2008), shows that BvFVE1 and BvCOL1 have roughly complementary expression profiles, possibly suggesting regulation by different circadian clock output pathways and/or opposing regulatory roles. Alternatively, circadian clock regulation of BvFVE1 and GRP7 may reflect the broader involvement of these genes in other biological processes (e.g. cold stress response; Harmer et al., 2000; Fowler et al., 2005; Franklin and Whitelam, 2007).
The present survey of autonomous pathway genes provides the first evidence for evolutionary conservation of homologues in beet as well as divergence and differential regulation of one gene. The results have further implications because (i) functional conservation of autonomous pathway genes outside Arabidopsis, with the exception of a few reports for monocots, has not yet been studied in detail; (ii) B. vulgaris, among dicot plants, is only a very distant relative of the model species A. thaliana, and belongs to a eudicot clade which is little understood at the molecular level; and (iii) the environmental requirements for floral transition differ markedly between beet and model plants, in particular Arabidopsis and rice. Thus, the findings for B. vulgaris expand the spectrum of evolutionarily diverse species for which molecular data are available and which are subject to differential environmental regulation of bolting and flowering. Finally, for sugar beet and other root and leaf crops, bolting and flowering are undesirable because they drastically reduce yield. The identification and genetic mapping of floral promoters provides tangible targets for marker-assisted or transgenic approaches towards crop improvement.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Sequence and structure of the autonomous pathway gene homologues BvLD and BvLDL1.
Figure S2. BvFLK promoter and 5' UTR.
Figure S3. Phylogenetic analysis of FLK (A) and FVE (B, C).
Figure S4. Multiple sequence alignment including A. thaliana FVE (AtFVE), BvFVE1, the putative translation product of the largest open reading frame in the B. vulgaris EST EG550040 (BvFVE2), and OsFVE.
Table S1. Primer sequences.
Table S2. Days to bolting and total number of leaves at bolting in 53 T2 families derived from transformation with BvFLK or FLK and non-transformed controls (Col-0 and flk-1).
Table S3. Unpaired t-test for number of days to bolting (DTB) and total number of leaves at bolting (TNL) between BvFLK or FLK transformants and non-transgenic A. thaliana plants within six T2 populations.
Table S4. Unpaired t-test for number of days to bolting (DTB) and total number of leaves at bolting (TNL) between BvFVE1 transformants and non-transgenic A. thaliana plants within four T2 populations.
Acknowledgments
The project is funded by the Deutsche Forschungsgemeinschaft under grant no. DFG JU 205/14-1. SFA-E is supported by a scholarship from the Ministry of Higher Education, Egypt. TC and EM-G are supported by grants from the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the British Beet Research Organisation (BBRO). We are grateful to Dr Britta Schulz (KWS Saat AG), Dr Georg Koch (formerly at Saatzucht Dieckmann GmbH & Co. KG), and Dr Axel Schechert (Strube Research GmbH & Co. KG) for providing the K1 and D2 mapping populations, and we thank Cay Kruse, Kevin Sawford, and Andrea Jennings for technical assistance.
References
- Abegg FA. A genetic factor for the annual habit in beets and linkage relationship. Journal of Agricultural Research. 1936;53:493–511. [Google Scholar]
- Albert VA, Soltis DE, Carlson JE, et al. Floral gene resources from basal angiosperms for comparative genomics research. BMC Plant Biology. 2005;5:5. doi: 10.1186/1471-2229-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers E, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amasino R. Take a cold flower. Nature Genetics. 2004;36:111–112. doi: 10.1038/ng0204-111. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Lee I, Weigel D, Amasino RM. The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. The Plant Journal. 1999;18:195–203. doi: 10.1046/j.1365-313x.1999.00442.x. [DOI] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nature Genetics. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- Baek IS, Park HY, You MK, Lee JH, Kim JK. Functional conservation and divergence of FVE genes that control flowering time and cold response in rice and Arabidopsis. Molecules and Cells. 2008;26:368–372. [PubMed] [Google Scholar]
- Bäurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bäurle I, Dean C. Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002733. e2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I, Smith L, Baulcombe DC, Dean C. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science. 2007;318:109–112. doi: 10.1126/science.1146565. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell. 2004;16 doi: 10.1105/tpc.015958. Supplement, S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry P, Wieber R, Saumitou-Laprade P, Pillen K, Van Dijk H, Jung C. Identification of RFLP markers closely linked to the bolting gene B and their significance for the study of the annual habit in beets (Beta vulgaris L.) Theoretical and Applied Genetics. 1994;88:852–858. doi: 10.1007/BF01253996. [DOI] [PubMed] [Google Scholar]
- Büttner B, Abou-Elwafa SF, Zhang W, Jung C, Muller AE. A survey of EMS-induced biennial Beta vulgaris mutants reveals a novel bolting locus which is unlinked to the bolting gene B. Theoretical and Applied Genetics. 2010;121:1117–1131. doi: 10.1007/s00122-010-1376-8. [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian-rhythm. Plant Physiology. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. Journal of Molecular Evolution. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X. Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Developmental Cell. 2003;4:53–66. doi: 10.1016/s1534-5807(02)00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia TYP, Müller A, Jung C, Mutasa-Göttgens ES. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. Journal of Experimental Botany. 2008;59:2735–2748. doi: 10.1093/jxb/ern129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Coneva V. Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiology. 2009;149:56–62. doi: 10.1104/pp.108.130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis D, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences, USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechyeva D, Schmidt T. Molecular organization of terminal repetitive DNA in Beta species. Chromosome Research. 2006;14:881–897. doi: 10.1007/s10577-006-1096-8. [DOI] [PubMed] [Google Scholar]
- De Pater S, Hensgens LAM, Schilperoort RA. Structure and expression of a light-inducible shoot-specific rice gene. Plant Molecular Biology. 1990;15:399–406. doi: 10.1007/BF00019157. [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Current Opinion in Plant Biology. 2009;12:178–184. doi: 10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development. 2007;134:2841–2850. doi: 10.1242/dev.02866. [DOI] [PubMed] [Google Scholar]
- Earley K, Haag J, Pontes O, Opper K, Juehre T, Song K, Pikaard C. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. The Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Tepfer M. Evaluation in tobacco of the organ specificity and strength of the rold promoter, domain-A of the 35S promoter and the 35S(2) promoter. Transgenic Research. 1995;4:388–396. doi: 10.1007/BF01973757. [DOI] [PubMed] [Google Scholar]
- El-Mezawy A, Dreyer F, Jacobs G, Jung C. High-resolution mapping of the bolting gene B of sugar beet. Theoretical and Applied Genetics. 2002;105:100–105. doi: 10.1007/s00122-001-0859-z. [DOI] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow ME. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiology. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nature Genetics. 2007;39:1410–1413. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proceedings of the National Academy of Sciences, USA. 1998;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusada N, Masuda T, Kuroda H, Shimada H, Ohta H, Takamiya K. Identification of a novel cis-element exhibiting cytokinin-dependent protein binding in vitro in the 5′-region of NADPH-protochlorophyllide oxidoreductase gene in cucumber. Plant Molecular Biology. 2005;59:631–645. doi: 10.1007/s11103-005-0579-x. [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH. Molecular light switches for plant genes. The Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A, Peacock WJ, Dennis ES, Trevaskis B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Annals of Botany. 2009;103:1165–1172. doi: 10.1093/aob/mcp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. The Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YH, Amasino RM. Role of chromatin modification in flowering-time control. Trends in Plant Science. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- He YH, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandiz C, et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiology. 2005;137:1420–1434. doi: 10.1104/pp.104.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light-entrained and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. The Plant Journal. 1994;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- Hennig L, Bouveret R, Gruissem W. MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends in Cell Biology. 2005;15:295–302. doi: 10.1016/j.tcb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Herwig R, Schulz B, Weisshaar B, et al. Construction of a ‘unigene’ cDNA clone set by oligonucleotide fingerprinting allows access to 25 000 potential sugar beet genes. The Plant Journal. 2002;32:845–857. doi: 10.1046/j.1365-313x.2002.01457.x. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Jacobs G, Telgmann A, Gaafar RM, Alam S, Jung C. A bacterial artificial chromosome (BAC) library of sugar beet and a physical map of the region encompassing the bolting gene B. Molecular Genetics and Genomics. 2003;269:126–136. doi: 10.1007/s00438-003-0821-7. [DOI] [PubMed] [Google Scholar]
- Hudson ME, Quail PH. Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiology. 2003;133:1605–1616. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YH, Park HY, Kim SK, Lee JH, Suh MC, Chung YS, Paek KH, Kim JK. Survey of rice proteins interacting with OsFCA and OsFY proteins which are homologous to the Arabidopsis flowering time proteins, FCA and FY. Plant and Cell Physiology. 2009;50:1479–1492. doi: 10.1093/pcp/pcp093. [DOI] [PubMed] [Google Scholar]
- Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine specific demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. The Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, et al. The SUMO E3 ligase, AtS1Z1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through effects on FLC chromatin structure. The Plant Journal. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Müller AE. Flowering time control and applications in plant breeding. Trends in Plant Science. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Kenzior AL, Folk WR. AtMSI4 and RbAp48 WD-40 repeat proteins bind metal ions. FEBS Letters. 1998;440:425–429. doi: 10.1016/s0014-5793(98)01500-2. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, et al. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nature Genetics. 2004;36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. The Plant Cell. 2006;18:2985–2998. doi: 10.1105/tpc.106.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Blankestijndevries H, Hanhart C, Soppe W, Peeters T. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome-5 that is not effective in the Landsbergerecta wild-type. The Plant Journal. 1994;6:911–919. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annals of Eugenetics. 1944;12:172–175. [Google Scholar]
- Krichevsky A, Gutgarts H, Kozlovsky SV, Tzfira T, Sutton A, Sternglanz R, Mandel G, Citovsky V. C2H2 zinc finger-SET histone methyltransferase is a plant-specific chromatin modifier. Developmental Biology. 2007;303:259–269. doi: 10.1016/j.ydbio.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. The Plant Cell. 1994;6:75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cho YS, Yoon HS, Suh MC, Moon J, Lee I, Weigel D, Yun CH, Kim JK. Conservation and divergence of FCA function between Arabidopsis and rice. Plant Molecular Biology. 2005;58:823–838. doi: 10.1007/s11103-005-8105-8. [DOI] [PubMed] [Google Scholar]
- Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. The Plant Cell. 2004;16:731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development. 2007;134:1901–1910. doi: 10.1242/dev.003103. [DOI] [PubMed] [Google Scholar]
- Lu Q, Xu ZK, Song RT. OsFY, a homolog of AtFY, encodes a protein that can interact with OsFCA-gamma in rice (Oryza sativa L.) Acta Biochimica et Biophysica Sinica. 2006;38:492–499. doi: 10.1111/j.1745-7270.2006.00188.x. [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Marquardt S, Boss PK, Hadfield J, Dean C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. Journal of Experimental Botany. 2006;57:3379–3386. doi: 10.1093/jxb/erl073. [DOI] [PubMed] [Google Scholar]
- Meier U. Growth stages of mono-and dicotyledonous plants. BBCH Monograph. Germany: Federal Biological Research Centre for Agriculture and Forestry; 2001. Phenological growth stages and BBCH-identification keys of beet. [Google Scholar]
- Mersereau M, Pazour GJ, Das A. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene. 1990;90:149–151. doi: 10.1016/0378-1119(90)90452-w. [DOI] [PubMed] [Google Scholar]
- Michaels SD. Flowering time regulation produces much fruit. Current Opinion in Plant Biology. 2009;12:75–80. doi: 10.1016/j.pbi.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. The Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Yu X, Shalitin D, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proceedings of the National Academy of Sciences, USA. 2004;101:12759–12764. doi: 10.1073/pnas.0404552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel P, Trehin C, Breuil-Broyer SP, Negrutiu I. Altering FVE/ MSI4 results in a substantial increase of biomass in Arabidopsis—the functional analysis of an ontogenesis accelerator. Molecular Breeding. 2009;23:239–257. [Google Scholar]
- Mouhu K, Hytonen T, Folta K, Rantanen M, Paulin M, Auvinen P, Elomaa P. Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant Biology. 2009;9:122. doi: 10.1186/1471-2229-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munerati O. L'eredità della tendenza alla annualità nella commune barbabietola, coltivata. Zeitschrift Züchtung, Reihe A, Pflanzenzüchtung. 1931;17:84–89. [Google Scholar]
- Murzina NV, Pei XY, Zhang W, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Tsunoda T, Obokata J. Photosynthesis nuclear genes generally lack TATA-boxes: a tobacco photosystem I gene responds to light through an initiator. The Plant Journal. 2002;29:1–10. doi: 10.1046/j.0960-7412.2001.01188.x. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proceedings of the National Academy of Sciences, USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps-Durr TL, Thomas J, Vahab P, Timmermans MCP. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. The Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends in Plant Science. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Dean C, Simpson GG. Regulated RNA processing in the control of Arabidopsis flowering. International Journal of Developmental Biology. 2005;49:773–780. doi: 10.1387/ijdb.051995vq. [DOI] [PubMed] [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson GG. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO Journal. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PA, He Y, Schmitz RJ, Amasino RM, Panella LW, Richards CM. Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris) Genetics. 2007;176:295–307. doi: 10.1534/genetics.106.069336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remay A, Lalanne D, Thouroude T, Le Couviour F, Hibrand-Saint Oyant L, Foucher F. A survey of flowering genes reveals the role of gibberellins in floral control in rose. Theoretical and Applied Genetics. 2009;119:767–781. doi: 10.1007/s00122-009-1087-1. [DOI] [PubMed] [Google Scholar]
- Ripoll JJ, Rodriguez-Cazorla E, Gonzalez-Reig S, et al. Antagonistic interactions between Arabidopsis K-homology domain genes uncover PEPPER as a positive regulator of the central floral repressor. FLOWERING LOCUS C. Developmental Biology. 2009;333:251–262. doi: 10.1016/j.ydbio.2009.06.035. [DOI] [PubMed] [Google Scholar]
- Salathia N, Davis SJ, Lynn JR, Michaels SD, Amasino RM, Millar AJ. FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biology. 2006;6:10. doi: 10.1186/1471-2229-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda SL, Amasino RM. Ecotype-specific expression of a flowering mutant phenotype in Arabidopsis thaliana. Plant Physiology. 1996;111:641–644. doi: 10.1104/pp.111.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi L, Wang YM, Stile MR, et al. The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. The Plant Journal. 2003;34:813–826. doi: 10.1046/j.1365-313x.2003.01767.x. [DOI] [PubMed] [Google Scholar]
- Schneider K, Kulosa D, Soerensen T, et al. Analysis of DNA polymorphisms in sugar beet (Beta vulgaris L.) and development of an SNP-based map of expressed genes. Theoretical and Applied Genetics. 2007;115:601–615. doi: 10.1007/s00122-007-0591-4. [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. The Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Cai D, Kleine M, Fan L, Wang S, Jung C. A complete physical map of a wild beet (Beta procumbens) translocation in sugar beet. Molecular Genetics and Genomics. 2006;275:504–511. doi: 10.1007/s00438-006-0108-x. [DOI] [PubMed] [Google Scholar]
- Simpson GG. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology. 2004;7:570–574. doi: 10.1016/j.pbi.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- Spensley M, Kim JY, Picot E, Reid J, Ott S, Helliwell C, Carre IA. Evolutionarily conserved regulatory motifs in the promoter of the Arabidopsis clock gene LATE ELONGATED HYPOCOTYL. The Plant Cell. 2009;21:2606–2623. doi: 10.1105/tpc.109.069898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schoning JC, Alfano JR, Staiger D. The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. The Plant Journal. 2008;56:239–250. doi: 10.1111/j.1365-313X.2008.03591.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annual Review of Plant Physiology and Plant Molecular Biology. 1995;46:445–474. [Google Scholar]
- Thum KE, Kim M, Morishige DT, Eibl C, Koop HU, Mullet JE. Analysis of barley chloroplast psbD light-responsive promoter elements in transplastomic tobacco. Plant Molecular Biology. 2001;47:353–366. doi: 10.1023/a:1011616400264. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Van Nocker S, Muszynski M, Briggs K, Amasino RM. Characterization of a gene from Zea mays related to the Arabidopsis flowering-time gene LUMINIDEPENDENS. Plant Molecular Biology. 2000;44:107–122. doi: 10.1023/a:1006472929800. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorrips RE. JoinMap® 3.0: software for the calculation of genetic linkage maps. Wageningen, The Netherlands: Plant Research International; 2001. [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research 0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veley KM, Michaels SD. Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiology. 2008;147:682–695. doi: 10.1104/pp.108.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Yu TS. Arabidopsis floral regulators FVE and AGL24 are phloem-mobile RNAs. Botanical Studies. 2010;51:17–26. [Google Scholar]
- Zhou DX. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends in Plant Science. 1999;4:210–214. doi: 10.1016/s1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.